Abstract
Extant terrestrial biodiversity arguably is driven by the evolutionary success of angiosperm plants, but the evolutionary mechanisms and timescales of angiosperm-dependent radiations remain poorly understood. The Scarabaeoidea is a diverse lineage of predominantly plant- and dung-feeding beetles. Here, we present a phylogenetic analysis of Scarabaeoidea based on four DNA markers for a taxonomically comprehensive set of specimens and link it to recently described fossil evidence. The phylogeny strongly supports multiple origins of coprophagy, phytophagy and anthophagy. The ingroup-based fossil calibration of the tree widely confirmed a Jurassic origin of the Scarabaeoidea crown group. The crown groups of phytophagous lineages began to radiate first (Pleurostict scarabs: 108 Ma; Glaphyridae between 101 Ma), followed by the later diversification of coprophagous lineages (crown-group age Scarabaeinae: 76 Ma; Aphodiinae: 50 Ma). Pollen feeding arose even later, at maximally 62 Ma in the oldest anthophagous lineage. The clear time lag between the origins of herbivores and coprophages suggests an evolutionary path driven by the angiosperms that first favoured the herbivore fauna (mammals and insects) followed by the secondary radiation of the dung feeders. This finding makes it less likely that extant dung beetle lineages initially fed on dinosaur excrements, as often hypothesized.
Keywords: Coleoptera, Scarabaeoidea, species diversity, time tree, phytophagy, coprophagy
1. Introduction
Terrestrial biodiversity is dominated by angiosperm plants and their herbivores, including mammals and insects that feed on leaves, soft shoots and fruits, or graze on low vegetation. The major radiation of the mammal crown groups originated fairly recently in the Palaeogene [1,2]. In particular, extant lineages of placental mammal lineages that contribute most of the herbivorous species, including the Proboscidea, Rodentia, Artiodactyla and Perissodactyla, arose after the crown groups of most lineages of angiosperms, their main food source, which can be placed into the Cretaceous [3–6]. Likewise, several herbivorous insect groups tracked the rise of the angiosperms, and coevolutionary interactions with their angiosperm hosts are regarded as a key factor promoting the extraordinary diversity of insects [7]. Other insect groups have diversified in conjunction with the rise of the mammals by using their dung, a greatly more nutritious resource than leaf litter and decaying plant material that are the diet of many early insect lineages.
Among the insects, the Coleoptera (beetles) are the most ecologically diverse order and include both phytophagous and coprophagous lineages. Phytophagy arose approximately 10 times in Coleoptera and may have promoted the largest radiation of beetles, the ‘Phytophaga’ [8,9] composed of Chrysomelidae, Cerambycidae and Curculionidae (leaf beetles, longhorns and weevils), with combined more than 100 000 described species. The second rank in total species diversity among phytophagous beetles goes to the Pleurosticti [10], a lineage of more than 25 000 described species in the superfamily Scarabaeoidea (=Lamellicornia). These beetles feed on leaves, flowers and pollen as adults, and on living roots, soil humus or decaying wood in the larval stages. The Scarabaeoidea also include the dung beetles (Scarabaeidae: Scarabaeinae, Aphodiinae), that together comprise ca 8300 species [11]. Scarab dung beetles and pleurosticts are considered to be sister lineages [12]. Their close relationship presents a unique opportunity to test hypotheses of species diversification driven by the interactions with angiosperm and mammal lineages and to use the evolution of insect lineages to track these major biotic changes in Earth history.
The origin of plant and dung feeding may be tractable because of their excellent fossil record that permits reliable dating. The fossil record of Scarabaeoidea comprises 224 dated fossil species [13], of which the oldest have been ascribed to the Lower Jurassic (Lower Lias) [14,15] and indicating a possible origin of the lineage in the Triassic [16–18]. Yet, the placement of these Jurassic fossils into the Scarabaeoidea has remained doubtful [11,15], and hence most scarab families were hypothesized to have originated only in the Cretaceous [15,18]. Since new Jurassic fossils were discovered recently whose assignment to Scarabaeoidea is well supported [19–21], the early diversification of Scarabaeoidea is again the subject of debate (e.g. [19]). Evidence of tunnelling in Early Mesozoic vertebrate coprolites has led some authors to believe that dung beetles were already associated with plant-eating dinosaurs [22], suggesting that coprophagy originated much earlier than evident from known fossils.
Here, we date the tree of Scarabaeoidea based on four nuclear and mitochondrial DNA markers and a comprehensive sample of scarabaeoid lineages. Using a variety of calibration priors, we assess the sensitivity of age estimates for phytophagous and coprophagous radiations to the choice of fossils for calibrations and gain an objective estimate of robustness of our time tree. This supersedes previous age estimates for scarab lineages from deep-level analyses of all Coleoptera [17,18], which used no crown-group fossils of Scarabaeoidea.
2. Material and methods
(a). Sampling and DNA sequencing
We selected 146 species to represent all major lineages of Scarabaeoidea and biogeographic regions and particular focus on the Scarabaeidae, the largest family. The nomenclature of family group names follows [23]. Additionally, we included taxa of 15 other beetle families as outgroups (electronic supplementary material, table S1). Given the uncertainty about the sister group of Scarabaeoidea [17,24,25], all trees were rooted with the distantly related carabid beetle Metrius contractus. Samples were preserved in 95% ethanol. DNA was extracted from thoracic leg muscle tissue using Promega WizardSV extraction plates. Vouchers are deposited at Natural History Museum, London and Zoologisches Forschungsmuseum A. Koenig Bonn. Four genes were PCR amplified, including the complete small (18S) and partial large subunit ribosomal RNA genes (28S), and partial mitochondrial 16S rRNA (rrnL) and cytochrome oxidase subunit 1 (cox1) genes (for primers, see the electronic supplementary material). Sequences were obtained with standard Sanger sequencing and Bigdye v. 3.1 technology (Applied Biosystems) on an ABI3730 automated sequencer. Sequences were edited manually using Sequencher v. 4.8 (Genecodes Corp., Ann Arbor, MI, USA). Specimen voucher details and GenBank sequence accession numbers are given in the electronic supplementary material, table S1.
(b). Phylogenetic analysis
Sequences were aligned with Mafft v. 5.8 [26,27], and ambiguous alignment positions were subsequently masked using Aliscore v. 0.2 under the default settings [28]. The length of the concatenated alignment was 4615 base pairs (bp), reduced to 3900 bp after masking the alignment with Aliscore. The length of each marker in the final matrix (after masking) was as follows: cox1, 826 (825) bp; rrnL, 484 (464) bp; 28S, 941 (686) bp; 18S, 2364 (1925) bp. Tree inference with Bayesian analysis on concatenated data matrices was conducted using parallel MrBayes v. 3.2 [29], conducting Markov chain Monte Carlo runs [30] after partitioning [31,32] the data for rrnL, 18S, 28S and three codon positions of cox1. Best-fitting models of sequence evolution were estimated using Bayes factors [31]. We used a GTR + I + G model as determined by Modeltest separately for each of the six partitions. Convergence was assessed for all parameters by evaluating stationarity of the Markov chain with Tracer v. 1.5 [33] and by sampling of Bayesian tree topologies with AWTY [34] (see the electronic supplementary material). Tree searches were conducted for 30 × 106 generations, using a random starting tree and two runs of three heated and one cold Markov chains (heating of 0.1). Chains were sampled every 1000th generation and the first 5 × 106 generations were discarded as burn-in.
