Abstract
We report the discovery of endogenous viral elements (EVEs) from Hepadnaviridae, Bornaviridae and Circoviridae in the speckled rattlesnake, Crotalus mitchellii, the first viperid snake for which a draft whole genome sequence assembly is available. Analysis of the draft assembly reveals genome fragments from the three virus families were inserted into the genome of this snake over the past 50 Myr. Cross-species PCR screening of orthologous loci and computational scanning of the python and king cobra genomes reveals that circoviruses integrated most recently (within the last approx. 10 Myr), whereas bornaviruses and hepadnaviruses integrated at least approximately 13 and approximately 50 Ma, respectively. This is, to our knowledge, the first report of circo-, borna- and hepadnaviruses in snakes and the first characterization of non-retroviral EVEs in non-avian reptiles. Our study provides a window into the historical dynamics of viruses in these host lineages and shows that their evolution involved multiple host-switches between mammals and reptiles.
Keywords: endogenous viral elements, hepadnavirus, bornavirus, circovirus, paleovirology, snakes, rattlesnake
1. Introduction
Endogenous viral elements (EVEs) are entire or fragmented viral genomes that have been integrated into the genome of their hosts and are therefore vertically inherited in a stable manner [1,2]. Although only retroviruses are known to require integration of their genomes into their host genome to complete their replication cycle, EVEs from all types of viruses have now been identified in eukaryotic genomes [3]. These elements result from endogenization events that occurred in the past, the genetic ‘fossils’ of which can be detected in whole genome sequence data millions of years later. Endogenization of viruses is not rare; in fact, it appears to be a recurrent and on-going process in the evolution of eukaryotic genomes [4,5].
The study of EVEs fossilized in their host genomes has opened new avenues to better understand the biology of viruses and the evolutionary history of host–virus interactions [6]. For example, EVE discovery can expand the past and current known host range of viral families [7–9], contribute to the characterization of the viral flora infecting a specific organism or lineage [10,11] and help to identify reservoir species and/or reconstruct complex transmission routes of zoonotic viruses [12–14]. Furthermore, endogenization events can be dated and used to calibrate long-term evolutionary patterns of modern virus families [15–18]. Finally, studies in palaeovirology are increasingly revealing how viruses have contributed to the gene repertoires of their hosts. Indeed, although relatively few EVE-derived host genes have been characterized in detail so far [19–22], numerous EVEs are transcribed and/or have evolved under purifying selection after endogenization [10,11,23–25], suggesting they may have been domesticated and now fulfil cellular functions in the host.
Non-retroviral EVEs have been identified in the genomes of species from several branches of the eukaryotic tree, including metazoans, fungi, plants, chromalveolates and excavates [1,3,11]. However, most branches have not been examined, either because of taxonomic biases in sequencing efforts or owing to a lack of screening. As a result, most vertebrate EVEs have been identified in mammalian genomes (in approx. 20 species), with only a few cases known in other host species; one EVE found in the genomes of two species of birds (Hepadnaviridae [8,18,26]), mononegavirales-like sequences found in two species of teleostean fishes and in the lamprey [12,24], and a circovirus- and nanovirus-like sequence reported in the frog Xenopus tropicalis [27]. No non-retroviral EVEs have been comprehensively characterized in non-avian reptiles (but see [28]), which is probably owing to there being only one whole genome sequence available until very recently (the Carolina anole, Anolis carolinensis [29]).
Among non-avian reptiles, snakes represent a large and diverse radiation of over 3000 species, for which genomic data are just starting to become available [30,31]. Among snakes, the rattlesnakes (which include two genera, Crotalus and Sistrurus) represent a clade of approximately 40 species of highly venomous snakes in the family Viperidae endemic to the Western Hemisphere. Owing partly to their unique morphology, ecology and physiology, as well as their cultural and medical importance, rattlesnakes have become one of the most extensively studied groups of non-avian reptiles [32]. Here, we search whole genome sequence data obtained from a speckled rattlesnake (Crotalus mitchellii) for endogenous viruses and identified multiple EVEs belonging to the Hepadnaviridae, Bornaviridae and Circoviridae families in this and several other species of pitvipers. In addition, we use the genomes of two additional snake species (Burmese python and king cobra [30,31]) to further estimate the timing of viral endogenization. Our results show that the three families of viruses that were previously not known to occur in the genomes of squamate reptiles have recurrently infiltrated the germline of snakes over the past 50 Myr.
2. Material and methods
(a). Whole genome sequencing and assembly of Crotalus mitchellii
The speckled rattlesnake (C. mitchellii) species complex includes species endemic to arid regions of western North America and Mexico [33,34]. The single specimen used for genome sequencing is shown on figure 1. It is a subadult female individual of C. m. pyrrhus (JMM 685; UTA R-60292) collected on 11 June 2012 from Palm Canyon, Kofa Mountains, La Paz County, AZ, USA (33.360°N, 114.106°W; 650 m).
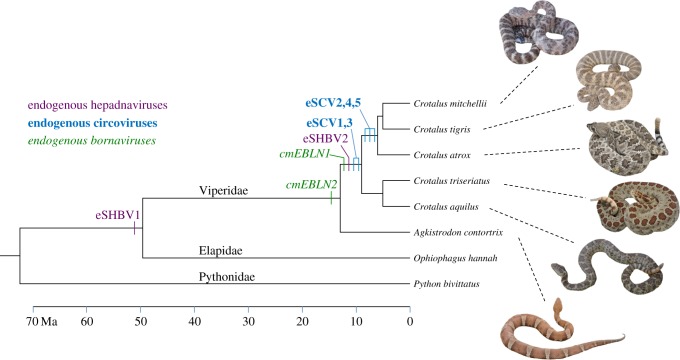
Figure 1.
Chronology of endogenization events during snake evolution. The branch lengths of the tree are proportional to time in million years. Snake divergence times were taken from Reyes-Velasco et al. [35], Castoe et al. [36], Burbrink & Pyron [37], and Hedges et al. [38]. The various endogenization events are placed on the tree based on the results of the cross-species screening for orthologous EVEs (table 1). The photographed C. mitchellii female specimen is the one for which whole genome sequences have been generated. Alignments of orthologous EVEs are provided in the electronic supplementary material, datasets S4–S12. (Online version in colour.)
Genomic DNA was extracted from snap-frozen liver tissue, stored at −80C, using standard lysis and proteinase digestion followed by phenol–chloroform–isoamyl alcohol methods. A genomic shotgun sequencing library was prepared by shearing genomic DNA using a Covaris ultrasonicator, and further preparation using an Illumina Truseq DNAseq library kit. This shotgun library was sequenced on a single lane of an Illumina HiSeq 2000 using 100 bp paired-end reads. Sequencing produced 408 million total paired reads, which were quality trimmed using the Trim Sequences tool in CLC Genomics Workbench (CLC Bio/Qiagen) with a quality score limit of 0.05 and a maximum of two ambiguities. Trimmed reads were assembled de novo using the De Novo Assembly tool in CLC Genomics Workbench with automatic word and bubble sizes, a minimum contig length of 200 bp, paired-read scaffolding, and reads mapped back to contigs for gap-filling (mixmatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.05, similarity fraction = 0.8). The total length of the resulting 478 598 contigs was 1.14 Gb, providing approximately 40× coverage of the genome, with an N50 scaffold size of 5.3 kb (GenBank accession number: JPMF00000000).
