Abstract
Nucleic acids have become a powerful tool in nanotechnology because of their conformational polymorphism. However, lack of a medium in which nucleic acid structures exhibit long-term stability has been a bottleneck. Ionic liquids (ILs) are potential solvents in the nanotechnology field. Hydrated ILs, such as choline dihydrogen phosphate (choline dhp) and deep eutectic solvent (DES) prepared from choline chloride and urea, are ‘green’ solvents that ensure long-term stability of biomolecules. An understanding of the behaviour of nucleic acids in hydrated ILs is necessary for developing DNA materials. We here review current knowledge about the structures and stabilities of nucleic acids in choline dhp and DES. Interestingly, in choline dhp, A–T base pairs are more stable than G–C base pairs, the reverse of the situation in buffered NaCl solution. Moreover, DNA triplex formation is markedly stabilized in hydrated ILs compared with aqueous solution. In choline dhp, the stability of Hoogsteen base pairs is comparable to that of Watson–Crick base pairs. Moreover, the parallel form of the G-quadruplex is stabilized in DES compared with aqueous solution. The behaviours of various DNA molecules in ILs detailed here should be useful for designing oligonucleotides for the development of nanomaterials and nanodevices.
INTRODUCTION
Single strands of nucleic acids recognize and bind to other nucleic acid sequences via highly specific base recognition interactions such as Watson–Crick base pairing and triplets with Watson–Crick and Hoogsteen base pairing. Nucleic acids with G-rich motifs form G-quartets stabilized by Hoogsteen base pairs under certain conditions (Figure 1a). Because of the specific interactions among the bases, nucleic acids spontaneously form secondary or higher-order structures such as duplexes, triplexes and G-quadruplexes (Figure 1b). Nucleic acids can thus be ‘programmed’ to adopt predefined structures by designing their primary sequence. These specific structures are the bases for the enormous potential of DNA-based devices in the fields of nanobiotechnology and biomedical technology.
Figure 1.
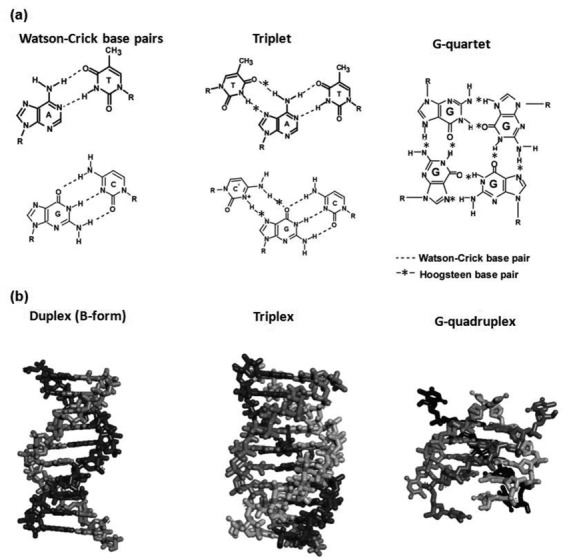
(a) Types of base pairs in nucleic acids. The triplet is drawn based on the assumption of acidic conditions; N3 of cytosine is protonated. (b) Structures of nucleic acids for duplex, triplex and G-quadruplex.
DNA can detect a complementary target sequence using Watson–Crick base pair formation; this sequence-based recognition has been applied to the development of sequence-sensing methods such as DNA microarrays of gene expression analyses (1) and single nucleotide polymorphism detection (2), chips for gene sequencing by hybridisation (3,4) and phylogenetic studies (5). Nanoscale self-assembly systems based on duplex formation are useful in functional molecule arrays (6) and in systems such as molecular transport devices (7), molecular computing devices (molecular switches, logical units and programmable molecular systems for massively parallel computing) (7) and ‘molecular motors’ (8) that depend on structural transitions of nucleic acids in response to chemical stimuli.
Nucleic acids are also able to form Hoogsteen base pairs, as shown in Figure 1b and c (9). In a triple helix, a third strand, also called the triplex-forming oligonucleotide, binds with sequence specificity via A*T and G*C Hoogsteen base pairs (the asterisk indicates the Hoogsteen base pair) in the major groove of a Watson–Crick base-paired DNA duplex (9–12). Development of DNA materials and diagnostic applications based on the formation of Hoogsteen base pairs has been challenging, given that these base pairs are stable only in certain sequence motifs and at low pH (13,14). In polypurine tracts, for example, found in human immunodeficiency virus-1 provirus and triplet repeat diseases, cytosines in the third strand must be protonated at N3 (pKa = 4.5) to form C-G*C+ base triplets (12,15). Given that the formation of Hoogsteen base pairs of mixed G and A sequences is not stable at neutral pH (12), biological applications have been limited. G-rich sequences are attractive building materials in the nanotechnology field because of their stable structure and their stimulus-dependent conformational polymorphism. Researchers have taken advantage of these characteristics to develop DNA-based materials such as sensors, logic devices, circuits and drugs (16–19). For example, we have developed logic devices that depend on structural transitions among duplexes, G-quadruplexes and i-motifs in response to input molecules. Methods for selective regulation of the structure and stability of nucleic acids have potential applications ranging from gene therapy to novel material creation.
Although DNA is reasonably stable in aqueous solution, non-physiological temperature, pH and ionic strength disrupt the DNA helix and cause denaturation. Conventionally, DNA is stored under refrigeration for short- and long-term applications, and the influence of storage temperature has been discussed in the literature (20,21). Nucleic acids are not stable in aqueous solutions at ambient temperatures for long periods (several days to 1 month) (21) because of degradation by contaminating nucleases (22) and because of inherent chemical instability. This instability is a bottleneck in the progress of nanotechnology based on nucleic acids (23). Furthermore, aqueous solutions are useless in small-volume technologies since small volumes of water vapourize immediately under open-air conditions or at high temperatures (24). To achieve functionality of nanodevices, solvents free of the limitations of aqueous buffers are required.
In the past 20 years, certain remarkable features of ionic liquids (ILs) have made them attractive alternatives to water in various applications including electrochemistry, separation science, chemical synthesis and materials science (25–28). Room-temperature ILs provide favourable environments for reactions because with a vapour pressure close to zero, ILs are non-volatile. In addition, ILs are green solvents that have replaced aqueous solutions for many reactions (24,25). The anions and cations in ILs are usually molecular ions with low symmetry, a feature that disfavours packing into crystals. With numerous possible combinations of such cations and anions, ILs have been called ‘designer solvents’ because they can be customized for a diverse range of reactions (29). As nucleic acids carry a negative charge on each residue, high IL concentrations may induce aggregation and precipitation of these biomolecules.
In the past 10 years, two types of ILs have been reported as biocompatible. The first type is hydrated IL. The hydrated IL of choline dihydrogen phosphate (choline dhp) (Figure 2a) formed by dissolution in a small amount of water (∼30 wt%; ∼16.7 M) supports the retention of the structure and activity of cytochrome c, lysozyme and ribonuclease A (27,30) and extends the shelf-lives of these proteins (28). The second type includes deep eutectic solvents (DESs), which are mixtures that form an eutectic system with a melting point much lower than that of either of the individual components (31). The deep eutectic phenomenon was first described in 2003 for a mixture of choline chloride (2-hydroxyethyl-trimethylammonium chloride) and urea in a 1:2 molar ratio (Figure 2b). Choline chloride and urea have a melting point of 302°C and 133°C, respectively. The eutectic mixture melts as low as 12°C. Lysozyme dissolved in DES retains enzyme activity (32). Because DES enhances the solubility of biomolecules, it has been used for preclinical studies (33). Moreover, bacteria freeze-dried in DES remain viable, opening interesting perspectives for the use of whole microorganisms in biocatalytic reactions carried out in non-aqueous solvents (34). Choline dhp and DESs are also markedly less toxic compared with organic solvents.
Figure 2.
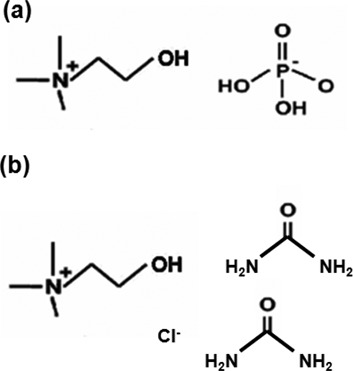
The chemical structure of (a) choline dhp and (b) the DES for a mixture of choline chloride (2-hydroxyethyl-trimethylammonium chloride) and urea in a 1:2 molar ratio.
Four factors mainly determine the structures formed by nucleic acids: hydrogen bonding, base stacking, conformational entropy (35–38) and cation binding (39–44). Because molecular ions bind not only to phosphate groups but also to bases via hydrogen bond formation and van der Waals interactions (45,46), ILs are expected to influence the structure and stability of nucleic acids. This review discusses the structure and stability of nucleic acids in choline dhp and DES and describes the advantages and unique properties imparted by these solvents. We expect that the properties of ILs will enable novel applications of nucleic acids in the nanobiotechnology field.
