Abstract
Different models of speciation predict contrasting patterns in the relationship between the dispersal ability of lineages and their diversification rates. This relationship is expected to be negative in isolation-limited models and positive in founder-event models. In addition, the combination of negative and positive effects of dispersal on speciation can result in higher diversification rates at intermediate levels of dispersal ability. Using molecular phylogenies to estimate diversification rates, and wing morphology to estimate dispersal ability, we analysed the influence of dispersal on diversification in the avifauna of Australasian archipelagoes. Contrary to expectations given the fragmented nature of island systems, the relationship between dispersal ability and diversification rate was monotonically negative. While multiple mechanisms could generate this pattern, they all share a phase of range expansion that is decoupled from speciation.
Keywords: intermediate dispersal model, diversification rates, ornithology, Melanesia, speciation
1. Introduction
The role of dispersal in controlling rates of diversification remains poorly understood. For most traditional modes of allopatric speciation, such as vicariance, an increase in dispersal ability should decrease speciation rates because barriers become less effective in limiting gene flow [1–3], resulting in a negative relationship between dispersal and diversification. However, if speciation is predominantly the result of a colonization event (founder-event speciation sensu [4]), then an increase in dispersal ability should raise speciation rates, because it increases the chances of colonization of new areas. Speciation after colonization can be triggered by founder effects [5,6] or ecological speciation due to exposure to new habitats and selective pressures [7–9]. In addition, the relationship between dispersal ability and diversification may not be monotonic; instead, speciation rates may be maximized at intermediate levels of dispersal, as in the intermediate dispersal model [10–12]. According to this model, lineages with the highest dispersal abilities have lower chances of speciation owing to high levels of gene flow between populations, whereas lineages with the lowest dispersal abilities have lower chances of speciation owing to lower rates of origination of barriers within their restricted geographical ranges. By contrast, lineages with intermediate dispersal abilities experience a combination of geographical expansion and subdivision that results in high speciation rates [1,10–13].
Empirical examination of the relationship between dispersal ability and diversification has been challenging because of the difficulties in quantifying both dispersal ability and diversification rates. Recent studies of birds have found negative, positive and unimodal relationships between dispersal and diversification. For example, in the continental radiation of the Furnariidae, a diverse family of suboscine passerines from the Neotropics, there is a predominantly negative relationship between dispersal ability, as inferred from wing shape, and speciation rates estimated using a calibrated phylogeny [12]. By contrast, a macroevolutionary analysis of all bird families recovered a positive relationship between diversification rates and an index of dispersal propensity based on ecological characteristics [14]. Finally, the avifauna of Northern Melanesia inspired the intermediate dispersal model by showing a unimodal relationship between an index of intraspecific differentiation and assessments of dispersal ability based on behaviour and biogeography [11].
The discrepancies between these studies may be caused by the differences in the methods used for estimating dispersal and/or diversification. For example, the index of diversification used for the study of the Melanesian avifauna was a count of the number of subspecies and allospecies within each species; because this index does not take clade age into account, it is a measure of clade diversity, not diversification rate. Alternatively, the varied findings across studies may reflect real differences in the relationship between dispersal and diversification. In particular, the degree of discontinuity of the geographical setting may determine the relationship between dispersal and speciation rate [12]. Within continents, dispersal may have a predominantly negative effect because even poor dispersers can colonize remote areas, and increased dispersal results in elevated levels of gene flow across weak or moderate barriers. By contrast, in highly discontinuous geographies like archipelagoes, because range expansion may be a limiting factor, long-distance dispersal may enhance speciation by allowing lineages to colonize new regions and subsequently diversify [11].
Here, we revisit the relationship between dispersal ability and diversification rates in the avifauna of Australasian archipelagoes with a focus on Northern Melanesia, using new estimates of dispersal ability and diversification rates. Because of the highly discontinuous geography of Australasian archipelagoes, we predict that colonization is a limiting factor controlling rates of diversification, resulting in either a positive or a unimodal relationship between dispersal ability and diversification rates [11,12].
2. Material and methods
The study region consists of archipelagoes east of Wallace's line, including Eastern Wallacea, Melanesia, Micronesia and Polynesia, but excluding islands on the Australopapuan continental shelf. We used a standardized methodology for defining the clades to be included in this study in order to avoid sampling biases and ensure that the clades had diversified mostly in island systems within the study region. We first identified species endemic to Northern Melanesia that had been included in molecular phylogenetic studies. For each of these endemics, we used phylogenetic information to identify the largest clade that included the endemic and did not contain more than one species distributed outside the study region. In cases of incomplete phylogenies, we also used taxonomic information such as generic limits or superspecies complexes to determine whether species not included in the published phylogeny belonged to the focal clade.
We estimated the net diversification rate of each clade as follows:
| 2.1 |
where N is the number of species in the clade, and A is the stem age for the clade [15]. Some assessments suggest that current taxonomies underestimate true species diversity in the region by considering basal evolutionary lineages as subspecies of widespread polytypic species [16–18]. Therefore, we assessed the robustness of our results to species limits by also estimating diversification rates using subspecies counts. The number of subspecies in each species was taken from the study of Dickinson [19].