Trees were also inferred using maximum likelihood (ML) performed in PhyML [35] using a GTR model (as selected by Modeltest using Akaike information criterion [36]) with all parameters estimated from the data and four substitution rate categories. Branch support was evaluated with the approximate likelihood ratio test (aLRT)/SH (Shimodaira–Hasegawa (SH))-like branch support [37]. Model parameters were estimated separately for each partition. Alternative tree hypotheses, exploring sister group relationship of the pleurostict chafers, were tested by site bootstrapping as implemented in Consel [38]. This software identifies the top ranking topology for alternative tree hypotheses under the likelihood criterion and assesses support for each topology calculating p-values for an approximately unbiased test, bootstrap probability tests (bootstrap probability of item/hypothesis (NP), non-scaled bootstrap probability (BP) and Bayesian posterior probability (PP)), SH test and weighted SH test. We used the default scaling factors of 0.5–1.4, with 10 000 pseudoreplicates for each. Individual site likelihoods used for the Consel analysis were calculated for the different topological hypothesis (constrained and unconstrained) in PhyML.
(c). Divergence time estimation
Fossil choice is of crucial importance for the tree calibration [39]. Fossil data were taken from numerous sources (table 1). Information on Scarabaeoidea fossils and their respective formations including issues related to their dating was compiled first by Krell [15] and significantly expanded recently [19–22,41,53]. As the assignment of many fossils to extant lineages is not always unambiguously possible, owing to poorly preserved or concealed synapomorphies, we followed a conservative approach for the choice of fossils and calibration dates, by selecting fossils with diagnostic features that the literature sources could assign unambiguously to extant taxa based on morphological synapomorphies. We only used fossils representing taxa that were recovered in our phylogenetic analysis and we further required that these taxa were represented by multiple terminals in the molecular analysis. If multiple fossils were available for one node calibration, we used the older one. Consequently, 14 fossils were retained for the calibration (table 1). The fossil date assigned to a given lineage was placed either on the crown or stem group node in various analyses (see Results). Divergence time estimation using a relaxed-clock Bayesian approach implemented in BEAST v. 1.7.2 [54] was conducted based on the concatenated datasets aligned with Mafft or after subsequent alignment masking with Aliscore. Model parameters were unlinked across partitions. The respective Bayesian inference (BI) trees were used as starting tree for the different BEAST runs. The Yule model was selected as tree prior and an uncorrelated lognormal model was used to estimate rate variation along branches. Two to four separate analyses (electronic supplementary material) were run for 4 × 107 generations until an effective sample size (ESS) of at least 150 was achieved for all parameters. ESS values were checked using Tracer v. 1.6. [33]. Trees were combined and sampled every 2000th to 4000th generation (electronic supplementary material) after removing a burn-in of 10% using LogCombiner v. 1.7.2 [54]. Exponential priors were set for all node constraints (table 1) with the minimum age of the fossil used as zero offset [55]. The exponential prior mean was chosen so that 95% of the probability is contained between the rigid lower bound and a ‘soft’ maximum bound of a dated fossil layer interval.
Table 1.
Calibration points (in Ma) used for the estimation of the divergence times of Scarabaeoidea. (In all cases, the time offset for minimum age is the upper bound of the fossil age range. §, node age applied to stem lineage or to crown group in a few calibrations; §§, node omitted in a few calibrations (see table 2); EPM, exponential prior mean.)
| node | clade | site | species reference | age | EPM |
|---|---|---|---|---|---|
| A§ | Lucanidae | Upper Jurassic Shara-Teg, Mongolia | Paralucanus mesozoicus [40] | 150.8–145.5a | 1.75 |
| B§ | Trogidae | Lower Cretaceous, Baysa, Russia | Trox sibericus [41] | 145.5–140.2a | 1.76 |
| C1 | Hybosoridae | Jurassic, Karatau-Mikhailovka, Kazakstan | Protohybosorus grandissimus [42] | 164.7–155.7a | 3.0 |
| D§ | Glaphyridae | Lower Cretaceous Baissa, Russia | Cretoglaphyrus spp. [43] | 145.5–140.2a | 1.76 |
| E | Scarabaeinae | Upper Cretaceous Lanxi, China | Prionocephale deplanate Lin [15] | 92–83.5b | 2.84 |
| F§§ | Aphodiinae | Upper Palaeocene, Menat, France | Aphodius charauxi [44,45] | 58.7–55.8a | 0.97 |
| G | Aegialia | Eocene, Green River, Wyoming, USA | Aegialia rupta [46] | 50.3–46.2a | 1.37 |
| H§§ | Aphodius | Oligocene, Florissant, USA | Aphodius aboriginalis [47] | 37.2–33.9a | 1.1 |
| I§ | Nearctic Sericini | Oligocene, Florissant, USA | Serica spp. [15] | 37.2–33.9a | 1.1 |
| J | Rhizotrogini | Eocene, White River, Green River Formation, USA. | Phyllophaga avus [48] | 50.3–46.2a | 1.37 |
| K | Cetoniinae | Mid Eocene, Eckfelder Maar, Germany | Cetoniinae undescribed [49] | 48.6–40.4a | 2.75 |
| L | Anomalini | Oligocene, Florissant, USA | Anomala scudderi [50] | 37.2–33.9a | 1.1 |
| M | Adroretini | Miocene, Shanwang, China | Adoretus spp. [15,51] | 16.0–11.6a | 1.47 |
| N | Dynastinae | Mid Eocene Clarno Formation, Oregon, USA | Oryctoantiquus borealis [52] | 48.6–37.2a | 3.81 |
aGateway to the Palaeobiology database: http://fossilworks.org/bridge.pl (accessed 14 April 2014).
bKrell [22].
3. Results
(a). Scarab relationships
ML and BI searches based on the full alignment and the Aliscore treated data were largely unanimous in recovery of monophyletic groups and within-clade topology of the principal scarabaeoid lineages (electronic supplementary material, table S3). We only describe the topology obtained by BI on the full alignment in greater detail (figure 1). Scarabaeoidea were recovered as monophyletic, and the basal splits within Scarabaeoidea separated Lucanidae (+Glaresidae in the Mafft alignment) from all others, followed by deep branches occupied by Trogidae + Bolboceratidae, while the latter was paraphyletic for the inclusion of Passalidae. Geotrupidae occupied the next-deepest branch. Contrary to expectations from the traditional classification, a sister relationship of dung scarabs (Aphodiinae + Scarabaeinae) and pleurostic scarabs (including the four major subfamilies Rutelinae, Cetoniinae, Melolonthinae and Rutelinae) was never recovered (figure 1). Instead, the dung scarabs were sister to a larger clade (PP = 0.5) that also includes Hybosoridae, Ochodaeidae and Glaphyridae, in addition to the pleurostict scarabs. (Glaphyridae (Ochodaeidae, Ermazinae)) were the immediate sister group to the pleurosticts, although support was weak (figure 1). These topologies are of major interest for the evolutionary diversification of scarab beetles and their co-radiation with angiosperms. They were confirmed by site bootstrapping, where alternative hypotheses such as the monophyly of Scarabaeidae, i.e. ((Aphodiinae, Scarabaeinae) Pleurosticti), or a sister group relation of Glaphyridae with Pleurosticti (electronic supplementary material, table S2) were found to be less likely compared with the unconstrained tree hypothesis (figure 1).
