Abstract
Various animals derive nutrients from symbiotic microorganisms with much-reduced genomes, but it is unknown whether, and how, the supply of these nutrients is regulated. Here, we demonstrate that the production of essential amino acids (EAAs) by the bacterium Buchnera aphidicola in the pea aphid Acyrthosiphon pisum is elevated when aphids are reared on diets from which that EAA are omitted, demonstrating that Buchnera scale EAA production to host demand. Quantitative proteomics of bacteriocytes (host cells bearing Buchnera) revealed that these metabolic changes are not accompanied by significant change in Buchnera or host proteins, suggesting that EAA production is regulated post-translationally. Bacteriocytes in aphids reared on diet lacking the EAA methionine had elevated concentrations of both methionine and the precursor cystathionine, indicating that methionine production is promoted by precursor supply and is not subject to feedback inhibition by methionine. Furthermore, methionine production by isolated Buchnera increased with increasing cystathionine concentration. We propose that Buchnera metabolism is poised for EAA production at certain maximal rates, and the realized release rate is determined by precursor supply from the host. The incidence of host regulation of symbiont nutritional function via supply of key nutritional inputs in other symbioses remains to be investigated.
Keywords: aphid, Buchnera aphidicola, essential amino acid synthesis, metabolic regulation, symbiosis
1. Introduction
Animals cannot synthesize various compounds required for their metabolism and growth, and most animals derive these essential nutrients from their diet. Certain animal groups, however, can use nutrient-deficient diets, because they bear symbiotic microorganisms that provide these compounds [1,2]. In principle, the host derives maximal benefit where the microbial supply of an essential nutrient is scaled to host demand. Undersupply would limit host growth and reproduction, whereas oversupply could be deleterious through allocation of resources to the production of non-required compounds or through toxicity. However, the extent to which microbial function varies with host demand and the underlying mechanisms are largely unknown.
This study concerns the symbiotic bacterium Buchnera aphidicola, which provides essential amino acids (EAAs: arginine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan and valine) to its aphid host [3,4]. Early physiological studies demonstrated reduced incorporation of radioactivity from 14C-substrates into EAAs in aphids reared on diets of high EAA content, suggesting that EAA production by Buchnera is responsive to dietary supply [5,6]. However, Buchnera has very limited capacity to vary gene expression. Its small genome (less than or equal to 0.64 Mb) has a dearth of recognizable regulatory sequences [7], and its gene expression is generally unresponsive to variation in dietary EAAs [8–11].
The purpose of this study was twofold: (i) to determine whether Buchnera supply of EAAs varies with aphid demand, and (ii) to investigate the relative importance of abundance of metabolic enzymes and metabolic processes in determining EAA production by Buchnera. Our experimental approach had three components: a budget analysis that compared EAA production by Buchnera reared on diets individually lacking each EAA and the complete diet containing all EAAs; a quantitative proteomics analysis of the impact of dietary EAAs on the level of Buchnera enzymes in EAA synthesis and, finally, an analysis of metabolic controls over EAA production that focused on Buchnera production of one EAA, methionine.
2. Material and methods
(a). Experimental material
The pea aphid Acyrthosiphon pisum clone CWR09/18 was derived from a single parthenogenetic female collected from an alfalfa field in Freeville, NY, USA in June 2009. This clone bore B. aphidicola and no secondary symbionts, as determined by diagnostic PCR assays and microscopy [12,13]. Routine cultures were maintained on pre-flowering Vicia faba cv. Windsor at 20°C with a 16 L : 8 D cycle. The experimental aphids were obtained by allowing apterous adults to larviposit over 24 h onto chemically defined diet (electronic supplementary material, Methods). The deposited larvae were retained on the diet for an additional 24 h, at which time they were described as 2-day-old larvae. To eliminate Buchnera from the aphids, apterous adults larviposited onto diet supplemented with 50 μg rifampicin ml−1 [14]; these aphids are called ‘aposymbiotic’, and the aphids from rifampicin-free diet are ‘symbiotic aphids’. The larvae were transferred to test diets at 2 days old, and experiments were terminated 5 days later, i.e. at 7 days. Test diets comprised either diets from which individual EAAs were deleted, or dietary methionine was replaced by [U-13C 15N1]-methionine (Cambridge Isotope Laboratories, Cambridge, MA, USA).