We estimated the stem age of each clade using a relaxed mitochondrial molecular clock. We obtained sequences of the cytochrome b (cytb) gene or NADH dehydrogenase subunit 2 (ND2) gene for each focal clade and a closely related outgroup from Genbank (www.genbank.gov). We estimated divergence times using Bayesian methods in the program BEAST [20]. Substitution rates were modelled using a GTR + Gamma model and rate heterogeneity across lineages was modelled using a relaxed lognormal clock [21]. The prior for the overall substitution rate was set to match the distribution of rates observed in mitochondrial sequences of a wide variety of avian groups ([22], lognormal distribution: log-mean = −4.6, log-standard deviation = 0.25). We determined burn-in and convergence by examining traces and effective sample size values for model likelihood and divergence time estimates using Tracer v. 1.5 [23].
Dispersal ability is notoriously difficult to estimate. Here, we assume that dispersal ability in birds is at least partially determined by their flight capabilities. Although behavioural factors may also have a strong influence on dispersal tendencies [24], behavioural predisposition for long-distance dispersal should be associated with strong flight capabilities, generating a correlation between behavioural, physiological and biomechanical aspects of dispersal. Therefore, we used the hand-wing index (HWI) [25], a proxy for the aspect ratio of the wing, as an index of dispersal ability. The advantage of focusing on the flight apparatus is that it can be measured using specimens. In particular, the aspect ratio of the wing is a key determinant of the efficiency of long-distance flight [12,26]. Moreover, there is empirical evidence linking HWI to the dispersal process: HWI is well correlated with migration distance among a variety of Palearctic birds [27], average distance flown over water estimated through dispersal experiments in tropical forest birds [12,28] and natal dispersal distances among British passerines [29]. The HWI is calculated as follows:
| 2.2 |
in which WL is the standard measure of wing length, and SL is a measure of the distance from the carpal joint to the tip of the first secondary feather [12]. For all species in our clades, three adult males—when available—were measured at the American Museum of Natural History. The average of each species was used for calculating an average HWI for each clade.
We used phylogenetic comparative methods and statistical modelling techniques to determine the function that best described the relationship between dispersal ability and diversification rate. In order to account for phylogenetic non-independence among clades, we used a phylogenetic generalized least-squares regression method [30], which uses a correlation structure derived from a lambda transformation of the phylogenetic tree of relationships among clades. To obtain the tree, we used sequences of cytb, ND2 and the recombination activating gene 1 for a representative of each clade. We then generated a maximum-likelihood tree in RAxML [31], and transformed the tree into a chronogram in which branch lengths are proportional to time using a maximum-likelihood approach [32].
Models of the relationship between the HWI and diversification rates were optimized using the pgls function in the R package Caper [33]. We used a logarithmic transformation of both the HWI and the diversification rate, because it resulted in a better fit to model assumptions. We compared constant, linear and quadratic models using Akaike information criterion (AIC) scores; reported p-values are calculated based on an F-test comparison with a model in which rates are constant, and all reported R2 values are adjusted values. For comparison, we also analysed the relationship between dispersal and diversification using Mayr & Diamond's [11] index of diversification: the number of subspecies and allospecies of each species. We then calculated an index for each clade as the average value of the contained species.
3. Results
A total of 28 clades from 21 families of birds distributed across Australasian archipelagoes satisfied our clade-selection criteria. The clades range from two to 22 species, 2.4–24.9 Myr in age, and represent a wide range of dispersal abilities (table 1). Wing morphology data were collected for 338 specimens representing 157 species (electronic supplementary material).
Table 1.
Clades that diversified in Australasian archipelagoes that were included in the analysis. (The reference listed for each family is the reference from which the phylogeny or the sequence data used to measure diversification rates were taken. The HWI listed is the average for the species in the clade.)