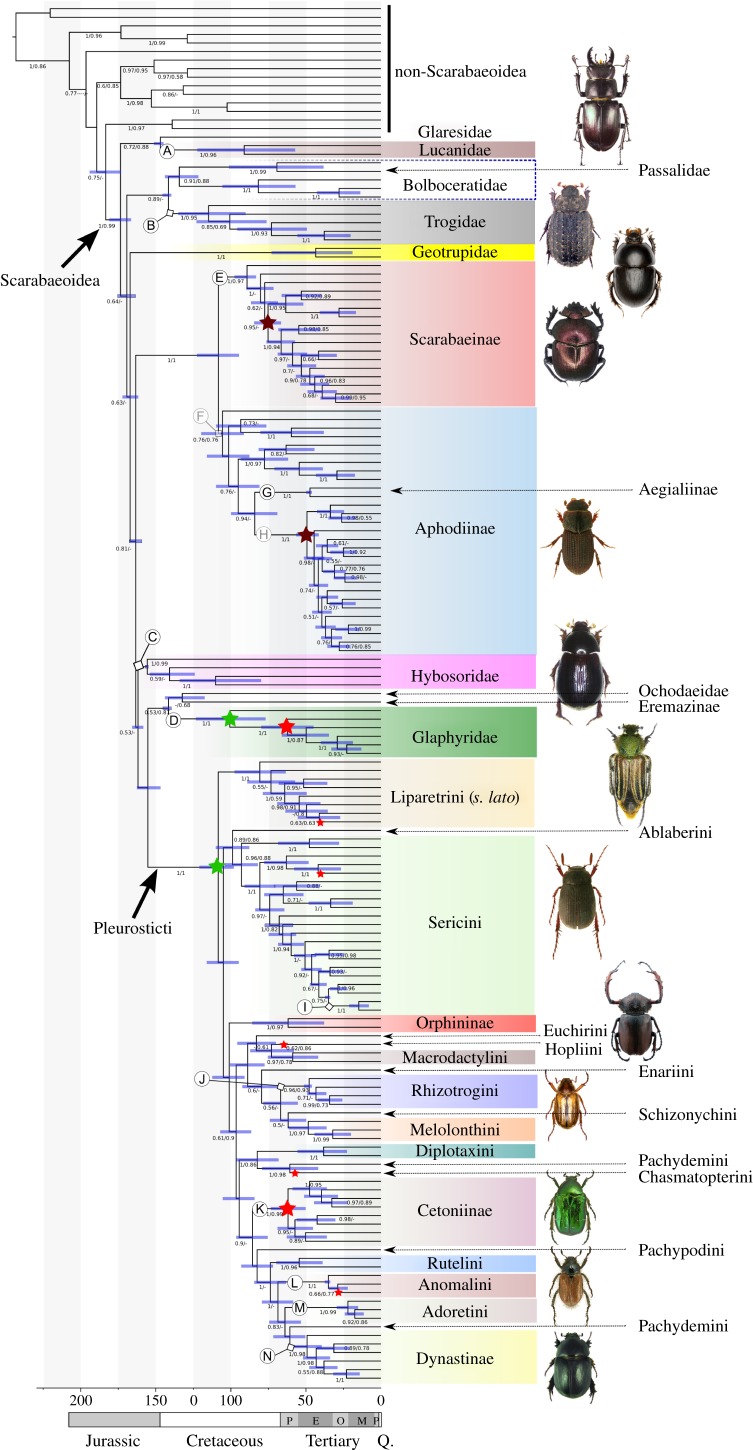
Figure 1.
Calibrated time tree of Scarabaeoidea (BEAST run 6) showing the branch support of nodes (above 0.5; posterior probabilities from BI/aLRT values from ML search) and the node priors A–N. The node bars depict the confidence interval of node ages (95% node height HD). Stars indicate origin of new feeding behaviours: green, phytophagy; brown, coprophagy; red, anthophagy.
Within the pleurosticts, the Melolonthinae was broadly paraphyletic by the inclusion of the four other subfamilies (Cetoniinae, Rutelinae, Dynastinae and Orphininae). The basal branching separates the ‘Southern Hemisphere Melolonthinae’ (PP = 1), here referred to as Liparetrini sensu lato (i.e. including several smaller tribes such as Automolini, Phyllotocini, Heteronychini, Maechidiini and Scitalini), followed by a branch of the well-supported Sericini + Ablaberini (PP = 0.91). The remainder of Melolonthinae, Cetoniinae and Rutelinae (the latter including Dynastinae) had low branch support (PP = 0.41). Interestingly, the Orphninae was always nested within the Pleurosticti (with low support; PP = 0.31), although based on morphology they were expected to be their sister [56,57]. The enigmatic, species-poor Euchirinae was sister to the clade Hopliini + Macrodactylini (PP = 0.64), although with low support (PP = 0.29). A large clade composed of Rutelinae including Dynastinae and the floricolous Cetoniinae is well supported (PP = 0.89), but this monophylum includes the genus Pachypus (=Pachypodiini) as sister group to Rutelinae (PP = 0.47). Among the Cetoniinae (PP = 1), Trichiini were monophyletic (PP = 0.89) being sister to a clade composed of Valgus and Osmoderma. The monophyly of Rutelinae was compromised by the position of Perissosoma (Pachydemini) + Dynastinae as sister to the ruteline Adoretini (PP = 0.84).
(b). Scarab divergence times
Dating was performed on the topology from Bayesian analysis obtained on the full alignment and the Aliscore treated data. Using 12 well-established fossil dates with ages from 15 to 150 Ma (table 1), the data resulted in a mean divergence time of Scarabaeoidea in the Lower Jurassic from 174 to 191 Ma under the various calibration priors (table 2). The base of the phytophagous pleurostict lineage was dated to the Lower Cretaceous between 109 and 128 Ma, whereas the crown lineage of the phytophagous Glaphyridae was placed between 101 and 141 Ma. The crown group of the Scarabaeinae was consistently placed to around 86 to 100 Ma, whereas the age of Aphodiinae varied under different calibrations from 65 to 125 Ma. In concordance with the fossil record [19,53], the origins of families Lucanidae and Trogidae generally were dated to a period in the Cretacous or Jurassic (table 2), whereas the crown group of Hybosoridae was placed into the Upper Jurassic.
Table 2.
Datasets and priors of BEAST runs and resulting crown-group divergence times (mean node heights) and confidence (mean standard deviation width of the 95% highest posterior density) for selected nodes (crown groups) in the Scarabaeoidea phylogeny.