DNA STRUCTURE IN ILs
Cations bind to phosphate to shield the negative charge on the phosphate of natural DNA. Cations can also bind non-specifically to several parts of DNA such as phosphates, bases and sugars at high concentrations. The non-specific binding induces aggregation or dissociation of DNA structures. Effects of ILs on nucleic acid structures have been investigated in several studies (Table 1). Circular dichroism (CD) has been used to monitor nucleic acid structure in most cases. MacFarlane et al. measured the CD spectrum of the long DNA duplex obtained from salmon testes in hydrated choline dhp (47). In choline dhp, these long DNA duplexes have the same B-form conformation structure as that in aqueous solution. Moreover, short strands of DNA and RNA (10- to12-mers) that form duplexes or triplexes also form the same structures in choline dhp and DES as those in aqueous solution (48,49). However, certain sequences form different structures in ILs than in aqueous solution. For example, a 32-bp DNA duplex of mixed GC/AT sequence and d(GG)8 form different structures in ILs than in an aqueous buffer (49). The 32-bp DNA duplex in a low-salt buffer (100 mM NaCl) exhibited a CD spectrum consistent with a B-form duplex. In contrast, in DES, the CD spectrum of the 32-bp DNA duplex is suggestive of an A-form helix. d(GG)8 forms a left-handed Z-form helix in ILs but forms a B-form duplex in a low-salt aqueous buffer (49). Duplexes undergo B- to A-form or B- to Z-form transitions when subjected to dehydrating conditions and high ionic strength, properties certainly characteristic of DES (49). The duplex formed by the sequence d(A4T4)4 is also of interest because it contains four A-tract sequence elements, defined as four or more consecutive A–T base pairs without a 5′-TpA-3′ step. These sequence elements are known to adopt an altered B-form helical structure, designated B*, with an unusually narrow minor groove and high base pair propeller twist, and have a propensity for cation localisation in the minor groove (49). For d(A4T4)4, any B*-form helical structure is predicted to be interspersed with a B-form helical structure because A tracts are disrupted by 5′-TpA-3′ steps. In a low-salt buffer, spectroscopic data are consistent with a mixed B-/B*-form structure; however, d(A4T4)4 has a markedly different CD spectrum in DES, indicating the change in its secondary structure (49).
Table 1. Selected recent studies of structures behaviour in ILs or DESs.
| Nucleic acid | Aqueous solution | IL | Reference | ||||
|---|---|---|---|---|---|---|---|
| Na+ solution | K+ solution | Choline dhp | Other IL | DES | DES with K+ | ||
| Duplex | |||||||
| DNA from salmon testes | B-form | B-form | B-form | – | – | – | 47 |
| 5′-TTATAACCTA-3′/5′-TAGGTTATAA-3′ | B-form | – | – | – | B-form | – | 48 |
| 5′-CGGCAAGCGC-3′/5′-GCGCTTGCCG-3′ | B-form | – | – | – | B-form | 48 | |
| 12-mer RNA duplex | A-form | – | – | – | A-form | – | 49 |
| 32-mer DNA duplex of mixed GC/AT | B-form | – | – | – | A-form | – | 49 |
| [d(GG)8]2 | B-form | – | – | – | Z-form | – | 49 |
| [d(A4T4) 4]2 | B*-form | – | – | – | n.d.a | – | 49 |
| Triplex | |||||||
| 5′-TTTTTTTCTTCT -3′/5′-AGAAGAA | A-like form | – | A-like form | – | – | ||
| AAAAA-3′/5′-TCTTCTTTTTTT-3′ | |||||||
| G-quadruplex | |||||||
| d AG3(T2AG3)3 | antiparallel | hybrid | antiparallel | n.d.a | – | parallel | 52,53 |
| d AG3(T2AG3)7 | antiparallel | hybrid | antiparallel | n.d.a | – | parallel | 52,53 |
| d (G3T4)3G3 | antiparallel | antiparallel | – | – | – | parallel | 52 |
| d (T4G4)4 | antiparallel | antiparallel | – | – | – | parallel | 52 |
| d (G4T2)3G4 | hybrid | hybrid | – | – | parallel | parallel | 52 |
| d (AG3TG3)2A2T2 | parallel | parallel | – | – | parallel | parallel | 52 |
| d (AG3)2CGCTG3AG2AG3 | parallel | parallel | – | – | – | parallel | 52 |
| d AG3CG2TGTG3(AGAG3)2G2AG2 | parallel | parallel | – | – | – | parallel | 52 |
| d G2T2G2TGTG2T2G2 | antiparallel | antiparallel | antiparallel | – | antiparallel | antiparallel | 49,52 |
an.d. indicates that nucleic acid structure in IL was different from that in aqueous solution, but the structure was not characterized.
The CD spectra of widely investigated G-quadruplex DNA in aqueous solutions and water-free DES have also been compared (Table 1). The human telomeric DNA (dAG3(T2AG3)3; Tel22) adopts a hybrid G-quadruplex structure in aqueous solution containing K+ (50) and an anti-parallel G-quadruplex structure in Na+ (51). Interestingly, Tel22 adopts a parallel G-quadruplex structure in DES containing 100 mM KCl (K+/DES) and does not form a stable G-quadruplex in DES alone (52). Similar to that in aqueous buffer, Tel22 adopts the anti-parallel G-quadruplex form in choline dhp (53). In K+/DES, a 46-mer of the human telomeric DNA sequence (dAG3(T2AG3)7) forms higher-order G-quadruplex structures composed of two individual parallel G-quadruplexes, indicating that DES provides a favourable environment for higher-order G-quadruplex structures. The structure of Tel22 has also been investigated in choline dhp. Similar to that in aqueous buffer, Tel22 adopted the anti-parallel G-quadruplex form in choline dhp (53).
Six other well-characterized G-quadruplex-forming DNAs, (d(G3T4)3G3 Oxytricha telomeric DNA; d(G4T2)3G4 Tetrahymena telomeric DNA; d(AG3TG3)2A2T2 c-myc; d(AG3)2CGCTG3AG2AG3 c-kit and dAG3CG2TGTG3(AGAG3)2G2AG2 KRAS), similar to Tel22, form parallel structures in K+/DES (Table 1) (52). However, the G-quadruplex structure formed by the thrombin-binding aptamer (d G2T2G2TGTG2T2G2, TBA) is an exception (49,52). It adopts an anti-parallel structure in K+/DES, similar to the structures formed in both K+ and Na+ water solutions (Table 1). TBA does not adopt a parallel quadruplex structure under any conditions tested, unlike the other G-quadruplex-forming oligonucleotides. The G-quadruplex form adopted in K+/DES was proposed to be driven by dehydration, in agreement with reports that the formation of the G-quadruplex is accompanied by water molecule release and that water depletion induced by the presence of crowding agents favours parallel-stranded G-quadruplexes in K+-containing solutions (54,55).
DNA STABILITY IN ILs
Hydrated ILs influence the stability of Watson–Crick base pairs in duplexes
The formation of Watson–Crick base pairs in a duplex is the basis for the transmission of genetic information. To understand the effects of choline dhp on the stability of Watson–Crick base pairs, the sequence dependence of DNA duplex stability in choline dhp was investigated by the evaluation of ultraviolet (UV) melting curves. The melting temperatures (Tms) of 10-mer DNA duplexes with different A–T base pair contents were measured in a solution containing 4 M NaCl or 4 M choline dhp (80 wt% choline dhp) (Table 2). The A–T base pairs in ODN1–ODN6 are consecutive. In ODN7, ODN8, ODN9 and ODN10, the sequences are random. Buffer containing 4 M NaCl was used, given that previous quantitative analyses of DNA duplex stabilities were performed in this solution (56). Typical UV melting curves are shown in Figure 3 (48). In NaCl-containing solution, Tm of DNA duplexes with consecutive A–T base pairs decreased from 45.3 to 30.7°C as the A–T content increased (Figure 3a and Table 2). In contrast, in choline dhp-containing solution, a reverse trend for the stabilities of DNA duplexes was observed. Tm of the DNA duplexes in the choline dhp-containing solution increased from 33.3 to 53.3°C as the A–T content increased (Figure 3b and Table 2). To confirm the effect of choline dhp, we also measured Tm of DNA duplexes ODN7, ODN8, ODN9 and ODN10 (Table 2). DNA duplexes with the highest A–T contents showed the highest Tms in choline dhp-containing solution. Intriguingly, the ODN7 duplex, composed only of A and T, was more stable in the choline dhp-containing solution than in the NaCl-containing solution, whereas ODN8 with only G–C base pairs was destabilized in the choline dhp-containing solution compared with the NaCl-containing solution. Finally, ODN7 was more stable than ODN8 in the choline dhp-containing solution, with Tm of 37.7 and 33.1°C, respectively.
Table 2. Sequences and melting temperatures for DNA duplex formation in 4 M NaCl and 4 M choline dhp.
| Sequence name | Sequence | Tm (°C)a | |
|---|---|---|---|
| 4 M NaCl | 4 M choline dhp | ||
| ODN1(10)b | 5′-AAAAAAAAAA-3′/5′-TTTTTTTTTT-3′ | 30.7 | 53.3 |
| ODN2(9) | 5′-AAAAAAAAAC-3′/5′-GTTTTTTTTT-3′ | 31.5 | 48.6 |
| ODN3(8) | 5′-AAAAAAAACC-3′/5′-GGTTTTTTTT-3′ | 36.0 | 46.6 |
| ODN4(7) | 5′-AAAAAAACCC-3′/5′-GGGTTTTTTT-3′ | 38.7 | 42.7 |
| ODN5(6) | 5′-AAAAAACCCC-3′/5′-GGGGTTTTTT-3′ | 40.0 | 36.3 |
| ODN6(5) | 5′-AAAAACCCCC-3′/5′-GGGGGTTTTT-3′ | 45.3 | 33.3 |
| ODN7(10) | 5′-AAATATATTT-3′/5′-AAATATATTT-3′ | 17.4 | 37.7 |
| ODN8(0) | 5′-GGGCGCGCCC-3′/5′-GGGCGCGCCC-3′ | 61.0 | 33.1 |
| ODN9(8) | 5′-TAGGTTATAA-3′/5′-TTATAACCTA-3′ | 26.7 | 34.8 |
| ODN10(2) | 5′-CGGCAAGCGC-3′/5′-GCGCTTGCCG-3′ | 47.2 | 30.5 |
aMelting temperature was calculated at a strand concentration of 5 μM.
bThe number of A–T base pairs in the DNA duplex is shown in parentheses.