| family | cladea | no. species | stem age (Myr) | diversification rate (log(N)/A) | HWI | reference |
|---|---|---|---|---|---|---|
| Accipitridae | Haliaeetus | 2 | 7.70 | 0.09 | 38.15 | [34] |
| Accipitridae | Henicopernis | 2 | 5.01 | 0.14 | 37.25 | [35] |
| Aegothelidae | Aegotheles | 2 | 6.90 | 0.10 | 29.95 | [36] |
| Alcedinidae | Ceyx | 12 | 4.93 | 0.50 | 18.94 | [37] |
| Cacatuidae | Cacatua | 5 | 8.49 | 0.19 | 28.79 | [38] |
| Cettiedae | Cettia | 5 | 3.63 | 0.44 | 14.78 | [39] |
| Columbidae | Alopecoenas | 5 | 3.36 | 0.48 | 27.4 | [40] |
| Columbidae | Reinwardtoena | 3 | 7.68 | 0.14 | 33.7 | [41] |
| Columbidae | Ptilinopus eugeniae | 2 | 3.23 | 0.21 | 27.35 | [42] |
| Columbidae | Ptilinopus roseicapilla | 16 | 2.93 | 0.95 | 25.5 | [42] |
| Columbidae | Henicophaps | 2 | 13.40 | 0.05 | 27.8 | [41] |
| Corvidae | Corvus | 4 | 3.92 | 0.35 | 31.9 | [43] |
| Dicruridae | Dicrurus | 2 | 6.09 | 0.11 | 25.75 | [44] |
| Meliphagidae | Meliarchus | 9 | 24.43 | 0.09 | 18.8 | [45] |
| Meliphagidae | Glycifohia | 3 | 24.97 | 0.04 | 21.4 | [45] |
| Monarchidae | Clytorhynchos | 14 | 6.26 | 0.42 | 17.9 | [46] |
| Monarchidae | Myiagra | 9 | 3.78 | 0.58 | 18.52 | [47] |
| Pachycephalidae | Pachycephala | 4 | 3.83 | 0.36 | 18.67 | [48] |
| Sylviidae | Phylloscopus | 3 | 2.96 | 0.37 | 12.92 | [49] |
| Pittidae | Pitta | 3 | 1.39 | 0.79 | 18.57 | [50] |
| Procellariidae | Pseudobulweria | 4 | 8.68 | 0.16 | 56.4 | [51] |
| Psittacidae | Eunymphicus | 2 | 2.83 | 0.25 | 36.05 | [52] |
| Rallidae | Gallirallus | 10 | 2.43 | 0.95 | 17.26 | [53] |
| Rhipiduridae | Rhipidura | 3 | 6.87 | 0.16 | 21.68 | [54] |
| Sturnidae | Mino | 2 | 4.76 | 0.15 | 19.14 | [55] |
| Turdidae | Zoothera | 2 | 3.14 | 0.22 | 26.82 | [56] |
| Tytonidae | Tyto | 5 | 10.87 | 0.15 | 33.33 | [57] |
| Zosteropidae | Zosterops | 22 | 3.59 | 0.86 | 16.4 | [58] |
aThe clade name corresponds to the focal genus or species (that is endemic to Melanesia); some clades contain multiple genera, see the electronic supplementary material, table S1 for a list of species included in the analysis.
The relationship between dispersal ability and diversification rates was monotonically negative (figure 1). A linear model (AIC = 67.4, p = 0.005, R2 = 0.17) fitted marginally better than a quadratic model (AIC = 68.6, p = 0.03), but both curves are very similar, with the quadratic model showing a monotonically decreasing relationship within the range of dispersal values (figure 1). A constant model had the lowest support (AIC = 71.7). The use of subspecies rather than species to estimate diversification rates produced similar results: a negative linear model (AIC = 66.4, p < 0.001, R2 = 0.28) was better than a quadratic model (AIC = 68.2, p = 0.004), which declined monotonically, and both were better than a constant rate model (AIC = 73.0; electronic supplementary material, figure S1).
Figure 1.
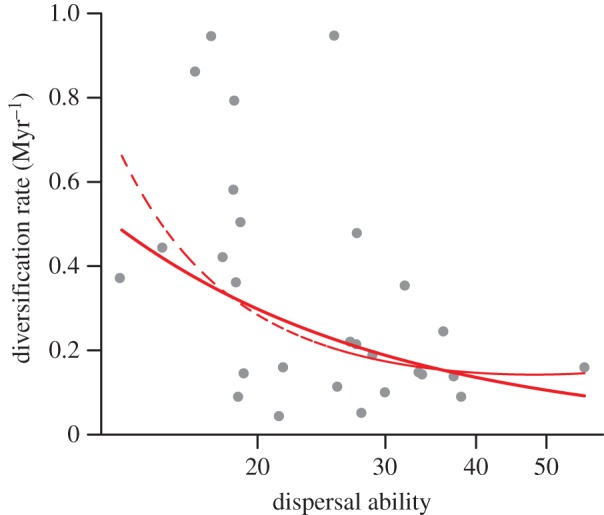
The relationship between dispersal ability and diversification rates for 28 clades of birds from Australasian archipelagoes. Dispersal abilities were estimated using the average HWI for each clade, and diversification rates were estimated using equation (2.1). The best-fitting model is a negative linear model (solid line, log(diversification rate) = 2.18–1.13 × log(dispersal)) followed by a quadratic model (dashed line, log(diversification rate) = 11.52–6.96 × log(dispersal) + 0.9 × log(dispersal)2). (Online version in colour.)
Using a simple count of species and allospecies as an index of diversification, as done by Mayr & Diamond [11], resulted in the best-fitting quadratic model assuming a unimodal shape, in which clades with intermediate dispersal abilities have the maximum diversification rates, consistent with the intermediate dispersal model (figure 2). However, quadratic (AIC = 62.7) and linear models (AIC = 62.4) were statistically indistinguishable and neither was better than a constant rate model (AIC = 61.3).
Figure 2.