| run 1 | run 2 | run 3 | run 4 | run 5 | run 6 | |
|---|---|---|---|---|---|---|
| origin crown group | A–N | A–N | A–E,G–N | A–E,G,I–N | A–E,G–H,J–N | C,E,G–H,J–N |
| origin stem lineage | — | — | — | — | I | A,B,D,I |
| Scarabaeoidea | 176.3 168.4–184.7 |
182.6 172.3–193.7 |
186.2 175.6–197.0 |
190.9 179.7–202.8 |
183.6 174.4–193.9 |
174.3 167.2–181.8 |
| Scarabaeinae | 86.8 83.5–93.1 |
86.6 83.5–92.2 |
92.6 83.5–103.9 |
100.2 86.7–114.8 |
90.7 83.5–100.4 |
89.6 83.5–98.1 |
| Scarabaeinae dung feeding | 72.7 63.1–82.1 |
72.6 63.3–81.1 |
78.7 68.0–90.1 |
85.5 73.5–98.4 |
76.6 66.7–87.2 |
75.5 66.8–84.9 |
| Cetoniinae | 58.2 45.0–71.8 |
56.8 42.6–69.9 |
64.4 51.1–77.1 |
71.9 57.4–86.4 |
62.4 50.7–74.9 |
62.2 50.4–73.7 |
| Aphodiinae | 65.5 57.3–73.3 |
69.2 58.4–79.9 |
111.8 94.8–129.7 |
124.7 108.0–142.7 |
109.0 94.0–127.4 |
106.3 91.7–120.3 |
| Aphodiinae dung feeding | 43.3 36.4–50.4 |
44.4 35.9–52.4 |
51.6 43.2–60.5 |
78.1 65.7–91.6 |
49.9 42.4–58.1 |
49.5 41.6–56.8 |
| Sericini | 99.9 85.8–114.4 |
96.6 81.3–111.3 |
103.0 89.9–116.8 |
111.3 97.4–124.7 |
95.4 83.1–108.2 |
93.4 82.8–104.4 |
| ‘Dynastinae’ | 47.0 37.2–56.9 |
46.0 37.2–55.2 |
51.9 40.9–62.5 |
57.6 46.1–69.4 |
49.8 39.0–59.6 |
49.4 39.6–58.9 |
| Glaphyridae | 141.6 140.2–144.3 |
141.3 140.2–143.6 |
141.3 140.2–143.5 |
141.4 140.2–143.6 |
141.3 140.2–143.4 |
101.1 77.1–123.9 |
| southern world Melolonthines | 77.4 55.2–98.4 |
82.0 60.5–104.4 |
88.7 68.5–108.7 |
95.8 75.1–115.7 |
83.7 65.7–103.0 |
81.1 68.7–98.0 |
| Pleurosticti (crown group) | 113.6 97.4–129.6 |
112.2 95.9–128.7 |
119.1 105.2–134.0 |
128.1 113.1–142.1 |
111.6 97.9–125.1 |
108.9 98.4–121.5 |
| Pleurosticti (steam lineage) | 155.4 133.3–171.1 |
163.2 156.1–170.6 |
165.1 158.9–172.9 |
167.6 159.8–175.9 |
164.0 157.6–170.6 |
156.0 147.5–163.3 |
| Trogidae | 141.7 140.2–144.8 |
141.8 140.2–144.9 |
141.8 140.2–144.9 |
141.9 140.2–145.3 |
141.8 140.2–144.6 |
115.4 90.5–135.6 |
| Lucanidae | 146.6 145.5–148.8 |
147.0 145.5–149.8 |
146.9 145.5–149.7 |
146.9 145.5–149.8 |
146.9 145.5–149.8 |
91.5 57.4–122.9 |
| Hybosoridae | 157.1 155.7–159.7 |
156.9 155.7–159.2 |
157.0 155.7–159.6 |
157.1 155.7–159.9 |
156.9 155.7–159.2 |
156.4 155.7–157.7 |
The calibration was not sensitive to the exclusion (masked data; run 1) or inclusion (unmasked data; runs 2–6) of alignment variable data, and therefore the consistency of fossil dates with the node ages was only tested with all data included. We found that node ages for fossil Aphodiinae and Aphodius (nodes F and H) were in conflict with the other settings during BEAST analysis. When these fossil calibrations were partly or entirely omitted from the search (runs 3–6), the main effect was the much older divergence time of Aphodiinae. In addition, dating was affected by four fossils (A, B, D and I; table 1) whose taxonomic assignment did not allow unambiguous placement to either the crown group or stem group nodes, because preserved morphological features of these fossils were not sufficient to place them in relation to the limited sampling of crown groups in our analysis. Moving these calibration points from the crown (run 5) to the stem group node (run 6) had little impact on most nodes, but resulted in much younger divergence times for four nodes (table 2). Specifically, crown-group age was reduced for Lucanidae from 147 to 91 Ma, for Scarabaeoidea from 186 to 174 Ma, for Trogidae from 141 to 115 Ma and for Glaphyridae from 141 to 101 Ma. The latter is of great interest because this date is very similar to that of Pleurosticts (109 Ma), which were affected only slightly by this alternative calibration.
The major transitions in feeding style can be traced to the branches by ancestral character state reconstruction scoring each terminal for phytophagy, coprophagy and anthophagy (see the electronic supplementary material). Hence, the minimal date of phytophagy is the origin of the Pleurosticti crown group in the Lower Cretaceous between 109 and 128 Ma. The second origin of phytophagy at the base (crown-group node) of Glaphyridae was dated to a minimum of 101 Ma, but up to 141 Ma depending on the calibration used (table 2). However, for the reduced (run 1) and full datasets (runs 2–6), the large node interval representing the stem lineage of Pleurosticti results in difficulties for the exact dating of the shift to phytophagy. The first appearance of a phytophagous ancestor to the Pleurosticti therefore may be assigned to a wide range up to the stem lineage age of 156 to 168 Ma (table 1). For the Glaphyridae, the greatest uncertainty comes from different possible crown-group ages (run 6, figure 1), whereas the stem group age in all cases was only slightly higher than the maximum crown-group age. Both phytophagous lineages gave rise to one major clade each feeding on flowers (pollen and petals) that occurred much later, whereby these traits appeared slightly earlier in Glaphyridae (63 to 79 Ma) than in Pleurosticti Cetoniinae (62 to 72 Ma). Several additional smaller clades also acquired flower-feeding, e.g. in the pleurostict tribes Hopliini, Chasmatopterini, Phyllotocini, but they arose only during the Eocene or Oligocene and hence comprised tip-level lineages (e.g. within Anomalini or Sericini). Dung feeding has at least two origins, defined by the basal nodes at which coprophagous lineages split from the saprophagous ancestors at 73 to 82 Ma in Scarabaeinae and 43 to 78 Ma in Aphodiinae. Depending on the choice of calibration, the shift to dung feeding in Scarabaeinae originated much (fossil calibration on node H included) or only slightly earlier (run 4, fossil calibration on node H excluded) than in Aphodiinae (table 2).
4. Discussion
Establishing the evolutionary timescale is a major step towards understanding the causes of diversification in organismal lineages. Here, we present a phylogenetic analysis of Scarabaeoidea based on a taxonomically comprehensive dataset and link them to recently described new fossil evidence. The topologies found in this study widely corroborate previous analyses based on partial nuclear rRNA data of Scarabaeoidea [58] and nuclear rRNA combined with mitochondrial data for Coleoptera-wide studies [17,59]. Key findings of this study—in accordance with these existing analyses including those with morphological data [25]—are the non-monophyly of Scarabaeidae, the monophyly of pleurostict chafers and the sister relationship between Scarabaeinae and Aphodiinae. The latter relationship was not supported by analyses of larval morphology [60] and three loci [17] but has been assumed in the earlier literature [56]. The separation of the dung scarabs and pleurostict chafers, currently grouped in the family Scarabaeidae was surprising, given the results from several studies using morphological characters [12,56,61]. Comparability of the current findings with these studies is, however, limited owing to their reduced taxon sampling that started from the assumption of a monophyletic Scarabaeidae (see the electronic supplementary material). The current data provided statistical support against this scenario, although more detailed studies of potentially confounding signal in both nuclear rRNA and mitochondrial data are required to confirm the non-monophyly of Scarabaeidae. The resolution of basal relationships of Scarabaeoidea otherwise largely conforms to expectations from the literature, which considers Glaresidae as a primitive, early branch [62], while Lucanidae and Trogidae (together with Passalidae) have been considered to form the earliest radiation of Scarabaeoidea [63]. Relationships within pleurostict chafers are widely congruent with the topology found by Ahrens & Vogler [64], although their study did not include 18S data.
(a). Scarab divergence times
Our ingroup-based fossil calibration of the tree confirmed a Jurassic origin of the Scarabaeoidea crown group (174 to 191 Ma). This is slightly younger than previous estimates of 195 and 191 Ma [17,18], respectively, based on penalized likelihood for tree linearization and different fossils for dating the crown-group divergence of Scarabaeoidea (Hunt et al. [17]: Holchorebus; McKenna & Farrell [18]: Juraclopus rohdendorfi, both using a minimum age of 152 Ma), in the course of dating the basal splits in the tree of Coleoptera. All of these studies are affected by the problem of an exact systematic placement of fossils owing to poor preservation of key morphological characters. Yet, the dating of basal nodes is widely consistent with the current fossil record and the fact that reliable fossil Scarabaeoidea are not older than the Middle Jurassic [15,19,21,22].