Figure 3.
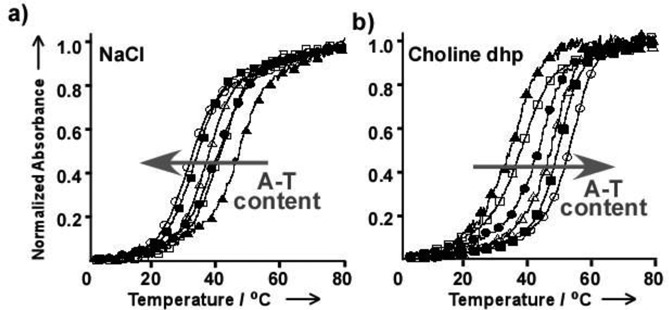
Normalized UV melting curves for ODN1 (open circles), ODN2 (closed squares), ODN3 (open triangles), ODN4 (closed circles), ODN5 (open squares) and ODN6 (closed triangles) in solution containing 50 mM MES (pH 6.0), 1 mM Na2EDTA and (a) 4 M NaCl or (b) 4 M choline dhp. DNA strand concentration was 5 μM. (Permission for use was received (48)).
To investigate how choline dhp alters the stabilities of DNA duplexes, the thermodynamic parameters for ODN9 and ODN10 were measured by conducting UV melting experiments (Table 3), as described previously (48,57–59). ODN9 and ODN10 showed two-state thermal melting transitions, and structures were not changed drastically by solution conditions (0.1, 0.5, 1, 2 and 4 M NaCl or choline dhp; data not shown). The free energy changes at 25°C (ΔG°25) of ODN9 and ODN10 in 4 M NaCl were −8.4 and −12.7 kcal mol−1, respectively. In 4 M choline dhp, however, the stabilities were reversed; ΔG°25 of ODN9 and ODN10 was −10.1 and −8.4 kcal mol−1, respectively. The higher A–T content duplex ODN9 was more stable in choline dhp-containing solution than in NaCl-containing solution because of a favourable enthalpic contribution, whereas ODN10 was destabilized in the choline dhp-containing solution compared with the NaCl-containing solution because of an unfavourable enthalpic contribution. Alkylammonium ion derivatives bind to single-stranded DNA, especially unpaired guanines, at high salt concentrations (42). ODN10 destabilisation in the choline dhp-containing solution was caused by unfavourable enthalpy, implying that choline ions decrease duplex stability either by destabilising hydrogen bond formation or by stabilising single-stranded DNA through preferential binding of choline ions to guanine bases in the single strand. Given that ODN9 stabilisation in the choline dhp-containing solution was enthalpically driven, choline ions increase duplex stability through their interaction with the duplex structure.
Table 3. Thermodynamic parameters for DNA duplex formation in 4 M NaCl or and 4 M choline dhpa.
| ΔH° (kcal mol−1) | TΔS° (kcal mol−1) | ΔG°25 (kcal mol−1) | Tmb (°C) | |
|---|---|---|---|---|
| 4 M NaCl | ||||
| ODN9 | −46.6 ± 2.8 | −38.2 ± 2.0 | −8.4 ± 0.3 | 38.6 |
| ODN10 | −54.7 ± 3.3 | −42.0 ± 2.9 | −12.7 ± 0.4 | 63.2 |
| 4 M choline dhp | ||||
| ODN9 | −66.8 ± 3.4 | −56.7 ± 2.7 | −10.1 ± 0.4 | 43.6 |
| ODN10 | −47.8 ± 3.7 | −39.4 ± 3.4 | −8.4 ± 0.5 | 38.2 |
aAll experiments were performed in buffer containing 50 mM MES (pH 6.0), 1 mM Na2EDTA and 4 M NaCl or 4 M choline dhp. Thermodynamic parameters were evaluated from the average values obtained from curve fitting and Tm−1 versus log(Ct/4) plots. Error estimates were obtained as described previously (59).
bMelting temperature was calculated at a strand concentration of 100 μM.
Choline dhp influences the stability of Watson–Crick and Hoogsteen base pairs in triplexes
The canonical DNA structure is a B-form duplex consisting of A–T and G–C Watson–Crick base pairs (the dash indicates the Watson–Crick base pair), and the G–C base pairs are more stable than the A–T base pairs. Nucleic acids can also form Hoogsteen base pairs. There are two types of triplexes: parallel and antiparallel triplexes. Parallel or triplexes of the ‘pyrimidine’ motif are pyrimidine-rich strands that bind parallel to the purine strand of the duplex, whereas antiparallel triplexes or triplexes of the ‘purine’ motif are purine-rich strands that bind antiparallel to the purine strand of the duplex (60,61). For example, in a triple helix, a third strand consisting of pyrimidine, called a triplex-forming oligonucleotide, binds with sequence specificity to A*T and G*C Hoogsteen base pairs in the major groove of a Watson–Crick base-paired DNA duplex. Pyrimidine motif triplexes are stabilized under acidic conditions because one of the two hydrogen bonds in the G*C+ Hoogsteen base pairs is formed only after protonation of N3 of cytosine (Figure 1a). pKa of N3 at cytosine is 4.5 (11). Thus, around neutral pH, G*C Hoogsteen base pairs are very unstable.
The effect of choline dhp on the stability of Hoogsteen base pairs was evaluated in the context of three oligonucleotides that form intermolecular pyrimidine motif triplexes (Ts1, Ts2 and Ts3) with different numbers of A*T Hoogsteen base pairs (Figure 4). iTs1, which has the same sequence as Ts1 except for the existence of the loop region, was also synthesized; this DNA can form an intramolecular pyrimidine motif triplex (Figure 4). Three intermolecular double-stranded DNAs (Ds1, Ds2 and Ds3) and an intramolecular double-stranded DNA (iDs1) of the same sequences of Watson–Crick base pairs as Ts1, Ts2, Ts3 and iTs1 were also prepared (Figure 4).
Figure 4.

Sequences and schematic structures of (a) triplexes (Ts1, Ts2, Ts3 and iTs1) and (b) duplexes (Ds1, Ds2, Ds3 and iDs1). Filled and open circles indicate Watson–Crick and Hoogsteen base pairs, respectively.
Figure 5 shows normalized UV melting curves at 260 nm for 30 μM pyrimidine motif triplexes. UV melting curves at 295 nm were also measured (data not shown) because the dissociation of Hoogsteen base pairs can be specifically monitored at this wavelength (62). When a melting curve for triplexes has two sigmoidal melting transitions, one corresponds to melting of the duplex and the other to the dissociation of the triplex strand. A transition at 260 nm occurring with the same Tm as that of the melting transition observed at 295 nm corresponds to the dissociation of Hoogsteen base pairs (Tm-H). A transition observed only at 260 nm corresponds to the dissociation of Watson–Crick base pairs (Tm-W). UV melting curves at 260 nm and 295 nm showing single transitions with very similar Tms indicate that the Hoogsteen and Watson–Crick base pairs dissociate at the same temperature (Tm-H&W). The melting temperatures of the duplexes and triplexes analyzed are shown in Table 4. The melting curves of Ts1 in 4 M NaCl at 260 nm and 295 nm showed single sigmoidal melting transitions with almost the same Tms (Figure 5 and Table 4), suggesting that Watson–Crick and Hoogsteen base pairs dissociated at the same time. The melting curve of Ts2 in 4 M NaCl at 260 nm had two sigmoidal melting transitions. The lower temperature transition corresponded to the dissociation of Hoogsteen base pairs because Tm was similar to that of the transition at 295 nm, whereas the higher temperature transition corresponded to the dissociation of Watson–Crick base pairs. Ts3 in 4 M NaCl showed one sigmoidal melting transition at 260 nm but no transitions at 295 nm, indicating that third strand binding via Hoogsteen base pairing did not occur under these conditions. Moreover, Tm of Ds3 in NaCl solution was almost identical to that of Ts3 in 4 M NaCl at 260 nm (Table 4). This finding indicated that Ts3, which has the highest A*T base pair content, does not form a pyrimidine motif triplex. The formation of pyrimidine motif triplexes was also confirmed by CD (data not shown), as reported previously (62).
Figure 5.
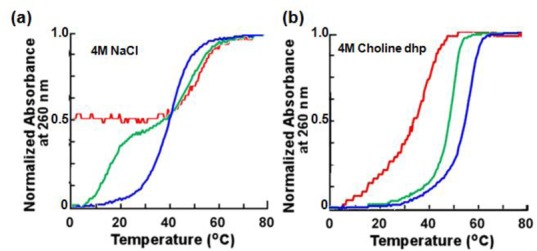
Thermal stability of DNA triplexes. Normalized UV melting curves at 260 nm for Ts1 (blue), Ts2 (green) and Ts3 (red) in (a) 4 M NaCl solution and (b) 4 M choline dhp solution. Solutions also contained 50 mM Tris (pH 7.0) and 1 mM Na2EDTA. Total DNA strand concentration was 30 μM .