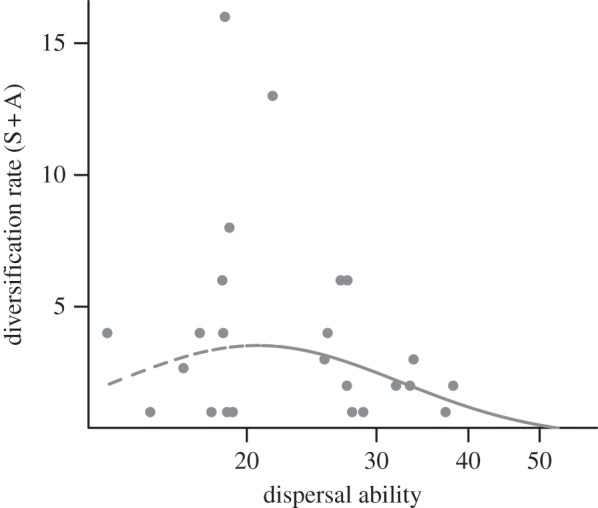
The relationship between a taxonomic index of diversification (the number of subspecies and allospecies per species (S + A)) and dispersal ability results in the best-fitting quadratic model assuming a unimodal shape, but the model is not significantly different from a constant model.
4. Discussion
We found a negative relationship between dispersal ability and diversification rates among bird clades that diversified in the discontinuous geography of Australasian archipelagoes. If opportunities for speciation were limited by dispersal across barriers, increased dispersal ability would have led to higher rates of speciation, resulting in a positive relationship between dispersal ability and diversification rate. Our discovery of a negative relationship implies that the limiting factor for speciation in this avifauna was not the chance of colonization of new islands, but the effectiveness of geographical barriers in limiting gene flow, which decreases as dispersal ability increases.
These results do not corroborate previous findings of an intermediate dispersal pattern in the avifauna of Australasian archipelagoes in which diversification was highest at intermediate levels of dispersal ability [10,11]. When we used Mayr & Diamond's [11] taxonomy-based index of diversification that does not take clade age into account, a unimodal pattern emerged (albeit not significant, figure 2). This suggests that the discrepancy between our results and Mayr & Diamond's [11] is not owing to differences in taxonomic sampling or dispersal ability estimates. Rather, it is the direct estimation of diversification rates that revealed a negative relationship. Taxonomy-based indices of speciation across clades that do not consider clade age are measures of total lineage richness rather than a proxy for diversification rates and can be misleading because they are sensitive to differences in clade age.
While the influence of extinction is not directly testable, we expect extinction to have weakened the negative relationship we found between dispersal ability and net diversification rates. This is because species that are poor dispersers are expected to experience higher rates of extinction (and thus, lower net diversification rates) owing to reduced range size [59–61] or reduced rates of recolonization in metapopulation dynamics [62]. Therefore, extinction is most likely dampening a potentially steeper negative relationship.
A negative relationship between dispersal ability and diversification rates suggests a predominant role for modes of speciation limited by isolation, rather than range expansion [12]. One such mode is vicariance, which is based on the subdivision of widespread ancestral biotas. Vicariance has not been considered to be a significant process of speciation in archipelagoes because many islands were never connected to other landmasses in the past (i.e. isolated volcanic islands). However, at least two factors make vicariance a plausible and potentially common mode of speciation in island settings. First, most islands have not been completely isolated throughout their history, but are part of tectonically dynamic archipelagoes with complex geological histories of fragmentation and collision; this is particularly true for Wallacean and Melanesian archipelagoes [63]. Second, fluctuations in sea level can result in subdivision and reconnection of islands separated by shallow-water gaps [64]. In addition, whereas a single long-distance dispersal event usually involves an individual lineage, a single vicariance event can affect entire biotas, potentially leading to multiple speciation events. As a consequence, even if not common, vicariance can be responsible for a substantial portion of speciation events in archipelagoes. For example, a detailed analysis of Aethopyga sunbirds from the Philippines revealed that intra-island and shallow-water barrier vicariant events may have contributed as much as dispersal over deep water to the generation of the group's diversity [65].
In addition to vicariance, evolutionary changes in dispersal ability could also generate a negative relationship between dispersal ability and diversification rates. Lineages may expand their geographical ranges and colonize new islands during evolutionary phases of high dispersal ability, and then differentiate and speciate during phases of reduced dispersal ability [10,58]. While this process may be important at smaller taxonomic scales, it cannot entirely explain the negative pattern found across clades because of the magnitude of dispersal ability differences between clades. Strong phylogenetic inertia in HWI values across clades (Pagel's λ = 0.99) and limited variation within clades (electronic supplementary material, table S1) suggests that dispersal ability is relatively conserved in this avifauna. However, it is still possible that changes in dispersal ability may play a role at smaller scales. For example, rails show pronounced variation in wing shape (electronic supplementary material, table S1). The widely distributed Gallirallus philippensis, with a relatively high aspect ratio (HWI = 27) compared to other rails and most passerines, may represent a lineage in the dispersive phase. Gallirallus philippensis belongs to a clade of mostly flightless rails [53] that has one of the highest diversification rates in the dataset. Analyses of wing shape evolution within clades and comparisons between continental and island clades may provide further insights into the macroevolutionary effects of changes in dispersal ability.