Based on the age estimates for phytophagous and coprophagous radiations, the origin of dung beetles showed a clear time lag relative to the herbivores. This timing suggests that both lineages were part of an evolutionary path driven by the angiosperms that first favoured the diversification of herbivores (mammals and insects) followed by the secondary radiation of the dung feeders. This finding also argues against the hypothesis that extant dung beetle lineages initially fed on dinosaur excrement, as often hypothesized ([22,65], but see [66]), because shift to dung feeding occurred at most 10 Ma before the dinosaurs went extinct. Multiple origins of dung feeding were in accordance with previous studies that had already proposed mycetophagy and saprophagy as the ancestral feeding mode of Scarabaeinae [63,67,68], which placed dung feeding at a time after the split from the common ancestor with the Aphodiinae. However, this hypothesis has not been tested with a comprehensive dataset that also includes the Aphodiinae [69]. Consistent with the findings here, the oldest fossils for these lineages are from the Upper Cretaceous and the Palaeocene for Scarabaeinae and Aphodiinae, respectively [22], i.e. 92 to 84 and 59 to 56 Ma, which is (except for run 4) earlier than the inferred origin of coprophagy and again supports a separate trajectory of both lineages and a long history prior to the origin of dung-producing mammals.
If species diversity of both radiations is plotted on the tree (figure 2), it appears that most of the lineage diversification postdated the rise of angiosperms and mammals, respectively. However, the stem lineage of pleurostict chafers coincides (figure 1) broadly with the appearance of crown-group angiosperms dated to 132–141 Ma using fossils [70] and to 140–180 Ma using molecules [71,72]. This leaves the possibility of a much earlier association of both lineages, but also confirms that extant lineages all derive from an ancestor that coincided with the time of major expansion of angiosperm diversity. The fossil record of Pleurosticti begins in the Tertiary [22], except for ‘Sericinae’ (e.g. Cretoserica and Lithanomala) and ‘Cretomelolonthinae’ (Cretomelolontha) [22,73], which date back to ca 145 Ma and hence precede by some distance the crown-group divergence time of Pleurosticti as estimated in this study. However, the systematic placement of these fossils was based on principally plesiomorphic character states and overall appearance. Owing to the limited number of preserved characters, the exact systematic position of these Lower Cretaceous fossils will hardly ever be inferred with certainty. Possibly these fossils might belong to the stem lineage of pleurosticts (a hypothesis that can be accommodated in our time tree; figure 1). A younger age of Pleurosticti is suggested by various biogeographical patterns, as recently derived lineages are absent from isolated southern landmasses (e.g. ‘Rutelinae’ and Cetoniinae in New Zealand [74]; Ablaberini and Sericini in the Australian region [57]), suggesting that the origin of several highly diverse extant lineages within Pleurosticti postdates the split of the Gondwanan supercontinent during the Jurassic.
Figure 2.
Crown-group diversification of Scarabaeiformia in relation to the increase of angiosperm and mammal diversity. The width of triangles roughly corresponds to the species numbers in major lineages. The increase of dominance of angiosperm diversity (green line; adapted from [4]) and the growth of extant mammal lineages (blue line; data from [1]) are superimposed on the tree. Star symbols indicate origin of derived feeding behaviours: green, phytophagy; brown, coprophagy; red, anthophagy.
(b). Scarab divergence in an evolutionary-ecological context
The divergence time estimates for the pleurostict crown group indicate an almost immediate tracking of the angiosperm diversification (figure 2) [4,5,71,72,75]. However, most of the extant lineages, such as Anomalini, Cetoniinae, Dynastinae, Rhizotrogini, Melolonthini and Adoretini (figure 2), diverged apparently much later during the Tertiary [22,63], i.e. a vast species diversity of pleurosticts arose during the later stages of the angiosperms' rise to dominance [4]. Similar time lags were also found for other angiosperm-related beetle radiations [76,77]. The time tree therefore links the diversification of pleurostict scarabs to the ecological changes imposed by the angiosperms since the Cretaceous. Key factors promoting diversification could have been the increased productivity and growth rates of angiosperms due to modification of leaf vein density [78]; the evolutionary rise of ectomycorrhiza enhancing chemical weathering of soils [79,80]; and the promotion of soil nutrient release by angiosperm litter that is easily decomposed [81]. This autocatalytic ‘litter revolution’ resulted not only in a boom of suitable pleurostict larval habitats, with abundant food resources under various ecological and biogeographical conditions, but also in a complex intestinal endosymbiont microflora [82,83]. This suggests a close link of the pleurostict species richness with general environmental change caused by the rise of the angiosperms. More specific analyses of shifts in species richness over time [84] and denser taxon sampling will be needed for refining and testing these hypotheses. However, because the feeding associations are not species-specific, the mechanisms of diversification are likely to differ from those suggested in the Phytophaga whose radiation presumably was driven by plant chemical defences and an evolutionary ‘arms race’ with the herbivore lineages [7,8].
For the dung scarabs, tracking of mammals began when several of the extant major lineages were present (figure 2), but the main diversification of dung beetles probably only occurred since the Miocene when more arid and fluctuating climate led to the spread of savannahs [85–87] and to the dominance of the Artiodactyla [88] as the main dung producers [67]. In fact, speciation rates in scarab dung beetles apparently increased [89] and most extant genera originated during the Miocene [90], linked to convergent switches between tunnelling and dung-rolling behaviour on all major continents (e.g. [68,69,91]). By contrast, the equally species-rich Aphodiinae lack the spectacular morphological and behavioural diversity of Scarabaeinae and, unable to transport dung, as ‘dwellers’ they remained confined to a peripheral niche space, but gained wider distribution outside of the tropics.
Apart from a number of species-rich lineages, which have been sampled in some detail for our analysis, the time tree also revealed a few older, yet species-poor lineages. These have often highly restricted distributions (e.g. Euchirini, Pachypodiini, Chasmatopterini and part of Pachydemini) and some are rare. Often these lineages are specialized, with adults showing highly derived feeding behaviour and morphology, and therefore their systematic placement is doubtful. Although some of these smaller lineages were not available for DNA analysis, the various dating approaches using multiple fossils in combination with the 4500 bp dataset provide a robust timescale for tracking the planet's mega-radiations. This provides the opportunity to explore wider questions in macroevolution as it refines the ecological context of diversifications in which herbivory (of insects and mammals) seems to be a key factor [92].
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to Silvia Fabrizi, Anna Papadoupoulou and Ruth Wild for help with the laboratory analysis, to Chris Barton for help with the GenBank submissions, to Paul Schoolmesters for helpful comments on species numbers, to Gerhard Burmeister, David Lees, Sergej Murzin, Guido Sabatinelli and Stefan Schmidt for collecting additional specimens and to Jonas Eberle for helping to compile the latest Consel version. For providing us with research and collection permits, we thank the various governmental institutions and departments in Eastern Cape (permit no.: WRO 122/07WR and WRO123/07WR), Gauteng (permit no.: CPF6 1281), Limpopo (permit no.: CPM-006-00001), Mpumalangma (permit no.: MPN-2009-11-20-1232) and Kwazulu-Natal (permit nos.: OP3752/2009, 1272/2007 and 3620/2006). We thank the two anonymous referees who helped to improve the final version of the manuscript.