Bacteriocytes were dissected from 7-day-old larval aphids into phosphate-buffered saline (PBS) using fine pins and a dissecting microscope at 10× to 40× magnification. For proteomics experiments, the bacteriocyte complement of 60 aphids was brought to 30 μl in PBS, mixed with 10 μl 4× SDS-PAGE loading buffer (125 mM Tris–HCl pH 6.8, 10% v/v ß-mercaptoethanol, 20% v/v glycerol, 4% w/v SDS), and incubated at 90°C for 5 min prior to separation by SDS-PAGE (10–14% acrylamide). For analysis of the free amino acid (FAA) pools, 15 replicate bacteriocyte samples (each from 30 aphids) and whole body samples (each comprising five aphids) were hand-homogenized on ice in PBS, centrifuged, flash-frozen and stored at −80°C, prior to analysis.
Buchnera preparations for metabolic experiments were obtained from dissected bacteriocytes exactly as in [15]. To initiate methionine release experiments, six replicate samples of Buchnera (2 × 108 cells ml−1 in 5 μl medium comprising 28 mM glucose, 8.6 mM NaCl, 1 mM MgSO4, 0.1 mM CaCl2, 0.25 M sucrose, 50 mM NaH2PO4, 13 mM K2H2PO4, pH 7.5) were combined with equal volume of medium supplemented 2 mM glutamate, glutamine, serine, aspartate and 2-oxobutanoate, together with cystathionine at 0–3 mM. At 5 min intervals over 30 min, one tube was centrifuged at 1000g for 70 s, and the supernatant was immediately flash-frozen in liquid nitrogen and stored at −80°C. The experiments were conducted at 22.5°C, and were repeated five times at different times on different sets of aphids.
(b). Budget analysis
EAA production by Buchnera was estimated by budget analysis, as used previously [3,6,16]. Protein growth of aphids on diets individually lacking each EAA was calculated over 2–7 days of larval development from the weight and protein density (mg protein g−1 weight) of the aphids and the increase in each EAA determined from the %EAA content of aphid protein (electronic supplementary material, Methods, tables S1 and S2). Protein growth of aposymbiotic aphids was adopted as a measure of endogenous reserves, and the difference between the values for symbiotic and aposymbiotic aphids represented Buchnera contribution to EAA growth. The Buchnera contribution to EAA growth of symbiotic aphids on the control diet was determined from the difference between the inputs from food (calculated from the volume of food ingested) and EAA outputs, comprising protein growth and elimination via honeydew (electronic supplementary material, Methods and table S3).
(c). Proteomics
The proteome was obtained for three independent biological replicates of bacteriocytes dissected from 7-day-old aphids reared on the complete diet and diets that lacked one of cysteine, isoleucine, leucine, lysine methionine, phenylalanine/tyrosine or valine. The three replicate datasets were obtained in three consecutive months. Each lane of the SDS-PAGE gel of bacteriocyte proteins was cut into 10 slices, and the proteins were reduced, alkylated and digested with trypsin and peptide extracted as in [17]. The extracted peptides were analysed by nanoLC-LTQ-Orbitrap (Thermo Electron) mass spectrometry using data-dependent acquisition and dynamic exclusion as detailed in the electronic supplementary material, Methods. Quantitative differences among samples were analysed using normalized values (NadjSPC) (derived as described in the electronic supplementary material, Methods) using principal components analysis (PCA) and multidimensional ANOVA, in the statistical platform ‘R’, following confirmation of high between-replicate reproducibility by pairwise correlation analysis (electronic supplementary material, table S4).
Mass spectrometry data matched to identified aphid and Buchnera proteins can be viewed in the Plant Proteome DataBase (PPDB) at http://ppdb.tc.cornell.edu/ under experimental identification numbers: #1136–1143 (Repl 1), #1150–1157 (Repl 2), 1158–1165 (Repl 3).
(d). Chemical analyses
For the metabolic analysis of methionine synthesis, the gas chromatography-mass spectrometry (GC-MS) analysis of [U-13C 15N1]-methionine content of 7-day-old aphids was conducted on protein hydrolysates (electronic supplementary material, Methods), using a Varian CP-3800 GC coupled to an CombiPal autosampler and a Varian 1200L triple quadrupole MS (Varian, Carey, NC, USA). Electron ionization (EI)-MS spectra were collected at 70 eV, and the mass of individual chromatographic peaks was compared to the Palisade spectral library and the retention times and mass spectra of labelled and unlabelled standards. Chemical ionization (CI)-MS data were used to assess parent ions of components that were labelled with more than one atom. Electronic supplementary material, figure S1 shows the fragment ions that were monitored to identify and measure the ion abundance of 13C-labelled and unlabelled methionine in the mass spectra.