What the scenarios of vicariance and changes in dispersal ability have in common is that lineages go through a phase of range expansion that is decoupled temporally from a phase of geographical isolation that can result in speciation. In the vicariance model, range expansion occurs when terrains are connected and speciation when terrains are divided by a barrier, usually the results of climatic, tectonic or geographical processes that operate over geological timescales. In the model of changes in dispersal ability, range expansion occurs when a lineage has high dispersal ability, and speciation occurs only after the evolution of a low dispersal phenotype. It is possible to conceive of other models with decoupled periods of range expansion and isolation. For example, lineages in an archipelago composed of islands that were never connected can go through a phase of range expansion during lower sea levels, when water gaps between islands become narrower, and a phase of isolation during high sea levels, when speciation is more likely [64]. This scenario provides a mechanism for the generation of a negative relationship between dispersal ability and diversification rates across oceanic islands that were never connected to other islands.
By contrast, in founder-event speciation, long-distance dispersal produces both range expansion and speciation, coupling the two processes. For example, if a rare phenomenon like a hurricane transports individuals from a lineage to a new island that they cannot reach under normal circumstances, the new population is immediately isolated from the source population, and speciation occurs soon thereafter. Whereas long-distance dispersal may be an important phenomenon determining patterns of distribution of taxa [66,67], our data suggest that it is not a dominant force in the generation of diversity, at least for the avifauna of Australasian archipelagoes.
Our data suggest that diversification in this region has predominantly occurred via modes of speciation in which phases of colonization are decoupled from periods of isolation. This decoupling could have occurred as a result of a dynamic geography (i.e. a vicariance model), evolutionary changes in dispersal ability, or fluctuations in the permeability of barriers (e.g. sea-level fluctuations). While we cannot distinguish between these mechanisms, our data do confirm that the limiting factor in speciation for these groups has been isolation, and that dispersal has inhibited diversification.
5. Conclusion
At a regional scale, and for a diverse group of birds, we found that dispersal ability is negatively related to diversification rates, suggesting that dispersal has inhibited avian diversification across Australasian archipelagoes. We attribute this negative relationship to a reduction in speciation rates caused by reduced efficacy of barriers to gene flow as dispersal ability increases. This also suggests that long-distance dispersal, although important for range expansion in Australasian archipelagoes, was not the limiting factor in the diversification of this avifauna. Instead, isolation has played a more important role in controlling diversification rates. The fact that dispersal has not stimulated diversification even in an extremely discontinuous geography such as Australasian archipelagoes, suggests a general inhibitory effect of dispersal on rates of global avian diversification; the expansion of empirical work beyond Australasia is needed to confirm this hypothesis, and promises to be an exciting avenue of future research.
Supplementary Material
Acknowledgements
We thank S. Naeem, J. Cracraft and two anonymous reviewers for extremely helpful comments on this paper.
Data accessibility
Data used in the analyses are included in the electronic supplemental material, along with analyses using subspecies rather than species, as referenced in the text.
Funding statement
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under grant no. (11–44155) awarded to B.C.W. S.C. is supported by a Chapman Fellowship, AMNH.
References
- 1.Mayr E. 1963. Animal species and evolution. Cambridge, MA: Belknap Press. [Google Scholar]
- 2.Coyne JA, Orr HA. 2004. Speciation, p. 545 Sunderland, MA: Sinauer Associates. [Google Scholar]
- 3.Bohonak AJ. 1999. Dispersal, gene flow, and population structure. Q. Rev. Biol. 74, 21–45. ( 10.1086/392950) [DOI] [PubMed] [Google Scholar]
- 4.Matzke NJ. 2013. Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Front. Biogeogr. 5, 242–248. [Google Scholar]
- 5.Wessel A, Hoch H, Asche M, von Rintelen T, Stelbrink B, Heck V, Stone FD, Howarth FG. 2013. Founder effects initiated rapid species radiation in Hawaiian cave planthoppers. Proc. Natl Acad. Sci. USA 110, 9391–9396. ( 10.1073/pnas.1301657110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Templeton AR. 2008. The reality and importance of founder speciation in evolution. Bioessays 30, 470–479. ( 10.1002/bies.20745) [DOI] [PubMed] [Google Scholar]