Data accessibility
Additional details are provided in the electronic supplementary material, provided from the Dryad Data repository (doi:10.5061/dryad.v99j8).
Funding statement
This study was supported by the Leverhulme Trust (F/00696/P) and the German Science Association (AH175/1-2, AH175/3).
References
- 1.Bininda-Emonds ORP, et al. 2007. The delayed rise of present-day mammals. Nature 557, 507–512. ( 10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 2.dos Reis M, Inoue J, Hasegawa M, Asher RJ, Donoghue PCJ, Yang Z. 2012. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc. R. Soc. B 279, 3491–3500. ( 10.1098/rspb.2012.0683) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lidgard S, Crane PR. 1988. Quantitative analyses of the early angiosperm radiation. Nature 331, 344–346. ( 10.1038/331344a0) [DOI] [Google Scholar]
- 4.Lidgard S, Crane PR. 1990. Angiosperm diversification and Cretaceous floristic trends: a comparison of palynofloras and leaf macrofloras. Paleobiology 16, 77–93. [Google Scholar]
- 5.Magallón SA, Sanderson MJ. 2005. Angiosperm divergence times: the effect of genes, codon positions and time constraints. Evolution 59, 1653–1670. ( 10.1554/04-565.1) [DOI] [PubMed] [Google Scholar]
- 6.Clarke JT, Warnock RCM, Donoghue PCJ. 2011. Establishing a time-scale for plant evolution. New Phytol. 192, 266–301. ( 10.1111/j.1469-8137.2011.03794.x) [DOI] [PubMed] [Google Scholar]
- 7.Ehrlich PR, Raven PH. 1964. Butterflies and plants: a study in coevolution. Evolution 18, 586–608. ( 10.2307/2406212) [DOI] [Google Scholar]
- 8.Farrell BD. 1998. ‘Inordinate Fondness’ explained: why are there so many beetles? Science 281, 555–559. ( 10.1126/science.281.5376.555) [DOI] [PubMed] [Google Scholar]
- 9.Marvaldi AE, Sequeira AS, O'Brien CW, Farrell BD. 2002. Molecular and morphological phylogenetics of weevils (Coleoptera: Curculionoidea): do niche shifts accompany diversification? Syst. Biol. 51, 761–785. ( 10.1080/10635150290102465) [DOI] [PubMed] [Google Scholar]
- 10.Erichson WF. 1847. Naturgeschichte der Insecten Deutschlands. Erste Abtheilung. Coleoptera, vol. 3, Lieferung 4. Nicolaische Buchhandlung, Berlin, pp. 481–640.
- 11.Scholtz CH, Grebennikov VV. 2005. Scarabaeoidea Latreille, 1802. In Coleoptera, Beetles. vol. 1: morphology and systematics (Archostemata, Adephaga, Myxophaga, Polyphaga partim). Handbook of zoology. vol. IV Arthropoda: insecta part 38 (eds Beutel RG, Leschen RAB.), pp. 367–426. New York, NY: W. de Gruyter-Berlin. [Google Scholar]
- 12.Browne J, Scholtz CH. 1999. A phylogeny of the families of Scarabaeoidea. Syst. Entomol. 24, 51–84. ( 10.1046/j.1365-3113.1999.00067.x) [DOI] [Google Scholar]
- 13.Krell F-T. 2007. Catalogue of fossil Scarabaeoidea (Coleoptera: Polyphaga) of the Mesozoic and Tertiary, v. 2007. Denver Museum of Nature and Science Technical Report 2007–8, 79 pp.
- 14.Crowson R. 1981. The biology of the Coleoptera, vol. xii, 802 pp. London, UK: Academic Press. [Google Scholar]
- 15.Krell F-T. 2000. The fossil record of Mesozoic and Tertiary Scarabaeoidea (Coleoptera: Polyphaga). Invert. Syst. 14, 871–905. ( 10.1071/IT00031) [DOI] [Google Scholar]
- 16.Morón Ríos MA. 1984. Escarabajos. 200 Millones de Años de Evolución, 132 pp. Distrito Federal, México: Instituto de Ecología. [Google Scholar]
- 17.Hunt T, et al. 2007. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 318, 1913–1916. ( 10.1126/science.1146954) [DOI] [PubMed] [Google Scholar]
- 18.McKenna DD, Farrell BD. 2009. Coleoptera. In The timetree of life (eds Hedges B, Kumar S.), pp. 278–289. Oxford, UK: Oxford University Press. [Google Scholar]
- 19.Bai M, Ahrens D, Yang X-K, Ren D. 2012. New fossil evidence of the early diversification of scarabs: Alloioscarabaeus cheni (Coleoptera: Scarabaeoidea) from the Middle Jurassic of Inner Mongolia, China. Insect Sci. 19, 159–171. ( 10.1111/j.1744-7917.2011.01460.x) [DOI] [Google Scholar]
- 20.Nikolajev GV, Ren D. 2010. The oldest fossil Ochodaeidae (Coleoptera: Scarabaeoidea) from the Middle Jurassic of China. Zootaxa 2553, 65–68. [Google Scholar]
- 21.Nikolajev GV, Ren D. 2011. The oldest species of the genus Glaphyrus Latr. (Coleoptera: Scarabaeoidea: Glaphyridae) from the Mesozoic of China. Paleontol. J. 45, 179–182. ( 10.1134/S0031030111010126) [DOI] [Google Scholar]
- 22.Krell F-T. 2006. Fossil record and evolution of Scarabaeoidea (Coleoptera: Polyphaga). Coleopt. Soc. Monogr. 5, 120–143. [Google Scholar]
- 23.Smith ABT. 2006. A review of the family-group names for the superfamily Scarabaeoidea (Coleoptera) with corrections to nomenclature and a current classification. Coleopt. Soc. Monogr. 5, 144–204. [Google Scholar]
- 24.Caterino MS, Hunt T, Vogler AP. 2005. On the constitution and phylogeny of Staphyliniformia (Insecta: Coleoptera). Mol. Phylogenet. Evol. 34, 655–672. ( 10.1016/j.ympev.2004.11.012) [DOI] [PubMed] [Google Scholar]
- 25.Lawrence JF, Ślipiński A, Seago AE, Thayer MK, Newton AF, Marvaldi AE. 2011. Phylogeny of the Coleoptera based on morphological characters of adults and larvae. Ann. Zool. 61, 1–217. ( 10.3161/000345411X576725) [DOI] [Google Scholar]
- 26.Katoh K, Kuma K, Toh H, Miyata T. 2005. Mafft version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. ( 10.1093/nar/gki198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh K, Misawa K, Kuma K, Miyata T. 2002. Mafft: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. ( 10.1093/nar/gkf436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misof B, Misof K. 2009. A Monte Carlo approach successfully identifies randomness in multiple sequence alignments: a more objective means of data exclusion. Syst. Biol. 58, 21–34. ( 10.1093/sysbio/syp006) [DOI] [PubMed] [Google Scholar]
- 29.Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17, 754–755. ( 10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Rannala B. 1997. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Mol. Biol. Evol. 14, 717–724. ( 10.1093/oxfordjournals.molbev.a025811) [DOI] [PubMed] [Google Scholar]
- 31.Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. 2004. Bayesian phylogenetic analysis of combined data. Syst. Biol. 53, 47–67. ( 10.1080/10635150490264699) [DOI] [PubMed] [Google Scholar]
- 32.Brandley MC, Schmitz A, Reeder TW. 2005. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst. Biol. 54, 373–390. ( 10.1080/10635150590946808) [DOI] [PubMed] [Google Scholar]
- 33.Rambaut A, Drummond AJ. 2009. Tracer v. 1.5 See http://beast.bio.ed.ac.uk/Tracer.