(b). Screening for endogenous viral elements in the genome of Crotalus mitchellii
To screen for the presence of EVEs in the rattlesnake genome, we used all viral genomes available in GenBank excluding retroviruses (n = 2048 as of November 2013) as queries to perform a TBLASTX search of C. mitchellii contigs with a 0.00001 e-value cut-off. Our study only focused on non-retroviral EVEs because endogenous retroviruses (ERVs) have been extensively characterized in the genomes of many vertebrates [39] and various studies have shown that snake genomes, like all vertebrate genomes, harbour dozens or hundreds of ERVs [30,40–42]. Our TBLASTX library did include the recently characterized snake arenaviruses, presumed to be the cause of inclusion body disease in various snake species [43], but no hits were recovered matching the genome of these viruses.
All hits longer than 50 amino acids (aa) resulting from the TBLASTX search were then used as queries to perform a reciprocal BLASTP search of the GenBank non-redundant protein database (accessed in November 2013). Based on preliminary TBLASTN searches, we found that snake genomes contain numerous ERVs, as found in all other vertebrates (data not shown). In addition to searching for non-retroviral endogenous viruses in C. mitchellii, we searched with the same library in two additional snake genomes that are publicly available: the king cobra, Ophiophagus hannah [31], and the Burmese python, Python molurus bivittatus [30].
(c). Estimating the timing of endogenization events
In order to estimate the timing of the endogenization events producing the various EVEs that we detected, we followed an approach previously used in palaeovirology studies [18,26,44]. We used EVE-specific PCR primers to screen for the presence or absence of orthologous EVEs in viperid snakes with various degrees of relatedness to C. mitchellii. When possible, primer pairs included a forward primer anchored in the upstream region flanking a given EVE and a reverse primer anchored in the downstream region flanking the EVE. This type of primer pair allowed amplification of orthologous loci, irrespective of whether the EVE was present or absent. Other primer pairs included one primer anchored within the EVE and another primer anchored either in the upstream or downstream region flanking the EVE. With these types of primer pairs, orthologous loci could only be amplified if the EVE was present. The primers are listed in the electronic supplementary material, table S1, and their sequences are provided in dataset S2.
For use in PCR reactions, snake DNA was extracted using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer's protocol. PCR reactions were conducted using the following thermal cycling profile: initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 30 s and elongation at 72°C for 1 min, ending with a 10 min elongation step at 72°C. Fragments from the PCR were visualized on a 1–2% agarose gel. Purified PCR products were directly sequenced using ABI BigDye sequencing mix (1.4 µl template PCR product, 0.4 µl BigDye, 2 µl manufacturer supplied buffer, 0.3 µl primer and 6 µl H2O). Sequencing reactions were ethanol precipitated and run on an ABI 3730 sequencer. The list of snake species screened using this approach, together with corresponding museum voucher numbers for these specimens, are provided in the electronic supplementary material, table S2. For the two publicly available snake genomes (the Burmese python and king cobra), we searched for EVEs using the same bioinformatic approach that was used for the C. mitchellii genome. All orthologous snake EVE sequences used in this study are available in the electronic supplemental material, dataset S3.
(d). Phylogenetic analyses of snake endogenous viral elements
In order to assess the phylogenetic relationships of the various EVEs identified in this study, we reconstructed maximum likelihood and Bayesian phylogenies of the Hepadnaviridae, Bornaviridae and Circoviridae (including new snake EVEs) using PhyML v. 3.0 [45] and MrBayes v. 3.2.1 [46], respectively. Alignments of snake EVEs, together with exogenous and endogenous representative members of each viral family, were performed manually at the aa level in BioEdit v. 5.0.6 [47]. Regions of ambiguous alignment and aligned regions with greater than 30% missing data were excluded from phylogenetic analyses. Best-fit models of aa evolution were selected in ProtTest [48] for the maximum-likelihood analysis using the Bayesian information criterion: JTT + G for bornaviruses, LG + I + G for the circovirus Rep, Blosum62 + G + F for the circovirus capsid and LG + I + G + F for hepadnaviruses. For the Bayesian analyses, we used a mixed aa model of evolution with a γ-shaped distribution of rates across sites; this model allows for model selection to be integrated into the analysis, and phylogenetic results to be integrated across all best-fit models. Two runs with four chains each were run for 11 000 000 generations with parameters and trees sampled every 10 000 generations. Convergence and proper mixing were confirmed using Tracer [49] and AWTY [50], and the first 1 000 000 generations were discarded as burn-in from each run before the posteriors were summarized. Alignments used for phylogenetic analyses are provided in the electronic supplementary material, dataset S1.
To estimate whether EVEs are evolving under selective constraints after they became integrated into their host genome, we conducted a series of selection analyses on orthologous EVEs detected in each species (table 1; electronic supplementary material, datasets S4–S12). These analyses are based on alignments of orthologous EVEs only and do not include exogenous viruses. The codon-based Z-test in MEGA 6 [51] was used with the Nei–Gojobori method and Jukes–Cantor correction and 500 bootstrap (BP) replicates. We also calculated long-term substitution rates of all three families following the approach used in [26] in order to compare them to short-term rates that have been calculated based on analyses of exogenous viruses [52,53]. As illustrated in the details in Box 2 of [3], fig. 4 of [5] and fig. 4 of [26], we first calculated the distance between each EVE and their closest extant virus relative (Kimura-2P distance). This distance corresponds to the sum of the mutations accumulated in EVEs at the host rate since endogenization and the mutations accumulated at the viral rate on the exogenous viral branch. In order to estimate the long-term viral rate, we subtracted the mutations accumulated at the host rate since endogenization, which we were able to estimate by calculating the distance between orthologous EVEs. We then divided the final distance, corresponding only to mutations accumulated at the viral rate, by the age of each EVE.
Table 1.