The samples used to determine the FAA content of honeydew, the cystathionine and methionine content of bacteriocytes, and whole body samples, and to quantify methionine release from Buchnera preparations were analysed using the AccQ Tag derivatization kit (Waters) by UPLC with PDA detector (Waters Acquity), as in [18] (electronic supplementary material, Methods).
3. Results
(a). Net production of most essential amino acids by Buchnera is elevated in aphids on diets lacking that essential amino acid
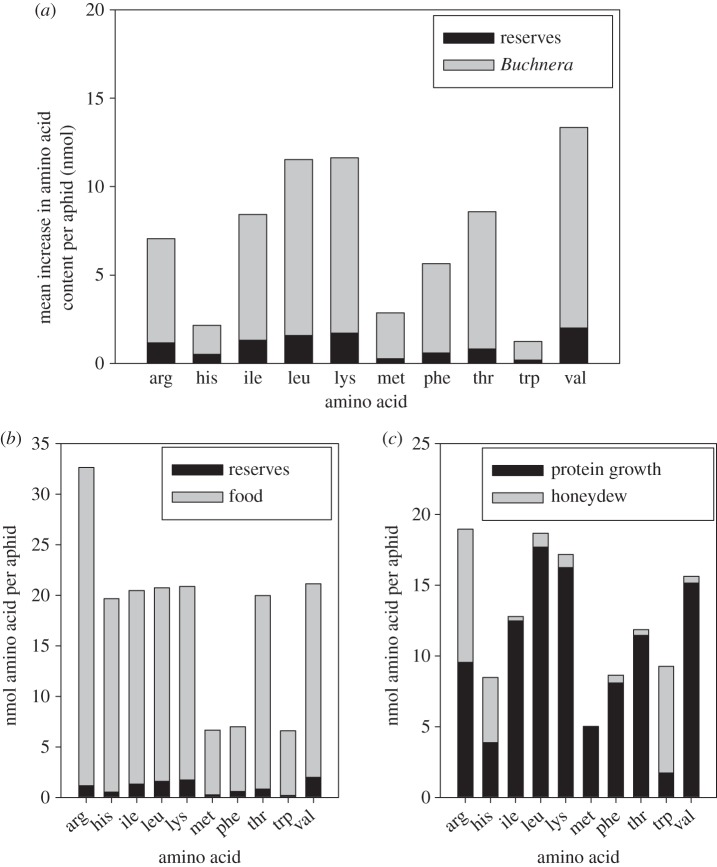
Protein synthesis is the principal fate of EAAs acquired by aphids [19]. We reasoned that the amount of each EAA contributing to protein growth of larval aphids on the diet lacking that EAA was derived from: (i) pre-existing reserves, as quantified by protein growth of aposymbiotic aphids (electronic supplementary material, table S2a), and (ii) Buchnera, quantified as the difference between the protein growth of symbiotic and aposymbiotic aphids (electronic supplementary material, table S2b). The calculated amount of each EAA provided by Buchnera varied more than 10-fold among the amino acids, from 1.03 nmol tryptophan to 11.35 nmol valine (figure 1a and electronic supplementary material, table S2b).
Figure 1.
EAA relations of 2–7-day-old pea aphids. (a) Contribution of reserves and de novo synthesis by Buchnera to protein growth on diets lacking each EAA. (b) Inputs from reserves and food to protein-EAAs in pea aphids on the complete diet. (c) Outputs to protein growth and honeydew of EAAs in pea aphids on the complete diet.
To establish whether the net production of EAAs by Buchnera differed between aphids reared on diets lacking each EAA and the complete diet (i.e. containing all EAAs), the budget for each EAA in aphids on the complete diet was constructed. The outputs comprised the EAA contribution to protein growth and EAA in the honeydew (electronic supplementary material, table S3a), and the known inputs comprised EAAs acquired from food and reserves (electronic supplementary material, table S3b). Net production of EAAs by Buchnera on the complete diet (indicated by a lower value of inputs than outputs) was observed for just two of the 10 EAAs, phenylalanine and tryptophan (figure 1b,c). For phenylalanine, net production on the complete diet comprised 1.66 nmol, depressed by 67% relative to the phenylalanine-free diet (5.05 nmol), but the production of tryptophan on the complete diet was 2.66 nmol, more than twofold greater than the value, 1.03 nmol, on the tryptophan-free diet (electronic supplementary material, table S5).
These data indicate that the net production of EAAs by Buchnera varies with diet; for nine of the 10 EAAs, production by Buchnera is reduced in aphids provided with that EAA, but tryptophan, exceptionally, displays the reverse relationship (figure 1b,c; electronic supplementary material, table S5). Possible reasons for the atypical response of tryptophan are addressed in the Discussion.