- 7.Price TD. 2007. Speciation in birds, 1st edn, p. 480 Greenwood Village, CO: Roberts & Co. Publishers. [Google Scholar]
- 8.Rosenzweig ML. 1995. Species diversity in space and time. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 9.Schluter D. 2001. Ecology and the origin of species. Trends Ecol. Evol. 16, 372–380. ( 10.1016/S0169-5347(01)02198-X) [DOI] [PubMed] [Google Scholar]
- 10.Diamond J, Gilpin ME, Mayr E. 1976. Species-distance relation for birds of the Solomon Archipelago, and the paradox of the great speciators. Proc. Natl Acad. Sci. USA 73, 2160–2164. ( 10.1073/pnas.73.6.2160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayr E, Diamond J. 2001. The birds of Northern Melanesia. New York, NY: Oxford University Press, Inc. [Google Scholar]
- 12.Claramunt S, Derryberry EP, Remsen JV, Jr, Brumfield RT. 2012. High dispersal ability inhibits speciation in a continental radiation of passerine birds. Proc. R. Soc. B 279, 1567–1574. ( 10.1098/rspb.2011.1922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price JP, Wagner WL. 2004. Speciation in Hawaiian angiosperm lineages: cause, consequence, and mode. Evolution 58, 2185–2200. ( 10.1111/j.0014-3820.2004.tb01597.x) [DOI] [PubMed] [Google Scholar]
- 14.Phillimore AB, Freckleton RP, Orme CD, Owens IPF. 2006. Ecology predicts large-scale patterns of phylogenetic diversification in birds. Am. Nat. 168, 220–229. ( 10.1086/505763) [DOI] [PubMed] [Google Scholar]
- 15.Kendall DG. 1949. Stochastic processes and population growth. J. R. Stat. Soc. Ser. B 11, 230–282. [Google Scholar]
- 16.Reddy S, Moyle RG. 2011. Systematics of the scimitar babblers (Pomatorhinus: Timaliidae): phylogeny, biogeography, and species-limits of four species-complexes. Biol. J. Linn. Soc. 102, 846–869. ( 10.1111/j.1095-8312.2010.01611.x) [DOI] [Google Scholar]
- 17.Moyle RG, Schilthuizen M, Rahman MA, Sheldon FH. 2005. Molecular phylogenetic analysis of the white-crowned forketail Enicurus leschenaulti in Borneo. J. Avian Biol. 36, 96–101. ( 10.1111/j.0908-8857.2005.03510.x) [DOI] [Google Scholar]
- 18.Cracraft J. 1983. Species concepts and speciation analysis. Curr. Ornithol. 1, 159–187. ( 10.1007/978-1-4615-6781-3_6) [DOI] [Google Scholar]
- 19.Dickinson E. 2003. The Howard and Moore complete checklist of the birds of the world, 3rd edn Princeton, NJ: Princeton University Press. [Google Scholar]
- 20.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88 ( 10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weir JT, Schluter D. 2008. Calibrating the avian molecular clock. Mol. Ecol. 17, 2321–2328. ( 10.1111/j.1365-294X.2008.03742.x) [DOI] [PubMed] [Google Scholar]
- 23.Rambaut A, Drummond AJ. 2010. Traver v. 1.5. Program distribution by the authors. Oxford, UK: University of Oxford. [Google Scholar]
- 24.Diamond JM. 1981. Flightlessness and fear of flying in island species. Nature 293, 507–508. ( 10.1038/293507a0) [DOI] [Google Scholar]
- 25.Kipp FA. 1958. Der Handflügel-Index als flugiologisches Maß. Vogelwarte 20, 77–86. [Google Scholar]
- 26.Pennycuick CJ. 2008. Modeling the flying bird. Burlington, MA: Academic Press. [Google Scholar]
- 27.Lockwood R, Swaddle JP, Rayner JMV. 1998. Avian wingtip shape reconsidered: wingtip shape indices and morphological adaptations to migration. J. Avian Biol. 29, 273–292. ( 10.2307/3677110) [DOI] [Google Scholar]
- 28.Moore RP, Robinson WD, Lovette IJ, Robinson TR. 2008. Experimental evidence for extreme dispersal limitation in tropical forest birds. Ecol. Lett. 11, 960–968. ( 10.1111/j.1461-0248.2008.01196.x) [DOI] [PubMed] [Google Scholar]
- 29.Dawideit BA, Phillimore AB, Laube I, Leisler B, Bohning-Gaese K. 2009. Ecomorphological predictors of natal dispersal distances in birds. J. Anim. Ecol. 78, 388–395. ( 10.1111/j.1365-2656.2008.01504.x) [DOI] [PubMed] [Google Scholar]
- 30.Freckleton RP, Harvey PH, Pagel M. 2002. Phylogenetic analysis and comparative data: a test and review of evidence. Am. Nat. 160, 712–726. ( 10.1086/343873) [DOI] [PubMed] [Google Scholar]
- 31.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. ( 10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 32.Paradis E. 2013. Molecular dating of phylogenies by likelihood methods: a comparison of models and a new information criterion. Mol. Phylogenet. Evol. 67, 436–444. ( 10.1016/j.ympev.2013.02.008) [DOI] [PubMed] [Google Scholar]
- 33.Orme D, Freckleton R, Thomas G, Petzoldt T, Isaac N, Pearse W. 2012. Caper: comparative analyses of phylogenetics and evolution in R. R package version 0.5 edn. See http://CRAN.R-project.org/package=caper.