Characteristics of the snake EVEs identified in this study. (+, positive PCR yielding a band of the size expected for presence of a specific EVE locus; −, positive PCR yielding a band of the size expected for absence of a specific EVE locus; neg. PCR, negative PCR, either because the primer pair does not allow amplification of empty site or because orthologous loci are not sufficiently conserved; (seq), the PCR product was sequenced (see the electronic supplementary material, datasets S4–S12). See also the electronic supplementary material, table S1, for more details.)
| name of EVE | Circoviridae (ssDNA) |
Bornaviridae (-ssRNA) |
Hepadnaviridae (dsDNA-RT) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| eSCV1 | eSCV2 | eSCV3 | eSCV4 | eSCV5 | cmEBLN1 | cmEBLN2 | eSHBV1 | eSHBV2 | ||
| results of BLASTP | Crotalus mitchellii contig | 224 958 | 224 959 | 9220 | 24 347 | 193 549 | 173 953 | 72 541 | 93 260 | 192 207 |
| length of EVE (aa) | 167 | 199 | 241 | 137 | 130 | 345 | 368 | 317 | 371 | |
| best BLASTP hit | porcine circovirus capsid | porcine circovirus capsid | bat circovirus Rep | bat circovirus Rep | bat circovirus Rep | avian bornavirus nucleoprot | avian bornavirus nucleoprot | parrot HBV Pol | parrot HBV Pol | |
| % aa similarity | 58 | 53 | 82 | 81 | 80 | 63 | 61 | 48 | 67 | |
| K2P nucleotide distance | 0.64 | 0.67 | 0.49 | 0.46 | 0.45 | 0.74 | 0.75 | 0.83 | 0.63 | |
| frameshifts/stops | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 5/2 | 1/0 | 2/1 | 0/0 | |
| screening for orthologous EVEs | Crotalus mitchellii | + (seq)a | + (seq)a | + (seq)a | + (seq)a | + (seq)a | + (seq)a | + (seq)a | + (seq)a | + (seq)a |
| Crotalus tigris | + (seq) | + (seq) | + (seq) | + (seq) | + (seq) | + | neg. PCR | not tested | + (seq) | |
| Crotalus atrox | + (seq) | + (seq)a | + (seq) | + (seq)a | + (seq)a | + | neg. PCR | not tested | neg. PCR | |
| Crotalus triseriatus | + (seq)a | − (seq) | + (seq)a | neg. PCR | — | + | + | not tested | + (seq)a | |
| Crotalus aquilus | + (seq) | — | + (seq) | neg. PCR | — | + (seq)a | + (seq) | not tested | + (seq) | |
| Agkistrodon contortrix | neg. PCR | — | neg. PCR | − (seq) | — | neg. PCR | + (seq)a | not tested | neg. PCR | |
| Ophiophagus hannah | locus absent | − (seq) | locus absent | − (seq) | locus absent | locus absent | locus absent | + (seq)a | locus absent | |
| Python bivittatus | locus absent | locus absent | locus absent | locus absent | locus absent | locus absent | locus absent | locus absent | locus absent | |
| age (Myr) K2P distance between orthologues inferred host rate (subst. site−1 year−1) b inferred viral rate (subst. site−1 year−1) c |
at least 8 0.014 8.7 × 10−10 7.8 × 10−08 |
between 6 and 8 0.005 4.1 × 10−10 1.1 × 10−07 |
at least 8 0.014 8.7 × 10−10 5.9 × 10−08 |
between 6 and 8 0.015 1.2 × 10−09 7.4 × 10−08 |
between 6 and 8 0.008 6.7 × 10−10 5.5 × 10−08 |
at least 8 0.04 2.5 × 10−09 8.7 × 10−08 |
at least 13 0.03 1.15 × 10−09 5.5 × 10−08 |
at least 50 0.23 2.3 × 10−09 1.3 × 10−08 |
at least 8 0.008 5 × 10−10 7.7 × 10−08 |
|
aSequences used to calculate K2P distances between orthologous snake EVEs.
bHost substitution rates were calculated by dividing the K2P distance between orthologous snake EVEs by the inferred age of the EVEs.
cSee methods for a detailed explanation of how long-term viral rates were inferred.
3. Results
(a). Multiple endogenous viral elements in the speckled rattlesnake
Our search for EVEs in the genome of the speckled rattlesnake (C. mitchellii) revealed that there are nine fragments with significant similarity to sequences from three extant virus families, each characterized by a different genome type: Circoviridae (ssDNA), Bornaviridae (-ssRNA) and Hepadnaviridae (dsDNA-RT). We named these EVEs eSCV1–5 for endogenous snake circovirus fragments 1 through to 5 and eSHBV1–2 for endogenous snake hepadnavirus fragments 1 and 2 (table 1). For bornavirus EVEs, we follow Horie et al. [28] and use EBLN1–2 for endogenous bornavirus-like N nucleoprotein fragments 1 and 2. The nucleotide sequences of all snake EVEs identified in this study are provided in the electronic supplementary material, dataset S3, and a schematic alignment of these sequences with a representative species of each family is illustrated in the electronic supplementary material, figure S1.
The eSCVs identified are 130–241 aa in length; two of them correspond to the viral capsid protein (53–58% aa similarity, K2P nucleotide distance = 0.64–0.67, over 85% of the length of the porcine circovirus 1 capsid), and three are homologous to the viral replication initiator (Rep) protein (80–82% aa similarity, K2P distance = 0.45–0.49 over 75% of the length of a bat circovirus Rep). Both circovirus genes have been found endogenized in various metazoan genomes [11,15]. Overall, eSCVs cover about 75% of the length of a typical circovirus genome (e.g. porcine circovirus 1 = 1768 bp).
Snake EBLNs are 345 and 368 aa long and 61–63% similar (aa level) to the avian bornavirus nucleoprotein (N) gene over its entire length (K2P distance = 0.74–0.75). In addition to a nucleoprotein, the genome of bornaviruses encodes five other proteins (a phosphoprotein, a matrix protein, a glycoprotein, an RNA-dependent RNA polymerase and an accessory protein). Several endogenous fragments of the matrix, RNA-polymerase and glycoprotein genes have been discovered in the genomes of a number of other vertebrate species [1,12,28,44]. Most endogenous bornaviruses reported to date correspond to the nucleoprotein, which may be owing to the existence of a 3′–5′ transcription gradient of bornavirus genomes, resulting in a higher abundance of nucleoprotein mRNA [28].
Previous studies on endogenous hepadnaviruses characterized cases in two bird species, the zebrafinch Taeniopygia guttata and the budgerigar Melopsittacus undulatus, each of which contained multiple partial copies (n > 10), as well as a full-length or nearly full-length copy of the hepadnaviral genome [8,18,26,54]. In snakes, the eSHBVs are 316–370 aa long, with 48–67% aa similarity (K2P distance = 0.63–0.83) to the avihepadnavirus polymerase and cover about 37% of the duck hepadnavirus genome, which is 3021 bp long.
EVE scans in the cobra and python genomes additionally yielded a 53 aa fragment in the cobra showing 60% similarity to the core protein of the parrot hepatitis B virus, in addition to eSHBV1. In the python, we also discovered a 58 aa fragment showing 72% similarity to the Rep protein of the bat circovirus and a 326 aa fragment showing 67% similarity to the nucleoprotein of the orangutan bornavirus (electronic supplementary material, table S1).
Importantly, all snake EVEs discovered in this study are flanked by different sequences, suggesting that they all derive from independent endogenization events. Furthermore, although it has been shown that endogenization could be catalysed by host retrotransposons [44], we did not detect any molecular signature (such as target-site duplications) indicating involvement of such mobile elements in snake EVEs endogenization.