(b). Dietary essential amino acid omission does not significantly alter the level of amino acid metabolism enzymes
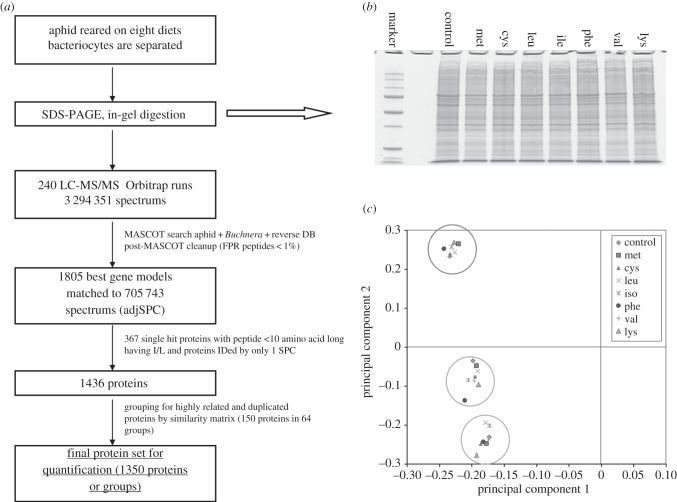
The effect of dietary EAA deletion on the abundance of enzymes in EAA biosynthetic pathways in bacteriocytes of 7-day-old aphids was investigated by quantitative proteomics analysis (figure 2a,b and electronic supplementary material, table S6). The workflow yielded 1350 proteins or protein groups (figure 2a) for quantitation, of which 946 were aphid proteins and 404 Buchnera proteins, including all Buchnera enzymes in EAA biosynthesis. PCA revealed that 99% of the variation in the dataset could be assigned to the first principal component (PC1), indicating a high degree of similarity across all the samples (figure 2c). The samples separated on PC2 by biological replicate, with minimal variation among diet treatments within each biological replicate. Multidimensional ANOVA revealed two of the 1435 proteins with statistically significant variation (p < 0.01) across diet treatments: ACYPI004142 (p = 10−6, heterogeneous nuclear ribonucleoprotein H, hnRNP H), which contributes to the hnRNP complex in mRNA processing, and ACYPI006133 (p = 0.008, adenylyl cyclase-associated protein 1: p = 0.009), contributing to cAMP signalling (electronic supplementary material, table S7). These genes are not known to be associated with EAA synthesis, and the differences are probably false positives.
Figure 2.
Quantitative proteomic analysis of bacteriocytes dissected from 7-day-old aphids that had been reared from day 2 on the complete diet and seven amino acid omission diets. (a) Experimental and bioinformatics workflow. (b) One-dimensional-SDS-PAGE gel lanes of the samples: complete diet (standard) and diets without cysteine (cys), isoleucine (ile), leucine (leu), lysine (lys), methionine (met), phenylalanine and tyrosine (phe + tyr) and valine (val). One of the three independent biological replicates is shown. (c) Principal components PC1 and PC2, with variance assigned to each axis in parentheses.
These data provided the strongest evidence that diet-dependent variation in EAA production by Buchnera in the pea aphid cannot be attributed to the regulation of the abundance of EAA biosynthesis enzymes or any other proteins in Buchnera or bacteriocytes.
(c). Precursor concentration regulates the rate of production of the essential amino acid methionine
To investigate the regulation of EAA synthesis further, we focused on a single EAA, methionine, which is synthesized from cystathionine via two reactions: aphid-mediated transformation of cystathionine to homocysteine, which is metabolized to methionine by a Buchnera reaction (homocysteine S-methyltransferase, MetE). We selected this EAA because metabolic analysis of its synthesis is facilitated by the fact that the host-derived precursor is not used in any other Buchnera reaction [18].