- 34.Wilgenbusch JC, Warren DL, Swofford DL. 2004. AWTY: a system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference See http://king2.scs.fsu.edu/CEBProjects/awty/awty_start.php.
- 35.Guidon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood . Syst. Biol. 52, 696–704. ( 10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 36.Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinform. Appl. Note 14, 817–818. ( 10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- 37.Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. ( 10.1080/10635150600755453) [DOI] [PubMed] [Google Scholar]
- 38.Shimodaira H. 2001. Consel: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246–1247. ( 10.1093/bioinformatics/17.12.1246) [DOI] [PubMed] [Google Scholar]
- 39.Parham JF, et al. 2012. Best practices for justifying fossil calibrations. Syst. Biol. 61, 1–14. ( 10.1093/sysbio/syr107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikolajev GV. 2000. New subfamily of stag beetles (Coleoptera: Scarabaeoidea: Lucanidae) from the Mesozoic of Mongolia, and its position in the system of the superfamily. Paleontol. J. 34(Suppl. 3), S327–S330. [Google Scholar]
- 41.Nikolaev GV. 2007. Mezozoiskii Etap Evolyutsii Plastinchatousykh (Insecta: Coleoptera: Scarabaeoidea), 222 pp. Almaty, Kazakhstan: Kazak Universiteti. [Google Scholar]
- 42.Nikolajev GV. 2010. On the Mesozoic taxa of scarabaeoid beetles of the Family Hybosoridae (Coleoptera: Scarabaeoidea). Paleontol. J. 44, 649–653. ( 10.1134/S0031030110060067) [DOI] [Google Scholar]
- 43.Nikolaev GV. 2005. Plastinchatousye zhuki podsemeystva Glaphyrinae (Coleoptera, Scarabaeidae) v nizhnem mele Zabaykal'ya. Zhivotnyy mir Dal'nego Vostoka 5, 69–78. [Google Scholar]
- 44.Piton L. 1940. Paléontologie du Gisement Éocène de Menat (Puy-de-Dôme) (Flore et Faune), 306 pp. Paris, France: Lechevalier. [Google Scholar]
- 45.Vincent P, Aubert M, Boivin P, Cantagrel J-M, Lenat J-F. 1977. Découverte d'un volcanisme paleocene en Auvergne: les maars de Menat et leurs annexes; etude géologique et geophysique. Bull. Soc. Geol. Fr. 7, 1057–1070. [Google Scholar]
- 46.Scudder SH. 1890. The Tertiary insects of North America. Report of the United States Geological Survey of the Territories 13, 743 pp., 28 pls., 1 map.
- 47.Wickham HF. 1912. A report on some recent collections of fossil Coleoptera from the Miocene shales of Florissant. Bull. Lab. Nat. Hist. Iowa 6, 3–38. [Google Scholar]
- 48.Cockerell TDA. 1921. Some Eocene insects from Colorado and Wyoming. Proc. US Natl Mus. 59, 29–39. ( 10.5479/si.00963801.59-2358.29) [DOI] [Google Scholar]
- 49.Wappler T. 2003. Die Insekten aus dem Mittel-Eozän des Eckfelder Maares, Vulkaneifel. Mainzer Naturwissenschaftliches Arch. 27, 1–234. [Google Scholar]
- 50.Wickham HF. 1914. New Miocene Coleoptera from Florissant. Bull. Mus. Comp. Zool. 58, 421–494. [Google Scholar]
- 51.Zhang J, Sun B, Zhang X. 1994. Miocene insects and spiders from Shanwang, Shandong, vol. 298, pp. 44 pls Beijing, China: Science Press. [Google Scholar]
- 52.Ratcliffe BC, Smith DM, Erwin D. 2005. Oryctoantiquus borealis, new genus and species from the Eocene of Oregon, U.S.A., the world's oldest fossil dynastine and largest fossil scarabaeid (Coleoptera: Scarabaeidae: Dynastinae). Coleopt. Bull. 59, 127–135. ( 10.1649/0010-065X(2005)059[0127:OBNGAS]2.0.CO;2) [DOI] [Google Scholar]
- 53.Bai M, Beutel RG, Shih C-K, Ren D, Yang X-K. 2012. Septiventeridae, a new and ancestral fossil family of Scarabaeoidea (Insecta: Coleoptera) from the Late Jurassic to Early Cretaceous Yixian Formation. J. Syst. Palaeontol. 11, 359–374. ( 10.1080/14772019.2012.660995) [DOI] [Google Scholar]
- 54.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 ( 10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ho SYW, Philipps MJ. 2009. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst. Biol. 58, 367–380. ( 10.1093/sysbio/syp035) [DOI] [PubMed] [Google Scholar]
- 56.Browne J, Scholtz CH. 1998. Evolution of the scarab hind wing articulation and wing base: a contribution toward the phylogeny of the Scarabaeidae (Scarabaeoidea: Coleoptera). Syst. Entomol. 23, 307–326. ( 10.1046/j.1365-3113.1998.00059.x) [DOI] [Google Scholar]
- 57.Ahrens D. 2006. The phylogeny of Sericini and their position within the Scarabaeidae based on morphological characters (Coleoptera: Scarabaeidae). Syst. Entomol. 31, 113–144. ( 10.1111/j.1365-3113.2005.00307.x) [DOI] [Google Scholar]
- 58.Smith ABT, Hawks DC, Heraty JM. 2006. An overview of the classification and the evolution of the major scarab beetle clades (Coleoptera: Scarabaeoidea) based on preliminary molecular analyses. Coleopts. Soc. Monogr. 5, 35–46. [Google Scholar]
- 59.Bocak L, Barton C, Crampton A, Chesters D, Ahrens D, Vogler AP. 2013. Building the Coleoptera tree-of-life for >8000 species: composition of public DNA data and fit with Linnaean classification. Syst. Entomol. 39, 97–110. ( 10.1111/syen.12037) [DOI] [Google Scholar]
- 60.Grebennikov VV, Scholtz CH. 2004. The basal phylogeny of Scarabaeoidea (Insecta: Coleoptera) inferred from larval morphology. Invert. Syst 18, 321–348. ( 10.1071/IS03013) [DOI] [Google Scholar]
- 61.Browne DJ, Scholtz CH. 1995. Phylogeny of the families of Scarabaeoidea (Coleoptera) based on characters of the hindwing articulation, hindwing base and wing venation. Syst. Entomol. 20, 145–173. ( 10.1111/j.1365-3113.1995.tb00089.x) [DOI] [Google Scholar]
- 62.Scholtz CH, Browne DJ, Kukalova-Peck J. 1994. Glaresidae, archaeopteryx of the Scarabaeoidea. Syst. Entomol. 19, 145–173. ( 10.1111/j.1365-3113.1994.tb00590.x) [DOI] [Google Scholar]
- 63.Scholtz CH, Chown SL. 1995. The evolution of habitat use and diet in the Scarabaeoidea: a phylogenetic approach. In Biology, phylogeny, and classification of Coleoptera: papers celebrating the 80th Birthday of Roy A. Crowson. (eds Pakaluk J, Slipinski SA.), pp. 355–374. Warszawa, Poland: Muzeum i Instytut Zoologii PAN. [Google Scholar]
- 64.Ahrens D, Vogler AP. 2008. Towards the phylogeny of chafers (Sericini): analysis of alignment-variable sequences and the evolution of segment numbers in the antennal club. Mol. Phylogenet. Evol. 47, 783–798. ( 10.1016/j.ympev.2008.02.010) [DOI] [PubMed] [Google Scholar]