(b). Estimates of the timing of endogenization events
The results of PCR and bioinformatics screens for orthologous EVEs are depicted in figure 1 and table 1 (see also the electronic supplementary material, table S1). In three instances (eSCV2, eSCV4 and eSCV5), we were able to amplify both the orthologous EVE-containing loci in species closely related to C. mitchellii and the empty EVE loci (i.e. EVEs absent) in more distantly related species, yielding both an upper and lower time limit for the endogenization events. For these three loci, we can therefore confidently conclude that endogenization took place in the ancestor of the (C. mitchellii + Crotalus tigris + Crotalus atrox) clade, between 6 and 8 Ma (figure 1). For the other loci, we were able to amplify only EVE-containing loci. This is because either our PCR design did not allow us to amplify empty sites (see Material and methods) or, irrespective of whether the orthologous loci contained a given EVE or not, they were not sufficiently conserved to allow amplification. In these instances, we could still estimate the lower bound date of endogenization, but only speculate on the upper limit based on the absence of amplification. Thus, our screening revealed that cmEBLN1, eSHBV2, eSCV1 and eSCV3 are at least 8 Myr old and that cmEBLN2 is at least 13 Myr old. Furthermore, we also found that eSHBV1 was present at the orthologous locus in the king cobra genome, showing that this EVE is at least 50 Myr old (electronic supplementary material, dataset S11).
To assess whether the rates of evolution of EVEs are consistent with genome-wide substitution rates in snakes, we calculated the distance between orthologous EVEs and divided this distance by the minimum age of each EVE to obtain a substitution rate (substitutions per site per year). This yielded rate estimates ranging from 4.1 × 10−10 to 2.5 × 10−9 subst. site−1 year−1, with an average of 1.16 × 10−9 subst. site−1 year−1 across EVEs (s.d. = 7.5 × 10−10; table 1). These rates are close to those recently estimated for fourfold degenerate third codon positions based on 44 genes in viperids (1.4 × 10−9 subst. site−1 year−1) and elapids (1.6 × 10−9 subst. site−1 year−1; [30]), suggesting that our inferences about the age of snake EVEs are consistent with what is currently known about the evolutionary dynamics in snake genomes, more broadly. These data also suggest that these elements are evolving at approximately the background neutral rate of the host genomes and are probably non-functional. Overall, the snake EVEs uncovered in this study provide evidence for multiple endogenization events in these lineages over at least the last 50 Myr.
(c). Phylogenetic analyses of snake endogenous viral elements
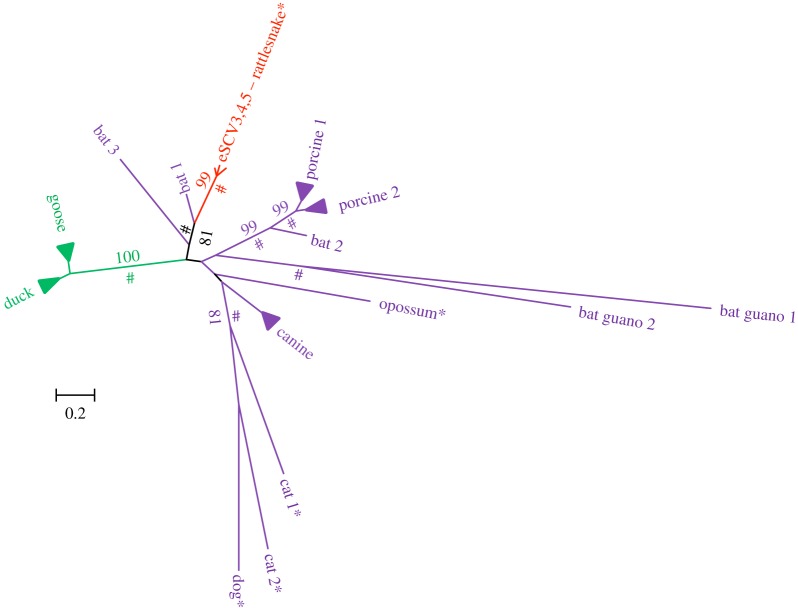
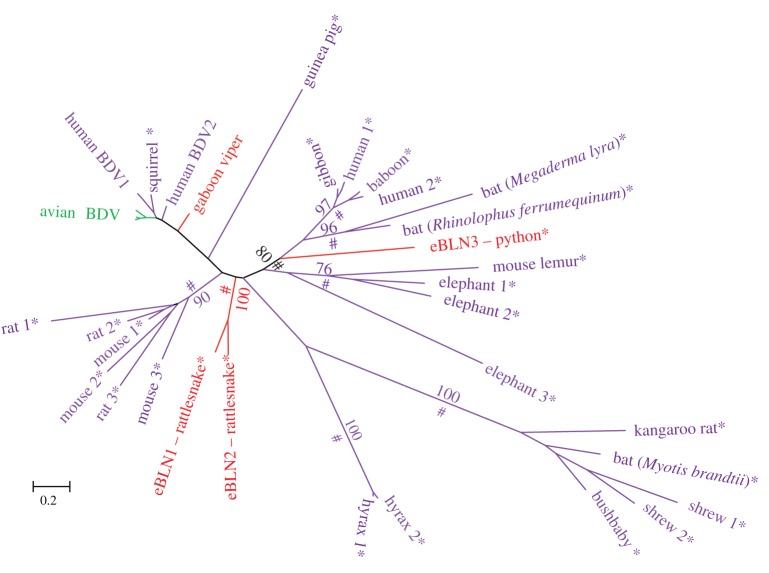
The phylogenetic relationships of all snake EVEs, together with their most closely related known exogenous and endogenous viruses, are shown in figures 2–4 (see also the electronic supplementary material, figure S2, for the circovirus capsid gene tree). In all trees, snake EVEs are characterized by relatively long branches and are distantly related to well-characterized exogenous viruses. Considering that the phylogenetic distance between snake EVEs and known exogenous viruses (e.g. between eSHBVs and other hepadnaviruses) is as large or larger than the distance separating extant viruses known to belong to distinct lineages infecting widely divergent hosts (e.g. mammalian hepadnaviruses versus avian hepadnaviruses), we contend that snake EVEs belong to new lineages that may be considered new viral species or genera. Interestingly, eSCV3, eSCV4 and eSCV5 are separated by very short branches in the tree, figure 2, reflecting minimal divergence between these sequences, potentially indicating that these EVEs result from repeated endogenization of the same virus. Together, these three EVEs form a fairly well-supported cluster (maximum-likelihood BP = 81; Bayesian posterior probability (PP) > 0.9) with an exogenous circovirus sequenced as part of a recent viral metagenomic study conducted on Rhinolophus ferrumequinum bats (figure 2 [55]). This grouping, also recovered in the capsid phylogeny (electronic supplementary material, figure S2), suggests circoviruses may have jumped between mammals and snakes during their evolutionary history. In the phylogeny of bornaviruses (figure 3), it is noteworthy that the python EBLN3 is more closely related to a subset of mammalian EBLNs (BP = 80; PP > 0.9) than it is to the rattlesnake EBLNs. Interestingly, none of the newly described snake EBLNs are closely related to the bornavirus transcript of the gaboon viper reported in [44] which seems to be more closely related to a clade including exogenous bird and human bornaviruses, as well as endogenous squirrel bornavirus (although this group is not well supported in our analysis). Overall, these results suggest repeated endogenization events of widely divergent bornaviruses in snakes and, potentially, multiple transfers between mammals and reptiles.