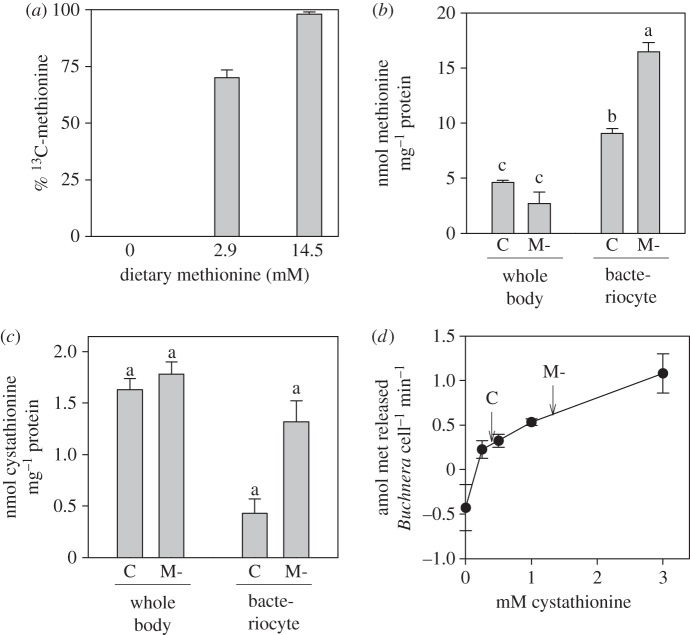
We first quantified the contribution of endogenous sources (i.e. endogenous reserves and Buchnera) to the methionine content of the aphids, to verify the conclusion from the budget analysis that methionine production by Buchnera is depressed in aphids reared on diets containing methionine. We fed the aphids on [13C 15N]-methionine diets from day 2 to day 7, and reasoned that all [13C 15N]-methionine recovered from the aphids would be unmodified from the diet, while unlabelled methionine would be derived from endogenous sources. On the complete diet which contains 2.9 mM methionine, 30% of the methionine in the aphids was identified as endogenous in origin (figure 3a). Our budgetary analysis indicated that pre-existing reserve of methionine (0.27 nmol: electronic supplementary material, table S2a) accounts for an estimated 5% of the aphid methionine content (4.9 nmol: electronic supplementary material, table S3a). By subtraction, 25% of the methionine acquired by the aphids over the experimental period is estimated to be derived from Buchnera. On diet with five times greater methionine concentration (14.5 mM [13C 15N]-methionine), the diet supplied 98% of the aphid methionine, indicating that the Buchnera contribution was negligible (figure 3a). These experiments demonstrate that the supply of methionine from Buchnera to aphid protein is reduced in aphids reared on diets containing that EAA.
Figure 3.
Metabolic basis of diet-dependent variation in methionine synthesis. (a) 13C-methionine content of aphids reared from day 2 to day 7 on diets with different methionine content, provided as 13C-methionine (4–6 replicates per treatment); 2.9 mM methionine is the concentration in the complete diet. (b,c) Concentration of methionine (b) and cystathionine (c) in whole body and bacteriocytes of aphids on complete diet (C) and methionine-free diet (M-), displayed as mean ± s.e. (10 replicates). ANOVA for methionine: tissue F1,36 = 244.82, p < 0.001; diet F1,36 = 22.16, p < 0.001; interaction F1,36 = 63.63, p < 0.001. ANOVA for cystathionine: tissue F1,36 = 306.37, p < 0.001; diet F1,36 = 59.85, p < 0.001; interaction F1,36 = 46.0, p < 0.001. (d) Variation in rate of methionine release by isolated Buchnera cells incubated with different concentrations of cystathionine, displayed as mean ± s.e. (five replicates). Arrows indicate the cystathionine concentration in bacteriocytes of aphids reared on the complete diet (C) and methionine-free diet (M-).
The metabolic controls over diet-dependent production of methionine by Buchnera could comprise reduced methionine (‘pull metabolism’) or increased cystathionine precursor (‘push metabolism’) in bacteriocytes of aphids on methionine-free diet. The concentrations of both cystathionine and methionine were significantly elevated in the bacteriocytes of aphids on the methionine-free diet, but the diet did not significantly affect their concentrations in the whole body (figure 3b,c). These results suggest that elevated precursor concentration in the bacteriocytes of aphids on methionine-free diet may drive increased rates of methionine synthesis.
To test the hypothesis that methionine synthesis rates by Buchnera vary with the supply of the cystathionine precursor, we made use of the fact that, although Buchnera cannot be cultured, it can be isolated from the symbiosis in a viable condition and releases EAAs at linear rates for at least 1 h [15]. Buchnera released no detectable methionine when incubated in cystathionine-free medium, and the rate of methionine release increased with increasing cystathionine concentration in the range 0.25–3 mM that includes the concentrations in bacteriocytes of aphids on the complete and methionine-free diets (figure 3d).
4. Discussion
Amino acid biosynthesis in free-living bacteria is regulated by repression of gene expression and feedback inhibition of enzyme activity, such that the amino acid supply is matched to demand for bacterial growth and proliferation [20]. The evolutionary transition to the symbiotic lifestyle in Buchnera has involved the overproduction of EAAs to support the biomass increase of both Buchnera cells and host tissues, with predicted changes in the regulation of EAA synthesis. The budgetary and methionine metabolism analyses in this study (figures 1 and 3a) demonstrate that this metabolic shift has been accompanied by a fundamental change in the regulation of symbiont metabolism, such that EAA production is scaled to host demand. In this way, EAA synthesis by Buchnera is analogous to food collection by a worker honeybee whose foraging intensity is dictated largely by the nutritional status of the hive (analogous to the aphid) and not its individual nutritional requirements [21–23].