Table 4. Melting temperatures for DNA triplexes and duplexes in 4 M NaCl and 4 M choline dhpa.
| Condition/Sequenceb | Tm (°C) at 260 nmb | ||||
|---|---|---|---|---|---|
| Tm-H | Tm-W | Tm-H&W | Tm-W | ||
| 4 M NaCl | |||||
| Ts1 (10)c | 39.4f | Ds1 (2)d | 43.8 | ||
| Ts2 (8) | 14.5 | 48.1 | Ds2 (4) | 47.5 | |
| Ts3 (6) | n.d.e | 51.6 | Ds3 (6) | 51.2 | |
| 4 M Choline dhp | |||||
| Ts1 (10)c | 55.5 | Ds1 (2)d | 51.4 | ||
| Ts2 (8) | 49.3 | Ds2 (4) | 43.2 | ||
| Ts3 (6) | 37.3f | Ds3 (6) | 40.2 | ||
aAll experiments were performed in buffer containing 50 mM Tris (pH 7.0), 1 mM Na2EDTA and 4 M NaCl or 4 M choline dhp.
bTotal DNA strand concentrations of triplexes and duplexes were 30 and 20 μM, respectively.
cThe number of A*T Hoogsteen base pairs is shown in parentheses.
dThe number of A–T Watson–Crick base pairs is shown in parentheses.
en.d. indicates that Tm-H was too low to be determined.
fIn case of Ds1 in 4 M NaCl and Ds3 in 4 M choline dhp where Tm-W is higher than Tm-H&W in Ts1 in 4 M NaCl and Ts3 in 4 M choline dhp, Tm-H&W would be underestimated because Tm-W and Tm-H were not consistent, although the melting curves of Ts1 in 4 M NaCl and Ts3 in 4 M choline dhp showed single sigmoidal transitions (Figure 5).
UV melting curves of Ts1, Ts2 and Ts3 in 4 M choline dhp solution had single sigmoidal melting transitions with almost the same Tms at 260 nm and 295 nm (Table 4). In the hydrated IL, as a consequence, Ts1, Ts2 and Ts3 formed triplexes even at pH 7.0, and the stability of Hoogsteen base pairs was comparable to that of Watson–Crick base pairs. The extent of stability differences in NaCl versus choline dhp depended on the A–T base pair content and likely resulted from specific interactions between DNA bases and choline ions (48). Tm-H&W of Ts1 and Ts2 in choline dhp solution was higher than Tm-W of Ds1 and Ds2 (Table 4). Ts3 formed a pyrimidine motif triplex in choline dhp solution, although Ts3 did not form a triplex in NaCl solution. Choline dhp stabilized the formation of Hoogsteen base pairs independent of the sequence, although the stability of Watson–Crick base-paired duplexes in choline dhp solution depended on the A–T content (48). The stabilisation of Hoogsteen base pairs in choline dhp solution was pronounced compared with that observed in previous studies with triplex-forming oligonucleotides with DNA backbone modifications.
To investigate how high choline dhp concentrations altered the stabilities of DNA structures, the thermodynamic parameters for the intramolecular pyrimidine motif triplex of iTs1 and duplex of iDs1 in 4 M NaCl and choline dhp were measured, estimated by UV melting curves (Table 5). ΔG°25 for the structure formation of iTs1 and iDs1 in 4 M NaCl was −13.1 and −8.8 kcal mol−1, respectively, suggesting that the three-stranded conformation of iTs is more stable than the iDs1 duplex. ΔG°25 for the structure formation of iTs1 and iDs1 in 4 M choline dhp was −17.5 and −8.9 kcal mol−1, respectively. ΔG°25 of iTs1 indicates that the iTs1 triplex was considerably more stable in choline dhp than in NaCl. Given that the stabilisation of the triplex in choline dhp was enthalpically driven, choline ions may increase triplex stability through their interaction with atoms in the triplex. Previous studies on triplexes in the presence of spermine (63), acridine (64), Hoechst 33258 (65), neomycin (64) and carbon nanotubes (66) indicate that these small molecules increase the stability of the pyrimidine motif triplex through enthalpic contributions. For example, triplex stabilisation by neomycin was up to 5 kcal mol−1 (ΔG°25). We thus demonstrated that choline dhp more effectively stabilized the triplex than small molecules evaluated previously (66,67).
Table 5. Thermodynamic parameters for the formation of DNA triplexes and duplexes in 4 M NaCl and 4 M choline dhpa.
| ΔH° (kcal mol−1) | TΔS° (kcal mol−1) | ΔG°25 (kcal mol−1) | Tmb (°C) | |
|---|---|---|---|---|
| 4 M NaCl | ||||
| iTs1 | −80.3 ± 8.7 | −67.3 ± 7.7 | −13.1 ± 2.5 | 78.5 |
| iDs1 | −57.2 ± 1.1 | −48.4 ± 0.9 | −8.8 ± 0.7 | 75.3 |
| 4 M choline dhp | ||||
| iTs1 | −96.3 ± 8.7 | −78.8 ± 7.7 | −17.5 ± 2.5 | 83.9 |
| iDs1 | −57.4 ± 1.0 | −48.5 ± 0.9 | −8.9 ± 0.7 | 77.3 |
aAll experiments were performed in buffer containing 50 mM Tris (pH 7.0), 1 mM Na2EDTA and 4 M NaCl or 4 M choline dhp. Thermodynamic parameters were evaluated from curve fitting.
bMelting temperature was calculated at a strand concentration of 30 μM.
Interaction between choline ions and the duplex or triplex evaluated using molecular dynamic simulations
To determine how choline ions stabilize DNA structures, 20-ns molecular dynamic (MD) simulations of DNA duplexes with A–T base pairs and triplexes were performed. Figure 6 shows the binding sites of choline ions around ODN1 drawn by MD trajectories (68). In general, sodium ions approach the phosphate groups of DNA strands to neutralize their negative charge. The distributions of choline ions around ODN1 were markedly different from those of sodium ions. Choline ions accumulated not only around the phosphate groups of DNA strands but also around bases and ribose sugars via hydrogen bond formation between the hydroxyl groups of choline ions and DNA (63). Figure 6 shows that the choline ions were buried in the minor groove of ODN1. These choline ions fit well into the minor groove of the A–T base pairs. The narrow groove of the A–T base pairs allows multiple hydrogen bonds between choline ions and DNA atoms. Choline ions stabilize the A–T base pairs in a DNA duplex because these ions bind preferentially to these base pairs in the minor groove (68). Shortly thereafter, Portella et al. investigated in more detail and found that choline ions are preferentially localized in the minor groove of the DNA duplex with A–T base pairs by MD simulations and nuclear magnetic resonance studies (69). Furthermore, free energy calculations by MD simulations showed that unpaired G and C bases exhibited more favourable solvation by choline ions than unpaired A and T bases, and the unpaired G and C bases were stabilized slightly compared with the A and T bases (69). The unique interaction of solvents with DNAs in the presence of choline ions could enable stability change of the DNA duplex in a sequence-specific manner.
Figure 6.
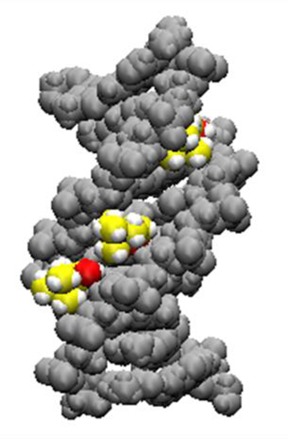
Choline ions buried in the minor groove of ODN1. DNA atoms are shown as grey spheres. Carbon atoms of choline ions are shown in yellow, oxygen atoms of choline ions are shown in red and hydrogen atoms are shown in white (68).
MD simulations of the DNA pyrimidine motif triplex of Ts1 with choline ions were also performed. To maintain the triplex structures during the simulation, N3 of cytosine in the third strand was protonated. For this analysis, the grooves of a pyrimidine motif triplex are defined as the major part of the major groove (ma-major groove), the minor part of the major groove (mi-major groove) and the minor groove (Figure 7) (10). Snapshots of Ts1 after 20-ns MD simulations in the absence and presence of choline ions are shown in Figure 8a and b, respectively. Choline ions are buried inside the minor groove (Figure 8c) and the ma-major groove (Figure 8d) and surround the third strand (Figure 8e). It has been reported that alkylammonium ions such as trimethyl ammonium ions bind to A–T base pairs in the minor groove of a DNA duplex to stabilize the duplex (68,70–71). As observed previously, both the minor and mi-major grooves of a triplex are rigid because of the existence of highly structured water molecules in both grooves (67). In contrast, the ma-major groove is flexible. Binding of small molecules in the ma-major groove probably enhances triplex stability independent of the sequence (66,67). Similarly, choline ions bridge the first and second strands or the first and third strands to strengthen and stabilize pyrimidine motif triplex formation.
Figure 7.

(a) The bases of the triplet of T–A*T. (b) The structure of Ts1 as depicted by the van der Waals model. First (5′-TTTTTTTCTTCT-3′), second (5′-AGAAGAAAAAAA-3′) and third (5′-TCTTCTTTTTTT-3′) strands are indicated by light blue, light green and pink, respectively.
Figure 8.