- 65.Halffter G. 1974. Eléments anciens de l'Entomofaune Néotropicale: Ses implications biogeographiques. Quaest. Entomol. 10, 223–262. [Google Scholar]
- 66.Arillo A, Ortuño VM. 2008. Did dinosaurs have any relation with dung-beetles? (The origin of coprophagy). J. Nat. Hist. 42, 1405–1408. ( 10.1080/00222930802105130) [DOI] [Google Scholar]
- 67.Cambefort Y. 1991. From saprophagy to coprophagy. In Dung beetle ecology (eds Hanski I, Cambefort Y.), pp. 22–35. Princeton, NJ: Princeton University Press. [Google Scholar]
- 68.Monaghan MT, Inward DG, Hunt T, Vogler AP. 2007. A molecular phylogenetic analysis of the Scarabaeinae (dung beetles). Mol. Phyl. Evol. 45, 674–692. ( 10.1016/j.ympev.2007.06.009) [DOI] [PubMed] [Google Scholar]
- 69.Philips TK. 2011. The evolutionary history and diversification of dung beetles. In Ecology and evolution of dung beetles (eds Simmons LW, Ridsdill-Smith TJ.), pp. 21–46. Oxford, UK: Blackwell Publishing Ltd. [Google Scholar]
- 70.Brenner G. 1996. The origin and evolution of the angiosperm carpel. In Flowering plant origin, evolution, and phylogeny (eds Taylor DW, Hickey LJ.), pp. 91–115. New York, NY: Chapman and Hall. [Google Scholar]
- 71.Bell CD, Soltis DE, Soltis PS. 2010. The age and the diversification of the angiosperms re-visited. Am. J. Bot. 97, 1296–1303. ( 10.3732/ajb.0900346) [DOI] [PubMed] [Google Scholar]
- 72.Bell CD, Soltis DE, Soltis PS. 2005. The age of the angiosperms: a molecular timescale without a clock. Evolution 59, 1245–1258. ( 10.1111/j.0014-3820.2005.tb01775.x) [DOI] [PubMed] [Google Scholar]
- 73.Nikolajev GV. 1998. Pleurostict lamellicorn beetles (Coleoptera, Scarabaeidae) from the Lower Cretaceous of Transbaikalia. Paleontol. J. 32, 513–521. [Google Scholar]
- 74.Watts CJ. 1984. A review of some New Zealand Scarabaeidae (Coleoptera). New Zealand Entomol. 8, 4–24. ( 10.1080/00779962.1984.9722456) [DOI] [Google Scholar]
- 75.Sanderson MJ, Thorne JL, Wikström N, Bremer K. 2004. Molecular evidence on plant divergence times. Am. J. Bot. 91, 1656–1665. ( 10.3732/ajb.91.10.1656) [DOI] [PubMed] [Google Scholar]
- 76.Gomez-Zurita J, Hunt T, Kopliku F, Vogler AP. 2007. Recalibrated tree of leaf beetles (Chrysomelidae) indicates independent diversification of angiosperms and their insect herbivores. PLoS ONE 2, e360 ( 10.1371/journal.pone.0000360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McKenna DD, Sequeira AS, Marvaldi AE, Farrell BD. 2009. Temporal lags and overlap in the diversification of weevils and flowering plants. Proc. Natl Acad. Sci. USA 106, 7083–7088. ( 10.1073/pnas.0810618106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Boer HJ, Eppinga MB, Wassen MJ, Dekker SC. 2012. A critical transition in leaf evolution facilitated the Cretaceous angiosperm revolution. Nat. Commun. 3, 1221 ( 10.1038/ncomms2217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taylor L, Banwart S, Leake J, Beerling DJ. 2011. Modeling the evolutionary rise of ectomycorrhiza on sub-surface weathering environments and the geochemical carbon cycle. Am. J. Sci. 311, 369–403. ( 10.2475/05.2011.01) [DOI] [Google Scholar]
- 80.Taylor LL, Banwart SA, Valdes PJ, Leake JR, Beerling DJ. 2012. Evaluating the effects of terrestrial ecosystems, climate and carbon dioxide on weathering over geological time: a global-scale process-based approach. Phil. Trans. R. Soc. B 367, 565–582. ( 10.1098/rstb.2011.0251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berendse F, Scheffer M. 2009. The angiosperm radiation revisited, an ecological explanation for Darwin's ‘abominable mystery’. Ecol. Lett. 12, 865–872. ( 10.1111/j.1461-0248.2009.01342.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang H, Jackson TA. 2008. Autochthonous bacterial flora indicated by PCR-DGGE of 16S rRNA gene fragments from the alimentary tract of Costelytra zealandica (Coleoptera: Scarabaeidae). J. Appl. Microbiol. 105, 1277–1285. ( 10.1111/j.1365-2672.2008.03867.x) [DOI] [PubMed] [Google Scholar]
- 83.Andert J, Marten A, Brandl R, Brune A. 2010. Inter- and intraspecific comparison of the bacterial assemblages in the hindgutof humivorous scarab beetle larvae (Pachnoda spp.). FEMS Microbiol. Ecol. 74, 439–449. ( 10.1111/j.1574-6941.2010.00950.x) [DOI] [PubMed] [Google Scholar]
- 84.FitzJohn RG, Maddison WP, Otto SP. 2009. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58, 595–611. ( 10.1093/sysbio/syp067) [DOI] [PubMed] [Google Scholar]
- 85.van der Hammen T. 1983. The palaeoecology and palaeogeography of savannas. In Tropical Savannas: ecosystems of the world, vol. 13 (ed. Bourliere F.), pp. 19–35. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 86.Jacobs BF. 2004. Palaeobotanical studies from tropical Africa: relevance to the evolution of forest, woodland and savannah biomes. Phil. Trans. R. Soc. Lond. B 359, 1573–1583. ( 10.1098/rstb.2004.1533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beerling DJ, Osborne CP. 2006. The origin of the savanna biome. Glob. Change Biol. 12, 2023–2031. ( 10.1111/j.1365-2486.2006.01239.x) [DOI] [Google Scholar]
- 88.Sinclair ARE. 1983. The adaptations of African ungulates and their effects on community functions. In Tropical Savannas: ecosystems of the world, vol. 13 (ed. Bourliere F.), pp. 401–425. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 89.Wirta H, Orsini L, Hanski I. 2008. An old adaptive radiation of forest dung beetles in Madagascar. Mol. Phylogenet. Evol. 47, 1076–1089. ( 10.1016/j.ympev.2008.03.010) [DOI] [PubMed] [Google Scholar]
- 90.Sole CL, Scholtz CH. 2010. Did dung beetles arise in Africa? A phylogenetic hypothesis based on five gene regions. Mol. Phylogenet. Evol. 56, 631–641. ( 10.1016/j.ympev.2010.04.023) [DOI] [PubMed] [Google Scholar]
- 91.Philips TK, Pretorius E, Scholtz CH. 2004. A phylogenetic analysis of dung beetles (Scarabaeinae, Scarabaeidae), unrolling an evolutionary history. Invertebr. Syst. 18, 53–88. ( 10.1071/IS03030) [DOI] [Google Scholar]
- 92.Price SA, Hopkins SSB, Smith KK, Roth VL. 2012. Tempo of trophic evolution and its impact on mammalian diversification. Proc. Natl Acad. Sci. USA 109, 7008–7012. ( 10.1073/pnas.1117133109) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional details are provided in the electronic supplementary material, provided from the Dryad Data repository (doi:10.5061/dryad.v99j8).