Figure 2.
Unrooted tree of the circovirus Rep protein. Asterisks indicate endogenous viruses. Numbers on branches correspond to boostrap support values more than 70%. Symbols # indicate posterior probabilities more than 0.9. Branches corresponding to closely related exogenous bird, canine and porcine circoviruses were collapsed for illustrative purposes. The sequence of bat exogenous circoviruses results from three viral metagenomic surveys: Bat 1 and 2 [55], Bat 3 [56], Bat guano 1 and 2 [57]. The alignment used to reconstruct this tree is provided in the electronic supplementary material, dataset S1. (Online version in colour.)
Figure 3.
Unrooted tree of the bornavirus nucleoprotein sequences. Asterisks indicate endogenous viruses. Numbers on branches correspond to boostrap support values more than 70%. Symbols # indicate posterior probabilities more than 0.9. Mammalian endogenous bornaviral sequences were taken from Horie et al. [44]. The alignment used to reconstruct this tree is provided in the electronic supplementary material, dataset S1. (Online version in colour.)
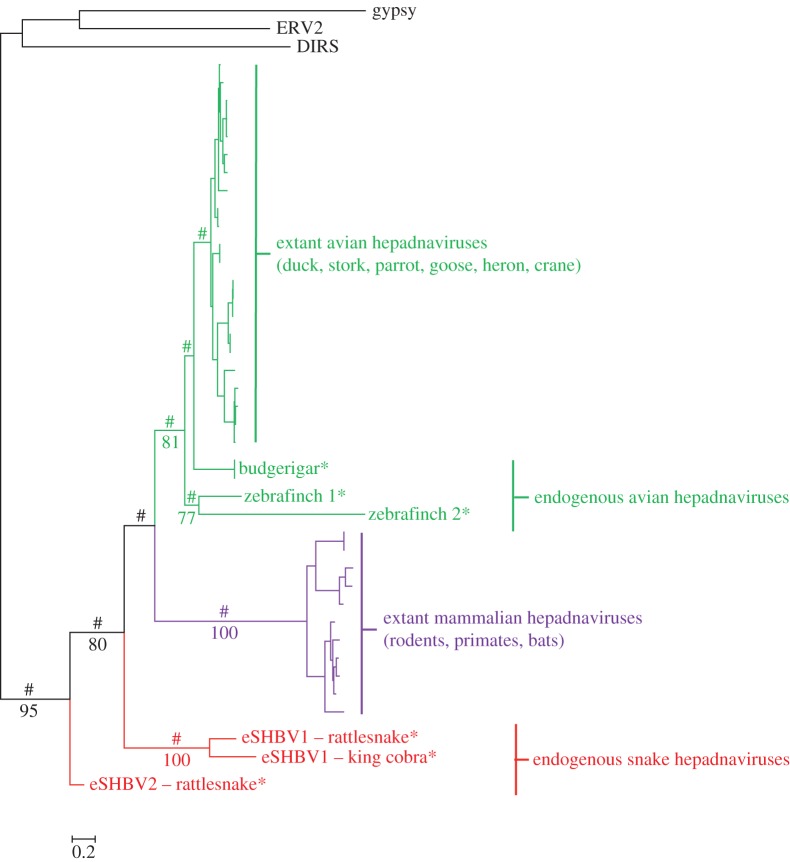
Figure 4.
Phylogenetic tree of the hepadnavirus polymerase. Asterisks indicate endogenous viruses. The tree was rooted using the reverse transcriptase domain of a DIRS and a gypsy retrotransposon, as well as that of an endogenous retrovirus. Numbers on branches correspond to BP support values more than 70%. Symbols # indicate PPs of more than 0.9. Bird endogenous hepadnaviral sequences were taken from Gilbert and Feschotte [26] and Cui & Holmes [8]. The alignment used to reconstruct this tree is provided in the electronic supplementary material, dataset S1. (Online version in colour.)
Interestingly, the cobra and rattlesnake eSHBV1 group tightly together (BP = 100; PP > 0.9), as expected given that the two sequences are orthologous. It is noteworthy that in this grouping, the length of the terminal branches of the two sequences represents the amount of evolution that took place after endogenization at the host genome's substitution rate. In the hepadnavirus tree (figure 4; rooted using the reverse transcriptase of three different retrotransposons), snake eHBVs appear paraphyletic, with eSHBV2 falling sister to a clade including eSHBV1 and mammalian exogenous hepadnaviruses and both avian exogenous and endogenous hepadnaviruses. Although the (eSHBV1 + mammalian hepadnaviruses + avian hepadnaviruses) clade receives moderate BP support (BP = 80) and the position of eSHBV1 within this clade is unresolved (BP < 70), the likely paraphyly of snake EVEs based on this tree indicates that the hepadnavirus phylogeny is not fully congruent with that of their hosts. This presents further evidence that, like bornaviruses and circoviruses, hepadnaviruses have probably jumped between snakes and other vertebrates during their evolution.
Our tests for selection on endogenized viral sequences did not reveal any evidence of purifying or positive selection (p > 0.5 in all tests), suggesting all EVEs have been evolving neutrally in snake genomes after endogenization. We infer long-term viral substitution rates ranging from 1.3 × 10−8 subst. site−1 year−1 for eSHBV1 to 1.1 × 10−7 subst. site−1 year−1 for eSCV2, with an average of 6.7 × 10−8 subst. site−1 year−1 (s.d. = 2.6 × 10−8) (table 1).
4. Discussion
A large number of viruses have been characterized in reptiles [58–60], including at least 11 families reported from snakes. Hepadnaviruses, bornaviruses and circoviruses, however, have not been reported in this group, with the exception of one transcript of a truncated bornavirus nucleoprotein gene (94 aa) found in a Gaboon viper transcriptome [44] and a TBLASTN hit corresponding to our python EBLN3 reported in a supplementary table of Horie et al. [28]. In fact, bornaviruses and hepadnaviruses have never been observed in non-avian reptiles, and only one member of the Circoviridae is known from a non-avian reptile (green sea turtle [61]). The multiple endogenization events we report across taxa, however, show that snakes have been repeatedly exposed to divergent members of these three viral families during the last 50 Myr (table 1). While these data do not imply that snakes are currently interacting with these viruses, they do indicate that they have interacted in the past. Our results broadly illustrate the potential of palaeovirology for shedding light on past host–virus interactions, for developing and testing hypotheses regarding host-switching, and as a guide for targeting plausible host taxa in future searches of circulating viruses. In addition to the identification of previously unreported endogenized viruses in snakes, our phylogenetic analysis reveals a clear lack of congruence between host and virus phylogenies, suggesting the three virus families have probably jumped between mammals and reptiles one or multiple times in the past (figures 2–4).