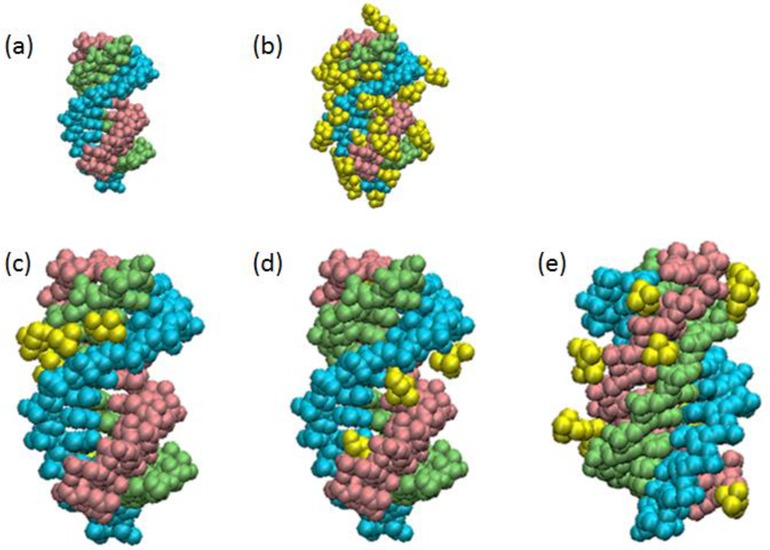
Choline ion binding to the triplex estimated by MD simulations. A snapshot of Ts1 after 20-ns MD simulations in the (a) absence or (b) presence of choline ions. First (5′-TTTTTTTCTTCT-3′), second (5′-AGAAGAAAAAAA-3′) and third (5′-TCTTCTTTTTTT-3′) strands in Ts1 are indicated by light blue, light green and pink, respectively. Ts1 and choline ions (yellow) are depicted as van der Waals models. The choline ions bound to the minor grove and major part of the major groove in Ts1 are highlighted in (c) and (d), respectively. The choline ions surrounding the third strand in Ts1 are highlighted in (e) .
Choline ions in the mi-major groove surround the phosphates of the third strand (Figure 8e). The mi-major groove appears to be too narrow for a choline ion to be buried inside. In general, the cations bind primarily to nucleotide phosphates and stabilize the ordered DNA structures by reducing the repulsive forces between the phosphate groups (43,72). Because the distances between phosphates across the mi-major groove are shorter than those across the other grooves, cations should accumulate in the mi-major groove. Sodium ions exchange rapidly with ions in the bulk solution (72). Because hydroxyl groups in choline ions are strongly polarized, these groups form hydrogen bonds and choline ions do not exchange rapidly (73).
Choline dhp and DES influence G-quadruplex stability
Among various non-canonical nucleic acid structures, G-quadruplex motifs have attracted great research attention as prospective targets for the chemical intervention of biological functions and as building materials in DNA nanotechnology. G-rich DNAs, such as dG3(T2AG3)3 derived from the human telomere sequence (HTS), dissolved in choline dhp can form stable G-quadruplex structures (53), although their stability is decreased relative to that in aqueous solutions. It has been reported that glycine betaine, a zwitterionic osmolyte with alkylammonium ion derivatives, destabilizes guanine-rich DNA structures by binding to guanine in single-stranded DNA at high salt concentrations (42,74). Similarly, choline ion, an alkylammonium ion derivative, could destabilize G-quadruplex structures. Ohno et al. reported on the interaction between hydrated ILs and G-quadruplexes. They found that the kosmotropicity of component ions was also important for the formation and stabilisation of a G-quadruplex in the hydrated ILs of choline dhp (53). Because only a small amount of free water was present, it would be difficult to preserve the G-quadruplex structure in aqueous solution containing high salt concentrations. The effect of hydrated ILs on RNase A stability was also regulated by the kosmotropicity of component ions (75). It was concluded that the water state affects the G-quadruplex structure, and the water state is the function of ion species (53).
In contrast, G-quadruplexes show ultrastability in DES compared with water (76). Qu et al. investigated the thermal stability of G-quadruplex DNAs by plotting UV melting curves. Tm of most of the G-quadruplexes was >90°C in DES in the presence of 100 mM KCl, although some of the G-quadruplexes were unstable in the absence of KCl. There is no direct evidence of the stability of G-quadruplexes at temperatures above 100°C because analyses cannot be performed in aqueous solution at these temperatures. DES has no vapour pressure and has high thermal and chemical stability and accordingly is not volatile and does not boil when heated to high temperature. These features make DES superior to water for reactions at high temperatures. For the Tetrahymena telomeric sequence, G-quadruplex structures are stable at 110°C in DES, even after being kept at this temperature for 30 min. The thermodynamic parameters for G-quadruplex formation were obtained at various DES concentrations. When the DES concentration increased from 0 to 100 wt%, ΔH°, ΔS° and ΔG°25 for G-quadruplex formation decreased from −56.7 to −146.0 kJ mol−1, −168.2 to −414.9 J mol−1 and −7.4 to −24.5 kJ mol−1, respectively. These changes indicate that the formation of the parallel G-quadruplex promoted by DES is enhanced by a favourable enthalpic contribution that exceeds an unfavourable entropic contribution. Undoubtedly, these ultrastable G-quadruplexes can be used for high-temperature biocatalytic reactions, biosensors and DNA architectures. It should also be noted that G-quadruplexes are stable in K+/DES at room temperature for at least 3 months (76).
UNIQUE BEHAVIOUR OF DNA IN ILs
Long-term structural and chemical stability of DNA in ILs
Finding a medium in which DNA is soluble without loss of its structure and in which it is stable for long periods of use at above room temperature has been a bottleneck in DNA nanotechnology. Hydrated ILs, such as choline dhp with 20% dissolved water, have been shown to be good solvents for proteins. To establish the chemical stability of DNA in ILs, MacFarlane et al. measured the CD spectra of the long DNA from salmon testes in hydrated choline dhp (4 M) over time (47). There was only a slight change in the intensity of CD spectra after the 6 months. DNA in aqueous solution would be substantially denatured after 6 months under these conditions (47). This finding suggests that the DNA was stabilized chemically by the hydrated IL medium, as has been observed for proteins (30). Prasad et al. studied DNA from salmon testes in the choline ion-based IL of choline-indole-3-acetate (chol-IAA) (77). DNA was solubilized in the IL at concentrations of up to 3.5% (wt/wt). No structural degradation of the molecule was observed for the sample solubilized in chol-IAA after 6 months of storage. Senapati et al. investigated the interaction between IL cations and a DNA duplex (CGCGAATTCGCG)2 by MD simulations. The results suggest that, in addition to the electrostatic association of cations in hydrated ILs with the DNA backbone, groove binding by IL cations through hydrophobic and polar interactions contributes strongly to DNA stability (78). Very interestingly, the IL ions disrupt the water cage around DNA, including the spine of hydration in the minor groove. This partial dehydration by ILs likely prevents hydrolytic reactions and stabilizes DNA for long periods.
Using ILs for DNA nanotechnology
Ohno et al. developed ion-conductive DNA films using ILs (79). Ion-conductive and flexible DNA films were prepared by both the direct mixing of DNA with salt containing polyethylene oxide and neutralisation of bases on the DNA with acid. The bases on the DNA were neutralized with several acids to convert these bases into IL. Addition of further IL (ethylimidazolium tetrafluoroborate, EtImBF4) allowed the preparation of highly ion conductive films. The highest ionic conductivity of 5.05 × 10−3 S cm−1 was found at 50°C when 93 wt% EtImBF4 was mixed with tetrafluoroboric acid neutralized DNA (79). Moreover, an IL domain was successfully prepared outside double-stranded DNA by fixing 1-alkyl-3-methyl-imidazolium cations on the phosphate groups of DNA (79). These reports will open a new field on the use of DNA as a biomass.
Liu et al. studied DNA-linked gold nanoparticles (AuNPs) in choline dhp and in ILs containing propylammonium nitrate, ethylammonium nitrate, methylammonium nitrate and dimethylammonium nitrate (80). DNA-functionalized AuNPs carry a high density of negative charges and thus may generate new physical properties in ILs. They showed that ILs transit from salts to increase or decrease DNA duplex stability depended on IL concentration. The onset of this transition depends on the structure of ILs: more hydrophobic cations destabilize DNA at lower IL concentrations. This trend is opposite to that observed with molecular solvents (e.g. ethanol, dimethoxysulfoxide (DMSO), dimethylformamide and acetonitrile) that destabilize DNA at low solvent concentrations (81). Specific DNA base pairing is disrupted at high DMSO concentrations, and AuNPs are held together by nonspecific interactions. In the other tested molecular solvents, DNA base pairs are maintained, although strong nonspecific interactions are also present. Several ILs can release protons and thus drastically change pH, also changing the melting temperature of DNA. Moreover, a method to efficiently separate DNA-staining dyes from DNA without disturbing DNA properties was reported using hydrophobic ILs (82). The study by Liu et al. also reveals the feasibility of using ILs as solvents for DNA-functionalized nanomaterials.
Studies of behaviour of DNA in ILs reveal its behaviour in cells
From a biological viewpoint, DNA behaviour in ILs may also be relevant to the cellular environment in which DNA is found. Osmolytes including choline ions and glycine betaine, an alkylammonium ion derivative, are in abundant supply in cells (83–85), and phosphatidyl choline is present in the nuclear membrane. It is likely that certain regions of DNA inside the nucleus form triplexes to regulate biological reactions such as transcription. Stability changes in nucleic acids also appeared in choline chloride solution, which is not a hydrated IL (48). If there is an interaction between osmolytes and DNA or nuclear membranes and DNA, stability changes in DNA will be induced in cells.