- 34.Wink M, Heidrich P, Fentzloff C. 1996. a mtDNA phylogeny of sea eagles (genus Haliaeetus) based on nucleotide sequences of the cytochrome b-gene. Biochem. Syst. Ecol. 24, 783–791. ( 10.1016/S0305-1978(96)00049-X) [DOI] [Google Scholar]
- 35.Barrowclough GF, Groth JG, Lai JE, Tsang SM. 2014. The phylogenetic relationships of the endemic genera of Australo-Papuan hawks. J. Raptor Res. 48, 36–43. ( 10.3356/JRR-13-33.1) [DOI] [Google Scholar]
- 36.Dumbacher JP, Pratt TK, Fleischer RC. 2003. Phylogeny of the owlet-nightjars (Aves: Aegothelidae) based on mitochondrial DNA sequence. Mol. Phylogenet. Evol. 29, 540–549. ( 10.1016/S1055-7903(03)00135-0) [DOI] [PubMed] [Google Scholar]
- 37.Andersen MJ, Oliveros CH, Filardi CE, Moyle RG. 2013. Phylogeography of the variable dwarf-kingfisher Ceyx lepidus (Aves: Alcedinidae) inferred from mitochondrial and nuclear DNA sequences. The Auk 130, 118–131. ( 10.1525/auk.2012.12102) [DOI] [Google Scholar]
- 38.White NE, Phillips MJ, Gilbert MT, Alfaro-Nunez A, Willerslev E, Mawson PR, Spencer PBS, Bunce M. 2011. The evolutionary history of cockatoos (Aves: Psittaciformes: Cacatuidae). Mol. Phylogenet. Evol. 59, 615–622. ( 10.1016/j.ympev.2011.03.011) [DOI] [PubMed] [Google Scholar]
- 39.Lecroy M, Barker FK. 2006. A new species of bush-warbler from Bougainville island and a monophyletic origin for Southwest Pacific Cettia. Am. Museum Novitates 3511, 1–20. ( 10.1206/0003-0082(2006)3511[1:ANSOBF]2.0.CO;2) [DOI] [Google Scholar]
- 40.Moyle RG, Jones RM, Andersen MJ. 2013. A reconsideration of Gallicolumba (Aves: Columbidae) relationships using fresh source material reveals pseudogenes, chimeras, and a novel phylogenetic hypothesis. Mol. Phylogenet. Evol. 66, 1060–1066. ( 10.1016/j.ympev.2012.11.024) [DOI] [PubMed] [Google Scholar]
- 41.Pereira SL, Johnson KP, Clayton DH, Baker AJ. 2007. Mitochondrial and nuclear DNA sequences support a Cretaceous origin of Columbiformes and a dispersal-driven radiation in the Paleogene. Syst. Biol. 56, 656–672. ( 10.1080/10635150701549672) [DOI] [PubMed] [Google Scholar]
- 42.Cibois A, Thibault J-C, Bonillo C, Filardi CE, Watling D, Pasquet E. 2014. Phylogeny and biogeography of fruit doves (Aves: Columbidae). Mol. Phylogenet. Evol. 70, 442–453. ( 10.1016/j.ympev.2013.08.019) [DOI] [PubMed] [Google Scholar]
- 43.Jonsson KA, Fabre P-H, Irestedt M. 2012. Brains, tools, innovation and biogeography in crows and ravens. BMC Evol. Biol. 12, 72 ( 10.1186/1471-2148-12-72) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasquet E, Pons J-M, Fuchs J, Cruaud C, Bretagnolle V. 2007. Evolutionary history and biogeography of the drongos (Dicruridae), a tropical Old World clade of corvoid passerines. Mol. Phylogenet. Evol. 45, 158–167. ( 10.1016/j.ympev.2007.03.010) [DOI] [PubMed] [Google Scholar]
- 45.Andersen MJ, Naikatini A, Moyle RG. 2013. A molecular phylogeny of Pacific honeyeater (Aves: Meliphagidae) reveals extensive paraphyly and an isolated Polynesian radiation. Mol. Phylogenet. Evol. 71, 308–315. ( 10.1016/j.ympev.2013.11.014) [DOI] [PubMed] [Google Scholar]
- 46.Filardi CE, Moyle RG. 2005. Single origin of a pan-Pacific bird group and upstream colonization of Australasia. Nature 438, 216–220. ( 10.1038/nature04057) [DOI] [PubMed] [Google Scholar]
- 47.Fabre P-H, Moltensen M, Fjeldsa J, Irestedt M, Lessard J-P, Jonsson KA. 2014. Multiple waves of colonization by monarch flycatchers (Myiagra, Monarchidae) across the Indo-Pacific and their implications for coexistence and speciation. J. Biogeogr. 41, 274–286. ( 10.1111/jbi.12202) [DOI] [Google Scholar]
- 48.Andersen MJ, Nyári AS, Mason I, Joseph L, Dumbacher JP, Filardi CE, Moyle RG. 2014. Molecular systematics of the world's most polytypic bird: the Pachycephala pectoralis/melanura (Aves: Pachycephalidae) species complex. Zool. J. Linnean Soc. 140, 566–588. ( 10.1111/zoj.12088) [DOI] [Google Scholar]
- 49.Olsson U, Alstrom P, Ericson PGP, Sundberg P. 2005. Non-monophyletic taxa and cryptic species: evidence from a molecular phylogeny of leaf-warblers (Phylloscopus, Aves). Mol. Phylogenet. Evol. 36, 261–276. ( 10.1016/j.ympev.2005.01.012) [DOI] [PubMed] [Google Scholar]