In the case of circoviruses, the absence of overlap in the geographical distribution of rattlesnakes (restricted to the Americas) and R. ferrumequinum bats (restricted to Europe and Asia) precludes any straightforward hypothesis regarding the mechanism of transfer between these hosts. Similarly, although eSCV3 and eSCV4 have been present in rattlesnake genomes for more than 6 Myr (figure 1), it is not possible to determine whether this transfer between hosts took place before or after endogenization. Our phylogenetic analyses indicate that, like mammals, snakes have also been exposed to widely divergent lineages of bornaviruses during their evolutionary history. In addition to the fact that human exogenous bornaviruses and squirrel endogenous bornaviruses are closer to bird exogenous bornaviruses than to other mammalian EBLNs, the polyphyly of snake EBLNs suggests that, since their origin > 93 Ma [1], bornaviruses have repeatedly jumped between mammals and reptiles (including birds).
It has been shown that the evolution of exogenous hepadnaviruses is characterized by frequent recent host-switching [62,63]. Our rooted tree indicates that snake hepadnaviruses are probably paraphyletic and that bird hepadnaviruses (endogenous + exogenous) are closer to mammalian than to snake hepadnaviruses, which is incongruent with the host phylogeny (in which birds are closer to snakes than to mammals [64]). This suggests that these viruses probably jumped between vertebrate hosts as well. Interestingly, eSHBVs appear to fall outside of the known diversity of bird and mammalian hepadnaviruses (although the position of eSHBV1 has poor BP support) and indicate that these viruses might be characterized as hepadna-like viruses belonging to a new family. However, a unique characteristic of Hepadnaviridae is that their genomes are extremely compact and code for at least two overlapping open reading frames over more than 60% of their length. This characteristic is shared by the two snake endogenous hepadnavirus sequences, both of which encode a polymerase in the +1 frame, as well as the expected overlapping surface protein in the +3 frame (electronic supplementary material, figure S1). Based on this observation, we propose snake eHBVs be considered basally diverging stem-group members of the Hepadnaviridae family.
There are multiple examples where it appears that EVE-derived genes have been exapted by their host, and currently fulfil a variety of host cellular functions [19–22,65]. Our results indicate that snake EVEs show no strong signatures of purifying or positive selection, but instead are evolving neutrally. This indicates that if they play or have played a functional role in the host, it is unlikely to be at the protein level. Thus, the absence of nonsense mutations in eSCV1–5 and eSHBV2 most likely reflects the recent integration of these EVEs into snake genomes, rather than functional conservation owing to purifying selection. A future challenge in palaeovirology studies will be to test whether EVEs could have played and/or still play a role at the RNA level, notably in antiviral immunity [66–69].
Finally, our study provides yet another stunning illustration of the fact that, in spite of the extremely high mutation rates characterizing most modern viruses, the fossils of ancient viral genomes can be identified in their host genomes even after millions, or tens of millions of years [1,12,15]. As observed in previous studies [17,26,70,71], the long-term substitution rates we have inferred for hepadnaviruses and circoviruses (between 1.2 × 10−8 and 7.7 × 10−8 subst. site−1 year−1 and between 5.9 × 10−8 and 7.6 × 10−8 subst. site−1 year−1, respectively), although clearly approximate, are three to five orders of magnitude slower than short-term rates calculated using sequences of circulating viruses (e.g. 1.2 × 10−3 subst. site−1 year−1 for circoviruses [53] and 7.9 × 10−5 subst. site−1 year−1 for hepadnaviruses [52]). It is known that many factors may influence viral substitution rates and their variation over time [72]. Various methodological biases probably influence the calculation of both short- and long-term rates and therefore could explain the large difference between these rates [3,5,73,74], which has also been observed in cellular organisms [75]. Specifically, it has been recently shown that pervasive purifying selection could be one of the major obstacles in accurately estimating the ancient age of recent pathogens [76], and the development of substitution models accounting for this phenomenon are beginning to yield more realistic results [76,77]. It is also important to consider that short-term rates based on pathogenic viruses (which most likely jumped relatively recently) may reflect the ongoing evolutionary arms race with their host, as opposed to their long-term evolutionary history [26]. Furthermore, viral evolution may be constrained owing to both intrinsic structural features of the virus and the diverse immunological strategies of the host [78]. We believe combining palaeovirology and metagenomic studies of modern viruses holds exciting promise for understanding diverse modes of viral evolution and to better grasp the dynamics of long- and short-term host virus interactions.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
C.G. thanks all the technical staff of the UMR CNRS 7267 for assistance in the laboratory and Richard Cordaux and Julien Thézé for insightful discussions. We thank two anonymous reviewers for helpful comments.
The specimen was collected in accordance with a scientific collecting permit (State of Arizona, M30207246) and registered Institutional Animal Care and Use Committee protocols (University of Texas at Arlington).
Data accessibility
Whole Genome Assembly (accession no. JPMF00000000). Voucher specimen: C. m. pyrrhus (JMM 685; UTA R-60292) deposited into the University of Texas at Arlington Amphibian and Reptile Diversity Research Center, Arlington, TX.
Funding statement
We would also like to gratefully acknowledge our funding sources, which include the University of Texas at Arlington faculty start-up funds (T.A.C.), Henry and Lorraine Darley (Nancy Ruth Fund) and the TI Foundation (J.M.M.), Milton L. Fischer Memorial Field Research Award and the Arch and Fran Diack Student Field Research Award (D.D.), Reed College start-up funds and sabbatical fellowship (S.S.), NSF Award MCB-1150213 (S.S.), the M.J. Murdock Charitable Trust (S.S.), the Fulbright Foundation (S.S.) and a fellowship from the Université Claude Bernard Lyon 1 (S.S.).