Hud et al. have shown that DNA with a nucleotide sequence derived from HTS adopts the parallel G-quadruplex fold in an anhydrous ChCl−urea DES (86). Previous studies had shown that cosolvents in aqueous solutions (e.g. acetonitrile, ethanol and polyethylene glycol) can shift the equilibrium structure of HTS DNA from the mixed parallel, antiparallel G-quadruplex folds that are favoured in aqueous solutions to the parallel fold that was first observed in the crystal state (81,87–88). Folding of HTS in the anhydrous ChCl−urea DES and mixed DES−water solvents has revealed diffusion-limited kinetics consistent with the Kramers rate theory (89). The kinetics of G-quadruplex properties is a function of viscosity (Figure 9). Moreover, future investigations of G-quadruplexes should take into account solvent friction effects, as in vivo folding is likely to be affected by solvent viscosity, given that the viscosity of the cellular milieu is as high as 140 cP (90). Thus, analysis of DNA behaviour in ILs from the physicochemical standpoint is very useful for understanding DNA behaviour in the cell.
Figure 9.
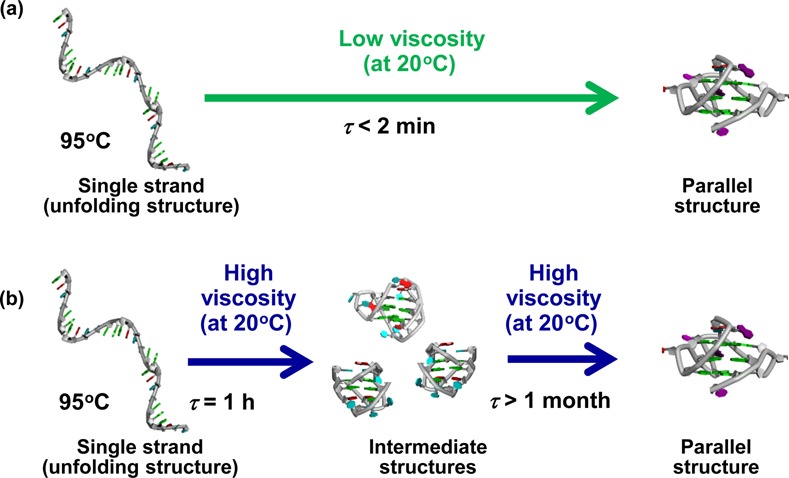
G-quadruplex folding depends on a solution with (a) low viscosity and (b) high viscosity (85).
CONCLUSIONS, CHALLENGES AND PERSPECTIVES
By harnessing interactions between ILs and DNA, we can control the stabilities and structures of nucleic acids. ILs have several properties that will enhance the functions of DNA nanodevices. Natural DNAs are not chemically stable in solution at ambient temperatures for long periods. MacFarlane et al. reported that DNA has long-term stability in choline dhp in the absence of nuclease (47). Nuclease degradation of DNA may be inhibited in 4 M choline dhp solution because a solution containing high salt concentration is not suitable for protein folding. Thus, choline dhp could be useful as a chemical DNA stabilizer and a nuclease inhibitor. Another important property of ILs is their low vapour pressure, making these liquids better solvents than water for low-volume devices. Thus, DNA devices should be generally reusable for multiple cycles in ILs.
Systems for sensing specific DNA sequences are important in the fields of medicine and nanobiosensing (91–94). Conventional methods for sensing a DNA sequence, including DNA microarrays, Southern blots and in situ hybridisation, are based on the formation of Watson–Crick base pairs and require the generation of single-stranded DNA prior to analysis. To simplify target detection, several approaches have been developed in which double-stranded targets are detected directly (15,95). As these systems use intercalating dyes and groove-binding ligands, they lack sequence specificity and are prone to false-positive detection (96,97). The triplex is a promising recognition motif for sequence-specific sensing of double-stranded DNA targets (15,95). Based on our finding of a significant stabilisation of Hoogsteen base pairs with choline dhp, sequence-specific sensing of double-stranded DNA will be possible without the need for denaturation of the duplex or complicated instrumentation. Thus, information in this review should facilitate the development of new DNA materials.
Acknowledgments
We thank Prof. Shigenari Tanaka and Dr Miki Nakano for useful comments on molecular dynamic simulation.
FUNDING
Scientific Research and MEXT (Ministry of Education, Culture, Sports, Science and Technology)-Supported Program for the Strategic Research Foundation at Private Universities, Japan, and the Hirao Taro Foundation of the Konan University Association for Academic Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Heller M.J. DNA microarray technology: devices, systems, and applications. Annu. Rev. Biomed. Eng. 2002;4:129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 2.Ott J., Hoh J. Set association analysis of SNP case-control and microarray data. J. Comput. Biol. 2003;10:569–574. doi: 10.1089/10665270360688192. [DOI] [PubMed] [Google Scholar]
- 3.Fedrigo O., Naylor G. A gene-specific DNA sequencing chip for exploring molecular evolutionary change. Nucleic Acids Res. 2004;32:1208–1213. doi: 10.1093/nar/gkh210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morishima C., Chung M., Ng K.W., Brambilla D.J., Gretch D.R. Strengths and limitations of commercial tests for hepatitis C virus RNA quantification. J. Clin. Microbiol. 2004;42:421–425. doi: 10.1128/JCM.42.1.421-425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeSantis T.Z., Dubosarskiy I., Murray S.R., Andersen G.L. Comprehensive aligned sequence construction for automated design of effective probes (CASCADE-P) using 16S rDNA. Bioinformatics. 2003;19:1461–1468. doi: 10.1093/bioinformatics/btg200. [DOI] [PubMed] [Google Scholar]
- 6.Chen J.H., Seeman N.C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 7.Phadke R.S. Biomolecular electronics in the twenty-first century. Appl. Biochem. Biotechnol. 2001;96:269–276. doi: 10.1385/abab:96:1-3:279. [DOI] [PubMed] [Google Scholar]
- 8.Yurke B., Turberfield A.J., Mills A.P., Jr, Simmel F.C., Neumann J.L. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 9.Beal P.A., Dervan P.B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 10.Chen H., Meena, McLaughlin L.W. A Janus-Wedge DNA triplex with A-W1-T and G-W2-C base triplets. J. Am. Chem. Soc. 2008;130:13190–131901. doi: 10.1021/ja804607v. [DOI] [PubMed] [Google Scholar]
- 11.Mergny J.L., Sun J.S., Rougee M., Montenay-Garestier T., Barcelo F., Chomilier J., Helene C. Sequence specificity in triple-helix formation: experimental and theoretical studies of the effect of mismatches on triplex stability. Biochemistry. 1991;30:9791–9798. doi: 10.1021/bi00104a031. [DOI] [PubMed] [Google Scholar]
- 12.Sugimoto N., Wu P., Hara H., Kawamoto Y. pH and cation effects on the properties of parallel pyrimidine motif DNA triplexes. Biochemistry. 2001;40:9396–9405. doi: 10.1021/bi010666l. [DOI] [PubMed] [Google Scholar]
- 13.Plum G.E., Park Y.W., Singleton S.F., Dervan P.B., Breslauer K.J. Thermodynamic characterization of the stability and the melting behavior of a DNA triplex: a spectroscopic and calorimetric study. Proc. Natl Acad. Sci. U.S.A. 1990;87:9436–9440. doi: 10.1073/pnas.87.23.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu P., Hara H., Kawamoto Y., Sugimoto N. Effect of cytosine protonation and cation on thermodynamic properties of parallel DNA triplex family. Nucleic Acids Res. Suppl. 2001:39–40. doi: 10.1093/nass/1.1.39. [DOI] [PubMed] [Google Scholar]
- 15.Bond J.P., Anderson C.F., Record M.T., Jr. Conformational transitions ofduplex and triplex nucleic acid helices: thermodynamic analysis of effects ofsalt concentration on stability using preferential interaction coefficients. Biophys J. 1994;67:825–836. doi: 10.1016/S0006-3495(94)80542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Y., Wang X., Xiao Y., Feng L., Zhao C., Ren J., Qu X. i-Motif quadruplex DNA-based biosensor for distinguishing single- and multiwalled carbon nanotubes. J. Am. Chem. Soc. 2009;131:13813–13818. doi: 10.1021/ja9051763. [DOI] [PubMed] [Google Scholar]
- 17.Peng Y., Li X., Ren J., Qu X. Single-walled carbon nanotubes binding to human telomeric i-motif DNA: significant acceleration of S1 nuclease cleavage rate. Chem. Commun. 2007:5176–5178. doi: 10.1039/b710950d. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y., Qu K., Zhao C., Wu L., Ren J., Wang J., Qu X. Nat Commun. 2012;3:1074. doi: 10.1038/ncomms2091. [DOI] [PubMed] [Google Scholar]
- 19.Miyoshi D., Inoue M., Sugimoto N. DNA logic gates based on structural polymorphism of telomere DNA molecules responding to chemical input signals. Angew. Chem. Int. Ed. Engl. 2006;45:7716–7719. doi: 10.1002/anie.200602404. [DOI] [PubMed] [Google Scholar]