- 50.Irestedt M, Fabre P-H, Batalho-Filho H, Jonsson KA, Roselaar CS, Sangster G, Ericson PGP. 2013. The spatio-temporal colonization and diversification across the Indo-Pacific by a ‘great speciator’ (Aves, Erythropitta erythrogaster). Proc. R. Soc. B 280, 20130309 ( 10.1098/rspb.2013.0309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gangloff B, Shirihai H, Watling D, Cruaud C, Couloux A, Tillier A, Pasquet E, Bretagnolle V. 2012. The complete phylogeny of Pseudobulweria, the most endangered seabird genus: systematics, species status and conservation implications. Conserv. Genet. 13, 39–52. ( 10.1007/s10592-011-0261-6) [DOI] [Google Scholar]
- 52.Boon W-M, Robinet O, Rawlence N, Bretagnolle V, Norman JA, Christidis L, Chambers GK. 2008. Morphological, behavioural and genetic differentiation within the horned parakeet (Eunymphicus cornutus) and its affinities to Cyanoramphus and Prosopeia. Emu 108, 251–260. ( 10.1071/MU07030) [DOI] [Google Scholar]
- 53.Kirchman JJ. 2012. Speciation of flightless rails on islands: a DNA-based phylogeny of the typical rails of the Pacific. The Auk 129, 56–69. ( 10.1525/auk.2012.11259) [DOI] [Google Scholar]
- 54.Nyári AS, Benz BW, Jonsson KA, Fjeldsa J, Moyle RG. 2009. Phylogenetic relationships of fantails (Aves: Rhipiduridae). Zool. Scripta 38, 553–561. ( 10.1111/j.1463-6409.2009.00397.x) [DOI] [Google Scholar]
- 55.Lovette IJ, Rubenstein DR. 2007. A comprehensive molecular phylogeny of the starlings (Aves: Sturnidae) and mockingbirds (Aves: Mimidae): congruent mtDNA and nuclear trees for a cosmopolitan avian radiation. Mol. Phylogenet. Evol. 44, 1031–1056. ( 10.1016/j.ympev.2007.03.017) [DOI] [PubMed] [Google Scholar]
- 56.Klicka J, Voelker G, Spellman GM. 2005. A molecular phylogenetic analysis of the 'true thrushes’ (Aves: Turdidae). Mol. Phylogenet. Evol. 34, 486–500. ( 10.1016/j.ympev.2004.10.001) [DOI] [PubMed] [Google Scholar]
- 57.Jonsson KA, Poulsen MK, Haryoko T, Reeve AH, Fabre P-H. 2013. A new species of masked-owl (Aves: Strigiformes: Tytonidae) from Seram, Indonesia. Zootaxa 3635, 051–061. ( 10.11646/zootaxa.3635.1.5) [DOI] [PubMed] [Google Scholar]
- 58.Moyle RG, Filardi CE, Smith CE, Diamond J. 2009. Explosive Pleistocene diversification and hemispheric expansion of a ‘great speciator’. Proc. Natl Acad. Sci. USA 106, 1863–1868. ( 10.1073/pnas.0809861105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stanley SM. 1990. The general correlation between rate of speciation and rate of extinction: fortuitous causal linkages. In Causes of evolution: a paleontological perspective (eds Ross RM, Allmon WD.), pp. 103–127. Chicago, IL: University of Chicago Press. [Google Scholar]
- 60.Reinhardt K, Kohler G, Maas S, Detzel P. 2005. Low dispersal ability and habitat specificity promote extinctions in rare but not in widespread species: the Orthoptera of Germany. Ecography 28, 593–602. ( 10.1111/j.2005.0906-7590.04285.x) [DOI] [Google Scholar]
- 61.Powell MG. 2007. Geographic range and genus ongevity of Late Paleozoic Brachiopods. Paleobiology 33, 530–546. ( 10.1666/07011.1) [DOI] [Google Scholar]
- 62.Hanski I. 1998. Metapopulation dynamics. Nature 396, 41–51. ( 10.1038/23876) [DOI] [Google Scholar]
- 63.Hall R. 2002. Cenozoic geological and plate tectonic evolution of SE Asia and the SW Pacific: computer-based reconstructions, models and animations. J. Asian Earth Sci. 20, 353–431. ( 10.1016/S1367-9120(01)00069-4) [DOI] [Google Scholar]
- 64.Ali JR, Aitchison JC. 2014. Exploring the combined role of eustasy and oceanic island thermal subsidence in shaping biodiversity on the Galapagos. J. Biogeogr. 41, 1227–1241. ( 10.1111/jbi.12313) [DOI] [Google Scholar]
- 65.Hosner PA, Nyári AS, Moyle RG. 2013. Water barriers and intra-island isolation contribute to diversification in the insular Aethopyga sunbirds (Aves: Nectariniidae). J. Biogeogr. 40, 1094–1106. ( 10.1111/jbi.12074) [DOI] [Google Scholar]
- 66.de Queiroz A. 2005. The resurrection of oceanic dispersal in historical biogeography. Trends Ecol. Evol. 20, 68–73. ( 10.1016/j.tree.2004.11.006) [DOI] [PubMed] [Google Scholar]
- 67.Gillespie RG, Baldwin BG, Waters JM, Fraser CI, Nikula R, Roderick GK. 2012. Long-distance dispersal: a framework for hypothesis testing. Trends Ecol. Evol. 27, 47–56. ( 10.1016/j.tree.2011.08.009) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in the analyses are included in the electronic supplemental material, along with analyses using subspecies rather than species, as referenced in the text.