References
- 1.Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet. 6, e1001191 ( 10.1371/journal.pgen.1001191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katzourakis A. 2013. Palaeovirology: inferring viral evolution from host genome sequence data. Phil. Trans. R. Soc. B 368, 20120493 ( 10.1098/rstb.2012.0493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 13, 283–296. ( 10.1038/nrg3199) [DOI] [PubMed] [Google Scholar]
- 4.Tarlinton RE, Meers J, Young PR. 2006. Retroviral invasion of the koala genome. Nature 442, 79–81. ( 10.1038/nature04841) [DOI] [PubMed] [Google Scholar]
- 5.Holmes EC. 2011. The evolution of endogenous viral elements. Cell Host Microbe. 10, 368–377. ( 10.1016/j.chom.2011.09.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel MR, Emerman M, Malik HS. 2011. Paleovirology: ghosts and gifts of viruses past. Curr. Opin. Virol. 1, 304–309. ( 10.1016/j.coviro.2011.06.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapoor A, Simmonds P, Lipkin WI. 2010. Discovery and characterization of mammalian endogenous parvoviruses. J. Virol. 84, 12 628–12 635. ( 10.1128/JVI.01732-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui J, Holmes EC. 2012. Endogenous hepadnaviruses in the genome of the budgerigar (Melopsittacus undulatus) and the evolution of avian hepadnaviruses. J. Virol. 86, 7688–7691. ( 10.1128/JVI.00769-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui J, Holmes EC. 2012. Evidence for an endogenous papillomavirus-like element in the platypus genome. J. Gen. Virol. 93, 1362–1366. ( 10.1099/vir.0.041483-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballinger MJ, Bruenn JA, Kotov AA, Taylor DJ. 2013. Selectively maintained paleoviruses in Holarctic water fleas reveal an ancient origin for phleboviruses. Virology 446, 276–282. ( 10.1016/j.virol.2013.07.032) [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Fu Y, Xie J, Cheng J, Ghabrial SA, Li G, Peng Y, Yi X, Jiang D. 2011. Widespread endogenization of densoviruses and parvoviruses in animal and human genomes. J. Virol. 85, 9863–9876. ( 10.1128/JVI.00828-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belyi VA, Levine AJ, Skalka AM. 2010. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 6, e1001030 ( 10.1371/journal.ppat.1001030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor DJ, Leach RW, Bruenn J. 2010. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol. Biol. 10, 193 ( 10.1186/1471-2148-10-193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niewiadomska AM, Gifford RJ. 2013. The extraordinary evolutionary history of the reticuloendotheliosis viruses. PLoS Biol. 11, e1001642 ( 10.1371/journal.pbio.1001642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belyi VA, Levine AJ, Skalka AM. 2010. Sequences from ancestral single-stranded DNA viruses in vertebrate genomes: the parvoviridae and Circoviridae are more than 40 to 50 million years old. J. Virol. 84, 12 458–12 462. ( 10.1128/JVI.01789-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thézé J, Bézier A, Periquet G, Drezen JM, Herniou EA. 2011. Paleozoic origin of insect large dsDNA viruses. Proc. Natl Acad. Sci. USA 108, 15 931–15 935. ( 10.1073/pnas.1105580108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefeuvre P, Harkins GW, Lett JM, Briddon RW, Chase MW, Moury B, Martin DP. 2011. Evolutionary time-scale of the begomoviruses: evidence from integrated sequences in the Nicotiana genome. PLoS ONE 6, e19193 ( 10.1371/journal.pone.0019193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suh A, Brosius J, Schmitz J, Kriegs JO. 2013. The genome of a Mesozoic paleovirus reveals the evolution of hepatitis B viruses. Nat. Commun. 4, 1791 ( 10.1038/ncomms2798) [DOI] [PubMed] [Google Scholar]
- 19.Arnaud F, et al. 2007. A paradigm for virus-host coevolution: sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog. 3, e170 ( 10.1371/journal.ppat.0030170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Y, Buckler-White A, Wollenberg K, Kozak CA. 2009. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc. Natl Acad. Sci. USA 106, 3259–3263. ( 10.1073/pnas.0900181106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herniou EA, Huguet E, Thézé J, Bézier A, Periquet G, Drezen JM. 2013. When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses. Phil. Trans. R. Soc. B 368, 20130051 ( 10.1098/rstb.2013.0051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T. 2013. Paleovirology of 'syncytins’, retroviral env genes exapted for a role in placentation. Phil. Trans. R. Soc. B 368, 20120507 ( 10.1098/rstb.2012.0507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballinger MJ, Bruenn JA, Taylor DJ. 2012. Phylogeny, integration and expression of sigma virus-like genes in Drosophila. Mol. Phylogenet. Evol. 65, 251–258. ( 10.1016/j.ympev.2012.06.008) [DOI] [PubMed] [Google Scholar]
- 24.Fort P, et al. 2012. Fossil rhabdoviral sequences integrated into arthropod genomes: ontogeny, evolution, and potential functionality. Mol. Biol. Evol. 29, 381–390. ( 10.1093/molbev/msr226) [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Fu Y, Jiang D, Li G, Xie J, Cheng J, Peng Y, Ghabrial SA, Yi X. 2010. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J. Virol. 84, 11 876–11 887. ( 10.1128/JVI.00955-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert C, Feschotte C. 2010. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol. 8, e1000495 ( 10.1371/journal.pbio.1000495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, et al. 2011. Widespread horizontal gene transfer from circular single-stranded DNA viruses to eukaryotic genomes. BMC Evol. Biol. 11, 276 ( 10.1186/1471-2148-11-276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horie M, Kobayashi Y, Suzuki Y, Tomonaga K. 2013. Comprehensive analysis of endogenous bornavirus-like elements in eukaryote genomes. Phil. Trans. R. Soc. B 368, 20120499 ( 10.1098/rstb.2012.0499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alföldi J, et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477, 587–591. ( 10.1038/nature10390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castoe TA, et al. 2013. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc. Natl Acad. Sci. USA 110, 20 645–20 650. ( 10.1073/pnas.1314475110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vonk FJ, et al. 2013. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl Acad. Sci. USA 110, 20 651–20 656. ( 10.1073/pnas.1314702110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klauber LM. 1997. Rattlesnakes: their habits, life histories and influence on mankind. Berkeley, CA: University of California Press. [Google Scholar]
- 33.Meik JM. 2008. Morphological analysis of the contact zone between the rattlesnakes Crotalus mitchellii stephensi and C. m. pyrrhus. In The biology of rattlesnakes (eds Hayes WK, Beaman KR, Caldwell MD, Bush SP.), pp. 39–46. Loma Linda, CA: Loma Linda University Press. [Google Scholar]
- 34.Campbell JA, Lamar WW. 2004. The venomous reptiles of the western hemisphere. Ithaca, NY: Comstock Pub. Associates. [Google Scholar]
- 35.Reyes-Velasco J, Meik JM, Smith EN, Castoe TA. 2013. Phylogenetic relationships of the enigmatic longtailed rattlesnakes (Crotalus ericsmithi, C. lannomi, and C. stejnegeri). Mol. Phylogenet. Evol. 69, 524–534. ( 10.1016/j.ympev.2013.07.025) [DOI] [PubMed] [Google Scholar]
- 36.Castoe TA, Gu W, de Koning AP, Daza JM, Jiang ZJ, Parkinson CL, Pollock DD. 2009. Dynamic nucleotide mutation gradients and control region usage in squamate reptile mitochondrial genomes. Cytogenet. Genome Res. 127, 112–127. ( 10.1159/000295342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burbrink FT, Pyron RA. 2008. The taming of the skew: estimating proper confidence intervals for divergence dates. Syst. Biol. 57, 317–328. ( 10.1080/10635150802040605) [DOI] [PubMed] [Google Scholar]
- 38.Hedges SB, Dudley J, Kumar S. 2006. TimeTree: a public knowledge base of divergence times among organisms. Bioinformatics 22, 2971–2972. ( 10.1093/bioinformatics/btl505) [DOI] [PubMed] [Google Scholar]
- 39.Gifford R, Tristem M. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26, 291–315. ( 10.1023/A:1024455415443) [DOI] [PubMed] [Google Scholar]
- 40.Anderson PR, Barbacid M, Tronick SR, Clark HF, Aaronson SA. 1979. Evolutionary relatedness of viper and primate endogenous retroviruses. Science 204, 318–321. ( 10.1126/science.219480) [DOI] [PubMed] [Google Scholar]
- 41.Huder JB, Boni J, Hatt J-M, Soldati G, Lutz H, Schupbach J. 2002. Identification and characterization of two closely related unclassifiable endogenous retroviruses in pythons (Python molurus and Python curtus). J. Virol. 76, 7607–7615. ( 10.1128/JVI.76.15.7607-7615.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castoe TA, et al. 2011. Discovery of highly divergent repeat landscapes in snake genomes using high throughput sequencing. Gen. Biol. Evol. 3, 641–653. ( 10.1093/gbe/evr043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, Reavill DR, Dunker F, DeRisi JL. 2012. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio 3, e00180 ( 10.1128/mBio.00180-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horie M, et al. 2010. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463, 84–87. ( 10.1038/nature08695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 46.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. ( 10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 47.Hall T. 2004. BioEdit version 5.0.6 See http://www.mbio.ncsu.edu/BioEdit/bioedit.html.