Any hypothesized mechanism that integrates EAA production by Buchnera with EAA demand of the aphid host must take into account, first, the small genetic repertoire of Buchnera including the dearth of recognizable signal transduction genes and regulatory sequences [7,24] and, second, the evidence that diet-driven variation in EAA production by Buchnera in the pea aphid is not accompanied by large changes in transcript [9,11] or protein levels (this study). The conclusion from the current study that the synthesis of the EAA methionine is determined by precursor supply is fully compatible with these constraints. The most parsimonious explanation for our findings is that the EAA biosynthetic capability of Buchnera is poised for maximal production of EAAs, with actual rates determined by the availability of precursors from the host. The very small metabolic network of Buchnera [15,18], with few or no competing reactions that would divert EAA precursors into alternative pathways seems adapted for regulation by substrate supply. An important topic for future research is the mechanisms by which host demand for individual EAAs results in changed precursor concentration in the bacteriocyte. We hypothesize that the processes operate at the whole insect level, potentially involving integration of the nutritional regulatory circuits across multiple organs (e.g. gut, fat body and bacteriocytes), and probably involving target of rapamycin signalling, which is strongly responsive to amino acids [25,26], together with regulation of the function of transporters on the bacteriocyte cell membrane [27].
In principle, the regulation of EAA production by precursor supply may be augmented by the efficient host removal of EAAs, by a combination of high affinity transporters and metabolism to other products. These processes would tend to reduce feedback inhibition in the symbiont and are known to contribute to high efflux in certain other symbioses, e.g. of ammonia from nitrogen-fixing rhizobia to the host cell in leguminous plants [28–30]. However, our finding that methionine levels are elevated in bacteriocytes from aphids reared on methionine-free diet (figure 3b) is not readily compatible with host-mediated alleviation of feedback inhibition. A further indication that metabolic removal of EAAs in aphid bacteriocytes is not central to the regulation of EAA production comes from the evidence that the protein synthesis machinery is not enriched in bacteriocytes [13,18,31], although certain aphid amino acid transporters in bacteriocytes may mediate efficient EAA export from the bacteriocyte [32].
The regulation of EAA production in the aphid symbiosis by precursor supply would enable more rapid changes in EAA flux than could be achieved by mechanisms dependent on change in gene expression. Arguably, this is advantageous for phloem sap feeding insects, because the concentration of individual EAAs in phloem sap can vary widely, including over small spatio-temporal scales [3,33,34]. The capacity of the symbiosis to integrate bacterial and dietary supply of EAAs with minimal time-lag would promote metabolic efficiency and support the very high growth and reproductive rates of these insects.
These considerations raise an important experimental issue: that the precise control over dietary inputs required for our experiments can be achieved with chemically defined diets but not the natural diet of plant phloem sap. Aphid diets differ from phloem sap in important ways. Their amino acid composition is fixed; they have a more balanced amino acid composition than phloem sap; and the individual EAA deletions used in this study are not representative of phloem sap variation. There is also evidence that some aspects of aphid metabolism differ between diet- and plant-reared aphids. In particular, diet-reared aphids produce honeydew with high concentrations of nitrogen-rich EAAs, especially arginine, histidine and tryptophan [35,36], and a proportion of the honeydew tryptophan can be synthesized by Buchnera [37]. These three nitrogen-rich amino acids have been interpreted as vehicles for elimination of excess nitrogen [19,35], a specific response to the nutrient-rich diets that are required to provide sufficient phagostimulatory cues to induce feeding by diet-reared aphids in the absence of other plant-associated cues. Even though the regulation of EAA production did not evolve in the context of the amino acid profiles in chemically defined diets, the core regulatory mechanisms are displayed, and can be investigated, in diet-reared aphids. As plants genetically manipulated to vary systematically in phloem amino acid composition become available, it will become increasingly possible to test the predictions of regulation by precursor supply on plant-reared aphids.