- 20.Legoff J., Tanton C., Lecerf M., Gresenguet G., Nzambi K., Bouhlal H., Weiss H., Belec L. Influence of storage temperature on the stability of HIV-1 RNA and HSV-2 DNA in cervicovaginal secretions collected by vaginal washing. J. Virol. Methods. 2006;138:196–200. doi: 10.1016/j.jviromet.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki Y., Miyoshi D., Sugimoto N. Regulation of DNA nucleases by molecular crowding. Nucleic Acids Res. 2007;35:4086–4093. doi: 10.1093/nar/gkm445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yilmaz L.S., Bergsven L.I., Noguera D.R. Systematic evaluation of single mismatch stability predictors for fluorescence in situ hybridization. Environ. Microbiol. 2008;10:2872–2885. doi: 10.1111/j.1462-2920.2008.01719.x. [DOI] [PubMed] [Google Scholar]
- 24.Armand M., Endres F., MacFarlane D.R., Ohno H., Scrosati B. Ionic-liquid materials for the electrochemical challenges of the future. Nat. Mater. 2009;8:621–629. doi: 10.1038/nmat2448. [DOI] [PubMed] [Google Scholar]
- 25.Welton T. Room-temperature ionic liquids. Solvents for synthesis and catalysis. Chem. Rev. 1999;99:2071–2084. doi: 10.1021/cr980032t. [DOI] [PubMed] [Google Scholar]
- 26.Trincao J., Johnson R.E., Wolfle W.T., Escalante C.R., Prakash S., Prakash L., Aggarwal A.K. Dpo4 is hindered in extending a G.T mismatch by a reverse wobble. Nat. Struct. Mol. Biol. 2004;11:457–462. doi: 10.1038/nsmb755. [DOI] [PubMed] [Google Scholar]
- 27.Fujita K., MacFarlane D.R., Forsyth M., Yoshizawa-Fujita M., Murata K., Nakamura N., Ohno H. Solubility and stability of cytochrome c in hydrated ionic liquids: effect of oxo acid residues and kosmotropicity. Biomacromolecules. 2007;8:2080–2086. doi: 10.1021/bm070041o. [DOI] [PubMed] [Google Scholar]
- 28.Fujita K., Ohno H. Enzymatic activity and thermal stability of metallo proteins in hydrated ionic liquids. Biopolymers. 2010;93:1093–1099. doi: 10.1002/bip.21526. [DOI] [PubMed] [Google Scholar]
- 29.Earle M.J., Seddon K.R. Ionic liquids. Green solvents for the future. Pure Appl. Chem. 2000;72:1391–1398. [Google Scholar]
- 30.Fujita K., MacFarlane D.R., Forsyth M. Protein solubilising and stabilising ionic liquids. Chem. Commun. (Camb.) 2005:4804–4806. doi: 10.1039/b508238b. [DOI] [PubMed] [Google Scholar]
- 31.Abbott A.P., Capper G., Davies D.L., Rasheed R.K., Tambyrajah V. Novel solvent properties of choline chloride/urea mixtures. Chem. Commun. (Camb.) 2003:70–71. doi: 10.1039/b210714g. [DOI] [PubMed] [Google Scholar]
- 32.Esquembre R., Sanz J.M., Wall J.G., del Monte F., Mateo C.R., Ferrer M.L. Thermal unfolding and refolding of lysozyme in deep eutectic solvents and their aqueous dilutions. Phys. Chem. Chem. Phys. 2013;15:11248–11256. doi: 10.1039/c3cp44299c. [DOI] [PubMed] [Google Scholar]
- 33.Morrison H.G., Sun C.C., Neervannan S. Characterization of thermal behavior of deep eutectic solvents and their potential as drug solubilization vehicles. Int. J. Pharm. 2009;378:136–139. doi: 10.1016/j.ijpharm.2009.05.039. [DOI] [PubMed] [Google Scholar]
- 34.Gutierrez M.C., Ferrer M.L., Yuste L., Rojo F., del Monte F. Bacteria incorporation in deep-eutectic solvents through freeze-drying. Angew. Chem. Int. Ed. Engl. 2010;49:2158–2162. doi: 10.1002/anie.200905212. [DOI] [PubMed] [Google Scholar]
- 35.Breslauer K.J., Frank R., Blocker H., Marky L.A. Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. U.S.A. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auffinger P., Westhof E. Melting of the solvent structure around a RNA duplex: a molecular dynamics simulation study. Biophys. Chem. 2002;95:203–210. doi: 10.1016/s0301-4622(01)00257-5. [DOI] [PubMed] [Google Scholar]
- 37.Auffinger P., Westhof E. Hydrophobic groups stabilize the hydration shell of 2’-O-methylated RNA duplexes. Angew. Chem. Int. Ed. Engl. 2001;40:4648–4650. doi: 10.1002/1521-3773(20011217)40:24<4648::aid-anie4648>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 38.Anderson C.F., Record M.T., Jr Salt-nucleic acid interactions. Annu. Rev. Phys. Chem. 1995;46:657–700. doi: 10.1146/annurev.pc.46.100195.003301. [DOI] [PubMed] [Google Scholar]
- 39.Feig M., Pettitt B.M. Sodium and chlorine ions as part of the DNA solvation shell. Biophys. J. 1999;77:1769–1781. doi: 10.1016/S0006-3495(99)77023-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feig M., Pettitt B.M. A molecular simulation picture of DNA hydration around A- and B-DNA. Biopolymers. 1998;48:199–209. doi: 10.1002/(SICI)1097-0282(1998)48:4<199::AID-BIP2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 41.Spink C.H., Chaires J.B. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry. 1999;38:496–508. doi: 10.1021/bi9820154. [DOI] [PubMed] [Google Scholar]
- 42.Nordstrom L.J., Clark C.A., Andersen B., Champlin S.M., Schwinefus J.J. Effect of ethylene glycol, urea, and N-methylated glycines on DNA thermal stability: the role of DNA base pair composition and hydration. Biochemistry. 2006;45:9604–9614. doi: 10.1021/bi052469i. [DOI] [PubMed] [Google Scholar]
- 43.Record M.T., Jr, Anderson C.F., Lohman T.M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 44.Rozners E., Moulder J. Hydration of short DNA, RNA and 2’-OMe oligonucleotides determined by osmotic stressing. Nucleic Acids Res. 2004;32:248–254. doi: 10.1093/nar/gkh175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong J., Capp M.W., Anderson C.F., Saecker R.M., Felitsky D.J., Anderson M.W., Record M.T., Jr Preferential interactions of glycine betaine and of urea with DNA: implications for DNA hydration and for effects of these solutes on DNA stability. Biochemistry. 2004;43:14744–14758. doi: 10.1021/bi049096q. [DOI] [PubMed] [Google Scholar]
- 46.Leng M., Felsenfeld G. The preferential interactions of polylysine and polyarginine with specific base sequences in DNA. Proc. Natl Acad. Sci. U.S.A. 1966;56:1325–1332. doi: 10.1073/pnas.56.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vijayaraghavan R., Izgorodin A., Ganesh V., Surianarayanan M., MacFarlane D.R. Long-term structural and chemical stability of DNA in hydrated ionic liquids. Angew. Chem. Int. Ed. Engl. 2010;49:1631–1633. doi: 10.1002/anie.200906610. [DOI] [PubMed] [Google Scholar]
- 48.Tateishi-Karimata H., Sugimoto N. A-T Base Pairs are More Stable Than G-C Base Pairs in a Hydrated Ionic Liquid. Angew. Chem. Int. Ed. Engl. 2012;51:1416–1419. doi: 10.1002/anie.201106423. [DOI] [PubMed] [Google Scholar]
- 49.Mamajanov I., Engelhart A.E., Bean H.D., Hud N.V. DNA and RNA in anhydrous media: duplex, triplex, and G-quadruplex secondary structures in a deep eutectic solvent. Angew. Chem. Int. Ed. Engl. 2010;49:6310–3314. doi: 10.1002/anie.201001561. [DOI] [PubMed] [Google Scholar]
- 50.Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim K.W., Ng V.C., Martin-Pintado N., Heddi B., Phan A.T. Structure of the human telomere in Na+ solution: an antiparallel (2+2) G-quadruplex scaffold reveals additional diversity. Nucleic Acids Res. 2013;41:10556–10562. doi: 10.1093/nar/gkt771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao C., Ren J., Qu X. G-quadruplexes form ultrastable parallel structures in deep eutectic solvent. Langmuir. 2013;29:1183–1191. doi: 10.1021/la3043186. [DOI] [PubMed] [Google Scholar]
- 53.Fujita K., Ohno H. Stable G-quadruplex structure in a hydrated ion pair: cholinium cation and dihydrogen phosphate anion. Chem. Commun. (Camb.) 2012;48:5751–5753. doi: 10.1039/c2cc30554b. [DOI] [PubMed] [Google Scholar]
- 54.Miyoshi D., Karimata H., Sugimoto N. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006;128:7957–7963. doi: 10.1021/ja061267m. [DOI] [PubMed] [Google Scholar]
- 55.Petraccone L., Spink C., Trent J.O., Garbett N.C., Mekmaysy C.S., Giancola C., Chaires J.B. Structure and stability of higher-order human telomeric quadruplexes. J. Am. Chem. Soc. 2011;133:20951–20961. doi: 10.1021/ja209192a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugimoto N., Nakano S., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 57.Schroeder S.J., Turner D.H. Optical melting measurements of nucleic acid thermodynamics. Methods Enzymol. 2009;468:371–387. doi: 10.1016/S0076-6879(09)68017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tateishi-Karimta H., Sugimoto N. Control of stability and structure of nucleic acids using cosolutes. Methods. 2014;67 doi: 10.1016/j.ymeth.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Bevington P. R. Data Reduction andError Analysis for the Physical Sciences. New York: McGraw-Hill; 1969. [Google Scholar]