- 48.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. ( 10.1093/bioinformatics/btr088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rambaut A, Suchard M, Drummond A. 2013. Tracer. See http://tree.bio.ed.ac.uk/software/tracer/.
- 50.Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL. 2008. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583. ( 10.1093/bioinformatics/btm388) [DOI] [PubMed] [Google Scholar]
- 51.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osiowy C, Giles E, Tanaka Y, Mizokami M, Minuk GY. 2006. Molecular evolution of hepatitis B virus over 25 years. J. Virol. 80, 10 307–10 314. ( 10.1128/JVI.00996-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Firth C, Charleston MA, Duffy S, Shapiro B, Holmes EC. 2009. Insights into the evolutionary history of an emerging livestock pathogen: porcine circovirus 2. J. Virol. 83, 12 813–12 821. ( 10.1128/JVI.01719-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu W, Pan S, Yang H, Bai W, Shen Z, Liu J, Xie Y. 2012. The first full-length endogenous hepadnaviruses: identification and analysis. J. Virol. 86, 9510–9513. ( 10.1128/JVI.01164-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He B, et al. 2013. Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel mammalian viruses. PLoS ONE 8, e61950 ( 10.1371/journal.pone.0061950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Z, et al. 2012. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 86, 10 999–11 012. ( 10.1128/JVI.01394-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH, Delwart E. 2010. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 84, 6955–6965. ( 10.1128/JVI.00501-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacobson ER. 2007. Infectious diseases and pathology of reptiles: color atlas and text, p. 736 Boca Raton, FL: CRC Press. [Google Scholar]
- 59.Ariel E. 2011. Viruses in reptiles. Vet. Res. 42, 100 ( 10.1186/1297-9716-42-100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marschang RE. 2011. Viruses infecting reptiles. Viruses 3, 2087–2126. ( 10.3390/v3112087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ng TF, Manire C, Borrowman K, Langer T, Ehrhart L, Breitbart M. 2009. Discovery of a novel single-stranded DNA virus from a sea turtle fibropapilloma by using viral metagenomics. J. Virol. 83, 2500–2509. ( 10.1128/JVI.01946-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Y, Holmes EC. 2007. Bayesian estimates of the evolutionary rate and age of hepatitis B virus. J. Mol. Evol. 65, 197–205. ( 10.1007/s00239-007-0054-1) [DOI] [PubMed] [Google Scholar]
- 63.Locarnini S, Littlejohn M, Aziz MN, Yuen L. 2013. Possible origins and evolution of the hepatitis B virus (HBV). Semin. Cancer Biol. 23, 561–575. ( 10.1016/j.semcancer.2013.08.006) [DOI] [PubMed] [Google Scholar]
- 64.Hugall AF, Foster R, Lee MS. 2007. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Syst. Biol. 56, 543–563. ( 10.1080/10635150701477825) [DOI] [PubMed] [Google Scholar]
- 65.Taylor DJ, Bruenn J. 2009. The evolution of novel fungal genes from non-retroviral RNA viruses. BMC Biol. 7, 88 ( 10.1186/1741-7007-7-88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bertsch C, Beuve M, Dolja VV, Wirth M, Pelsy F, Herrbach E, Lemaire O. 2009. Retention of the virus-derived sequences in the nuclear genome of grapevine as a potential pathway to virus resistance. Biol. Direct 4, 21 ( 10.1186/1745-6150-4-21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flegel TW. 2009. Hypothesis for heritable, anti-viral immunity in crustaceans and insects. Biol. Direct 4, 32 ( 10.1186/1745-6150-4-32) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koonin EV. 2010. Taming of the shrewd: novel eukaryotic genes from RNA viruses. BMC Biol. 8, 2 ( 10.1186/1741-7007-8-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aswad A, Katzourakis A. 2012. Paleovirology and virally derived immunity. Trends Ecol. Evol. 27, 627–636. ( 10.1016/j.tree.2012.07.007) [DOI] [PubMed] [Google Scholar]
- 70.Wertheim JO, Worobey M. 2009. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput. Biol. 5, e1000377 ( 10.1371/journal.pcbi.1000377) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Worobey M, et al. 2010. Island biogeography reveals the deep history of SIV. Science 329, 1487 ( 10.1126/science.1193550) [DOI] [PubMed] [Google Scholar]
- 72.Duffy S, Shackelton LA, Holmes EC. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9, 267–276. ( 10.1038/nrg2323) [DOI] [PubMed] [Google Scholar]
- 73.Gifford RJ. 2012. Viral evolution in deep time: lentiviruses and mammals. Trends Genet. 28, 89–100. ( 10.1016/j.tig.2011.11.003) [DOI] [PubMed] [Google Scholar]
- 74.Holmes EC. 2003. Molecular clocks and the puzzle of RNA virus origins. J. Virol. 77, 3893–3897. ( 10.1128/JVI.77.7.3893-3897.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ho SYW, Phillips MJ, Cooper A, Drummond AJ. 2005. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22, 1561–1568. ( 10.1093/molbev/msi145) [DOI] [PubMed] [Google Scholar]
- 76.Wertheim JO, Kosakovsky Pond SL. 2011. Purifying selection can obscure the ancient age of viral lineages. Mol. Biol. Evol. 28, 3355–3365. ( 10.1093/molbev/msr170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wertheim JO, Chu DK, Peiris JS, Kosakovsky Pond SL, Poon LL. 2013. A case for the ancient origin of coronaviruses. J. Virol. 87, 7039–7045. ( 10.1128/JVI.03273-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meyerson NR, Sawyer SL. 2011. Two-stepping through time: mammals and viruses. Trends Microbiol. 19, 286–294. ( 10.1016/j.tim.2011.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole Genome Assembly (accession no. JPMF00000000). Voucher specimen: C. m. pyrrhus (JMM 685; UTA R-60292) deposited into the University of Texas at Arlington Amphibian and Reptile Diversity Research Center, Arlington, TX.