A defining feature of the aphid symbiosis is the small genome content of the Buchnera symbiont, largely attributable to genomic deterioration [24,38]. As a result of its very limited genetic repertoire, Buchnera is metabolically fastidious, and its growth is inferred from analyses in silico to be dependent on host supply of 33 metabolites [18]. This nutritional fastidiousness is paralleled by a loss of regulatory capability [7,9,11], such that the host dictates the Buchnera metabolic function in real time by controlling key inputs to the Buchnera metabolic network (this study). Symbiotic bacteria with much-reduced genomes occur in various insects and other animals [2,24], and we predict that, as in Buchnera, their provisioning of nutrients to their hosts is regulated by the concentration of host precursors.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Funding statement
We thank National Science Foundation, USA (grant no. IOS-0919765) and graduate fellowship to C.W.R. from Sarkaria Institute of Insect Physiology and Toxicology for financial support.
References
- 1.Buchner P. 1965. Endosymbioses of animals with plant microorganisms. Chichester, UK: John Wiley & Sons. [Google Scholar]
- 2.Douglas AE. 2010. The symbiotic habit. Princeton, NJ: Princeton University Press. [Google Scholar]
- 3.Gunduz E, Douglas AE. 2009. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc. R. Soc. B 276, 987–991. ( 10.1098/rspb.2008.1476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douglas AE. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43, 17–37. ( 10.1146/annurev.ento.43.1.17) [DOI] [PubMed] [Google Scholar]
- 5.Febvay G, Rahbe Y, Rynkiewicz M, Guillaud J, Bonnot G. 1999. Fate of dietary sucrose and neosynthesis of amino acids in the pea aphid, Acyrthosiphon pisum, reared on different diets. J. Exp. Biol. 202, 2639–2652. [DOI] [PubMed] [Google Scholar]
- 6.Douglas AE, Minto LB, Wilkinson TL. 2001. Quantifying nutrient production by the microbial symbionts in an aphid. J. Exp. Biol. 204, 349–358. [DOI] [PubMed] [Google Scholar]
- 7.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86. ( 10.1038/35024074) [DOI] [PubMed] [Google Scholar]
- 8.Moran NA, Dunbar HE, Wilcox JL. 2005. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J. Bacteriol. 187, 4229–4237. ( 10.1128/JB.187.12.4229-4237.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reymond N, et al. 2006. Different levels of transcriptional regulation due to trophic constraints in the reduced genome of Buchnera aphidicola APS. Appl. Environ. Microbiol. 72, 7760–7766. ( 10.1128/AEM.01118-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinuelas J, Febvay G, Duport G, Colella S, Fayard JM, Charles H, Rahbe Y, Calevro F. 2011. Multimodal dynamic response of the Buchnera aphidicola pLeu plasmid to variations in leucine demand of its host, the pea aphid Acyrthosiphon pisum. Mol. Microbiol. 81, 1271–1285. ( 10.1111/j.1365-2958.2011.07760.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson AC, Dunbar HE, Davis GK, Hunter WB, Stern DL, Moran NA. 2006. A dual-genome microarray for the pea aphid, Acyrthosiphon pisum, and its obligate bacterial symbiont, Buchnera aphidicola. BMC Genomics 7, 50 ( 10.1186/1471-2164-7-50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas AE, Francois CLMJ, Minto LB. 2006. Facultative ‘secondary’ bacterial symbionts and the nutrition of the pea aphid, Acyrthosiphon pisum. Physiol. Entomol. 31, 262–269. ( 10.1111/j.1365-3032.2006.00516.x) [DOI] [Google Scholar]
- 13.Poliakov A, Russell CW, Ponnala L, Hoops HJ, Sun Q, Douglas AE, van Wijk KJ. 2011. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Mol. Cell. Proteomics 10, M110007039 ( 10.1074/mcp.M110.007039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahbé Y, Delobel B, Febvay G, Chategrel B. 1994. Aphid specific triglycerides in symbiotic and aposymbiotic Acyrthosiphon pisum. Insect Biochem. Mol. Biol. 24, 95–101. ( 10.1016/0965-1748(94)90127-9) [DOI] [Google Scholar]