- 60.Keppler M.D., Fox K.R. Relative stability of triplexes containing different numbers of T.AT and C+.GC triplets. Nucleic Acids Res. 1997;25:4644–4649. doi: 10.1093/nar/25.22.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gowers D.M., Fox K.R. DNA triple helix formation at oligopurine sites containing multiple contiguous pyrimidines. Nucleic Acids Res. 1997;25:3787–3794. doi: 10.1093/nar/25.19.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miyoshi D., Nakamura K., Tateishi-Karimata H., Ohmichi T., Sugimoto N. Hydration of Watson-Crick base pairs and dehydration of Hoogsteen base pairs inducing structural polymorphism under molecular crowding conditions. J. Am. Chem. Soc. 2009;131:3522–3531. doi: 10.1021/ja805972a. [DOI] [PubMed] [Google Scholar]
- 63.Antony T., Thomas T, Shirahata A., Sigal L.H., Thomas T.J. Selectivity of spermine homologson triplex DNA stabilization. Antisense Nucleic Acid Drug Dev. 1990;9:221–231. doi: 10.1089/oli.1.1999.9.221. [DOI] [PubMed] [Google Scholar]
- 64.Arya D.P. New approaches toward recognition of nucleic acid triple helices. Acc. Chem. Res. 2011;44:134–146. doi: 10.1021/ar100113q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willis B., Arya D.P. Triple recognition of B-DNA by a neomycin-Hoechst 33258-pyrene conjugate. Biochemistry. 2010;49:452–469. doi: 10.1021/bi9016796. [DOI] [PubMed] [Google Scholar]
- 66.Zhao C., Qu K., Xu C., Ren J., Qu X. Triplex inducer-directed self-assembly of single-walled carbon nanotubes: a triplex DNA-based approach for controlled manipulation of nanostructures. Nucleic Acids Res. 2011;39:3939–3948. doi: 10.1093/nar/gkq1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas T., Thomas T.J. Selectivity of polyamines in triplex DNA stabilization. Biochemistry. 1993;32:14068–14074. doi: 10.1021/bi00213a041. [DOI] [PubMed] [Google Scholar]
- 68.Nakano M., Tateishi-Karimata H., Tanaka S., Sugimoto N. Choline ion interactions with DNA atoms explain unique stabilization of A-T base pairs in DNA duplexes: a microscopic view. J. Phys. Chem. B. 2014;118:379–389. doi: 10.1021/jp406647b. [DOI] [PubMed] [Google Scholar]
- 69.Portella G., Germann M.W., Hud N.V., Orozco M. MD and NMR analyses of choline and TMA binding to duplex DNA: on the origins of aberrant sequence-dependent stability by alkyl cations in aqueous and water-free solvents. J. Am. Chem. Soc. 2014;136:3075–3086. doi: 10.1021/ja410698u. [DOI] [PubMed] [Google Scholar]
- 70.Melchior W.B., Jr, Von Hippel P.H. Alteration of the relative stability of dA-dT and dG-dC base pairs in DNA. Proc. Natl Acad. Sci. U.S.A. 1973;70:298–302. doi: 10.1073/pnas.70.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stellwagen E., Dong Q., Stellwagen N.C. Quantitative analysis of monovalent counterion binding to random-sequence, double-stranded DNA using the replacement ion method. Biochemistry. 2007;46:2050–2058. doi: 10.1021/bi062132w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bloomfield V.A. DNA condensation. Curr. Opin. Struct. Biol. 1996;6:334–341. doi: 10.1016/s0959-440x(96)80052-2. [DOI] [PubMed] [Google Scholar]
- 73.Cardoso L., Micaelo N.M. DNA molecular solvation in neat ionic liquids. Chemphyschem. 2011;12:275–277. doi: 10.1002/cphc.201000645. [DOI] [PubMed] [Google Scholar]
- 74.Felitsky D.J., Cannon J.G., Capp M.W., Hong J., Van Wynsberghe A.W., Anderson C.F., Record M.T., Jr The exclusion of glycine betaine from anionic biopolymer surface: why glycine betaine is an effective osmoprotectant but also a compatible solute. Biochemistry. 2004;43:14732–14743. doi: 10.1021/bi049115w. [DOI] [PubMed] [Google Scholar]
- 75.Weingartner H., Cabrele C., Herrmann C. How ionic liquids can help to stabilize native proteins. Phys. Chem. Chem. Phys. 2012;14:415–426. doi: 10.1039/c1cp21947b. [DOI] [PubMed] [Google Scholar]
- 76.Zhao C., Qu X. Recent progress in G-quadruplex DNA in deep eutectic solvent. Methods. 2013;64:52–58. doi: 10.1016/j.ymeth.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 77.Mukesh C., Mondal D., Sharma M., Prasad K. Rapid dissolution of DNA in a novel bio-based ionic liquid with long-term structural and chemical stability: successful recycling of the ionic liquid for reuse in the process. Chem. Commun. (Camb.) 2013;49:6849–6851. doi: 10.1039/c3cc42829j. [DOI] [PubMed] [Google Scholar]
- 78.Chandran A., Ghoshdastidar D., Senapati S. Groove binding mechanism of ionic liquids: a key factor in long-term stability of DNA in hydrated ionic liquids? J. Am. Chem. Soc. 2012;134:20330–20339. doi: 10.1021/ja304519d. [DOI] [PubMed] [Google Scholar]
- 79.Nishimura N., Ohno H. Design of successive ion conduction paths in DNA films with ionic liquids. J. Mater. Chem. 2005;12:2299–2304. [Google Scholar]
- 80.Menhaj Arsalan Beg, Smith Brendan D., Liu Juewen. Exploring the thermal stability of DNA-linked gold nanoparticles in ionic liquids and molecular solvents. Chem. Sci. 2012;3:3216–3220. [Google Scholar]
- 81.Marchand A., Ferreira R., Tateishi-Karimata H., Miyoshi D., Sugimoto N., Gabelica V. Sequence and solvent effects on telomeric DNA bimolecular G-quadruplex folding kinetics. J. Phys. Chem. B. 2013;117:12391–12401. doi: 10.1021/jp406857s. [DOI] [PubMed] [Google Scholar]
- 82.Khimji I., Doan K., Bruggeman K., Huang P.J., Vajha P., Liu J. Extraction of DNA staining dyes from DNA using hydrophobic ionic liquids. Chem. Commun. (Camb.) 2013;49:4537–4539. doi: 10.1039/c3cc41364k. [DOI] [PubMed] [Google Scholar]
- 83.Courtenay E.S., Capp M.W., Anderson C.F., Record M.T., Jr Vapor pressure osmometry studies of osmolyte-protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of “osmotic stress” experiments in vitro. Biochemistry. 2000;39:4455–4471. doi: 10.1021/bi992887l. [DOI] [PubMed] [Google Scholar]
- 84.Zeisel S.H., da Costa K.A. Choline: an essential nutrient for public health. Nutr. Rev. 2009;67:615–623. doi: 10.1111/j.1753-4887.2009.00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ueland P.M. Choline and betaine in health and disease. J. Inherit. Metab. Dis. 2011;34:3–15. doi: 10.1007/s10545-010-9088-4. [DOI] [PubMed] [Google Scholar]
- 86.Lannan F.M., Mamajanov I., Hud N.V. Human telomere sequence DNA in water-free and high-viscosity solvents: G-quadruplex folding governed by Kramers rate theory. J. Am. Chem. Soc. 2012;134:15324–15330. doi: 10.1021/ja303499m. [DOI] [PubMed] [Google Scholar]
- 87.Heddi B., Phan A.T. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 88.Miller M.C., Buscaglia R., Chaires J.B., Lane A.N., Trent J.O. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3′ sequence of d(AG3(TTAG3)3) J. Am. Chem. Soc. 2010;132:17105–17107. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- 89.Kramers H.A. Brownian motion in a field of force and the diffusion model of chemical reactions. Physica. 1940;7:284–304. [Google Scholar]
- 90.Kuimova M.K., Yahioglu G., Levitt J.A., Suhling K. Molecular rotor measures viscosity of live cells via fluorescence lifetime imaging. J. Am. Chem. Soc. 2008;130:6672–6673. doi: 10.1021/ja800570d. [DOI] [PubMed] [Google Scholar]
- 91.Drummond T.G., Hill M.G., Barton J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 92.Lee J.B., Campolongo M.J., Kahn J.S., Roh Y.H., Hartman M.R., Luo D. DNA-based nanostructures for molecular sensing. Nanoscale. 2010;2:188–197. doi: 10.1039/b9nr00142e. [DOI] [PubMed] [Google Scholar]
- 93.Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 94.Lockhart D.J., Dong H., Byrne M.C., Follettie M.T., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 95.Kuhn H., Demidov V.V., Coull J.M., Fiandaca M.J., Gildea B.D., Frank-Kamenetskii M.D. Hybridization of DNA and PNA molecular beacons to single-stranded and double-stranded DNA targets. J. Am. Chem. Soc. 2002;124:1097–1103. doi: 10.1021/ja0041324. [DOI] [PubMed] [Google Scholar]
- 96.Ihmels H., Meiswinkel A., Mohrschladt C.J., Otto D., Waidelich M., Towler M., White R., Albrecht M., Schnurpfeil A. Anthryl-substituted heterocycles as acid-sensitive fluorescence probes. J. Org. Chem. 2005;70:3929–3938. doi: 10.1021/jo047841z. [DOI] [PubMed] [Google Scholar]
- 97.Persil O., Hud N.V. Harnessing DNA intercalation. Trends Biotechnol. 2007;25:433–436. doi: 10.1016/j.tibtech.2007.08.003. [DOI] [PubMed] [Google Scholar]