- 15.Russell CW, Bouvaine S, Newell PD, Douglas AE. 2013. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl. Environ. Microbiol. 79, 6117–6123. ( 10.1128/AEM.01543-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas GH, Zucker J, Macdonald SJ, Sorokin A, Goryanin I, Douglas AE. 2009. A fragile metabolic network adapted for cooperation in the symbiotic bacterium Buchnera aphidicola. BMC Syst. Biol. 3, 24 ( 10.1186/1752-0509-3-24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zybailov B, Friso G, Kim J, Rudella A, Rodriguez VR, Asakura Y, Sun Q, van Wijk KJ. 2009. Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol. Cell. Proteomics 8, 1789–1810. ( 10.1074/mcp.M900104-MCP200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macdonald SJ, Lin GG, Russell CW, Thomas GH, Douglas AE. 2012. The central role of the host cell in symbiotic nitrogen metabolism. Proc. R. Soc. B 279, 2965–2973. ( 10.1098/rspb.2012.0414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douglas AE. 2003. The nutritional physiology of aphids. Adv. Insect Physiol. 31, 73–140. ( 10.1016/S0065-2806(03)31002-1) [DOI] [Google Scholar]
- 20.Umbarger HE. 1978. Amino acid biosynthesis and its regulation. Annu. Rev. Biochem. 47, 532–606. ( 10.1146/annurev.bi.47.070178.002533) [DOI] [PubMed] [Google Scholar]
- 21.Ament SA, Wang Y, Robinson GE. 2010. Nutritional regulation of division of labor in honey bees: toward a systems biology perspective. Wiley interdisciplinary reviews. Syst. Biol. Med. 2, 566–576. ( 10.1002/wsbm.73) [DOI] [PubMed] [Google Scholar]
- 22.Jarau S, Hrncir M. 2009. Food exploitation by social insects: ecological, behavioral and theoretical approaches. Boca Raton, FL: CRC Press. [Google Scholar]
- 23.Seeley TD. 1995. The wisdom of the hive: the social physiology of honey bee colonies. Cambridge, MA: Harvard University Press. [Google Scholar]
- 24.McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10, 13–26. [DOI] [PubMed] [Google Scholar]
- 25.Loewith R, Hall MN. 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201. ( 10.1534/genetics.111.133363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nobukuni T, et al. 2005. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl Acad. Sci. USA 102, 14 238–14 243. ( 10.1073/pnas.0506925102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price DR, Feng H, Baker JD, Bavan S, Luetje CW, Wilson AC. 2014. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc. Natl Acad. Sci. USA 111, 320–325. ( 10.1073/pnas.1306068111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masalkar P, Wallace IS, Hwang JH, Roberts DM. 2010. Interaction of cytosolic glutamine synthetase of soybean root nodules with the C-terminal domain of the symbiosome membrane nodulin 26 aquaglyceroporin. J. Biol. Chem. 285, 23 880–23 888. ( 10.1074/jbc.M110.135657) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mouritzen P, Rosendahl L. 1997. Identification of a transport mechanism for NH4+ in the symbiosome membrane of pea root nodules. Plant Physiol. 115, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udvardi M, Poole PS. 2013. Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 781–805. ( 10.1146/annurev-arplant-050312-120235) [DOI] [PubMed] [Google Scholar]
- 31.Hansen AK, Moran NA. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc. Natl Acad. Sci. USA 108, 2849–2854. ( 10.1073/pnas.1013465108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price DR, Duncan RP, Shigenobu S, Wilson AC. 2011. Genome expansion and differential expression of amino acid transporters at the aphid/Buchnera symbiotic interface. Mol. Biol. Evol. 28, 3113–3126. ( 10.1093/molbev/msr140) [DOI] [PubMed] [Google Scholar]
- 33.Gattolin S, Newbury HJ, Bale JS, Tseng HM, Barrett DA, Pritchard J. 2008. A diurnal component to the variation in sieve tube amino acid content in wheat. Plant Physiol. 147, 912–921. ( 10.1104/pp.108.116079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kallarackal J, Bauer SN, Nowak H, Hajirezaei MR, Komor E. 2012. Diurnal changes in assimilate concentrations and fluxes in the phloem of castor bean (Ricinus communis L.) and tansy (Tanacetum vulgare L.). Planta 236, 209–223. ( 10.1007/s00425-012-1600-7) [DOI] [PubMed] [Google Scholar]
- 35.Prosser WA, Douglas AE. 1991. The aposymbiotic aphid: an analysis of chlortetracycline-treated pea aphid, Acyrthosiphon pisum. J. Insect Physiol. 37, 713–719. ( 10.1016/0022-1910(91)90104-8) [DOI] [Google Scholar]
- 36.Sasaki T, Hayashi H, Ishikawa H. 1991. Growth and reproduction of the symbiotic and aposymbiotic pea aphids, Acyrthosiphon pisum maintained on artificial diets. J. Insect Physiol. 37, 749–756. ( 10.1016/0022-1910(91)90109-D) [DOI] [Google Scholar]
- 37.Douglas AE, Prosser WA. 1992. Synthesis of the essential amino acid tryptophan in the pea aphid (Acyrthosiphon pisum) symbiosis. J. Insect Physiol. 38, 565–568. ( 10.1016/0022-1910(92)90107-O) [DOI] [Google Scholar]
- 38.Moran NA. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl Acad. Sci. USA 93, 2873–2878. ( 10.1073/pnas.93.7.2873) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.