Abstract
Genetic recombination is important for generating diversity and to ensure faithful segregation of chromosomes at meiosis. However, few crossovers (COs) are formed per meiosis despite an excess of DNA double-strand break precursors. This reflects the existence of active mechanisms that limit CO formation. We previously showed that AtFANCM is a meiotic anti-CO factor. The same genetic screen now identified AtMHF2 as another player of the same anti-CO pathway. FANCM and MHF2 are both Fanconi Anemia (FA) associated proteins, prompting us to test the other FA genes conserved in Arabidopsis for a role in CO control at meiosis. This revealed that among the FA proteins tested, only FANCM and its two DNA-binding co-factors MHF1 and MHF2 limit CO formation at meiosis.
INTRODUCTION
One prominent feature of eukaryotic sexual reproduction is meiosis, a specific type of cell division where two rounds of chromosome segregation follow a single round of DNA replication. This produces haploid spores from a diploid mother cell. At the first division, correct chromosome segregation relies on physical connections between homologues which are provided by crossovers (COs). COs are reciprocal exchanges of genetic material between homologues. These events are initiated by the formation of DNA double-strand breaks (DSBs) which will be repaired by homologous recombination as COs or non-crossovers (NCOs). At least two pathways to CO formation exist with different genetic requirements. Species exist with only one of these pathways; however Arabidopsis, humans and budding yeast, for example, have both (1). The first pathway, which is prominent in most species, is dependent on a group of proteins collectively referred to as ZMMs (for Zip1, Zip2, Zip3 and Zip4, Mer3 and Msh4–Msh5) and on the Mlh1–Mlh3 heterodimer, first identified in Saccharomyces cerevisiae and conserved in a large range of eukaryotes (2,3). The COs that arise from this pathway are sensitive to a phenomenon known as CO interference where one CO reduces the probability of another CO occurring at adjacent loci (4). The second pathway of CO formation involves the endonuclease MUS81 and produces COs that are not sensitive to interference (1). Interestingly, CO number is relatively low in most eukaryotes, being very close to the one, obligatory, CO per chromosome pair, despite a large excess of recombination precursors (5). This suggests that active mechanisms limit CO frequency, whose molecular factors remain largely unknown. The helicase Fanconi Anemia Complementation Group M (FANCM) has been found to be a major meiotic anti-CO factor in Arabidopsis, limiting MUS81-dependent CO formation, a normally minor pathway of CO formation in Arabidopsis thaliana (6). This function seems to be evolutionarily conserved as Fml1, the fission yeast FANCM ortholog, also directs NCO formation (7).
Fanconi Anemia (FA) is a rare heritable human disease that is characterized by early onset of bone marrow failure and susceptibility to certain cancers. The FA pathway, which implicates at least 16 proteins in human cells, appears to be present in all eukaryotes and promotes genome stability by resolving blocked replication forks (8,9). The FA genes have been initially identified as preventing FA in humans. The FA proteins can be categorized into three groups, according to their biochemical function. (i) The core complex is the first recruited to DNA stalled replication forks, using FANCM as a landing pad. Two newly discovered co-factors of FANCM, namely MHF1 and MHF2, have been shown to stimulate FANCM DNA-binding activity and its targeting to chromatin (10,11). (ii) The FA-ID complex is recruited and ubiquitinated by the core complex at the damage site. (iii) The downstream partners are thought to act independently of the first two groups but have strong links with the homologous recombination machinery, and mutation of any leads to development of the disease in human (8). Implication of FANCM in the control of meiotic CO formation raises the question whether other FA proteins limit meiotic COs, or if the FANCM meiotic function is unique among FA proteins. Here, using both forward and reverse genetic screens, we show that from a series of FA proteins conserved in Arabidopsis, only AtMHF1 and AtMHF2 were identified as CO-limiting factors. We propose that FANCM and its direct DNA-binding cofactors MHF1 and MHF2 prevent meiotic CO formation, without the other FA proteins being involved.
MATERIALS AND METHODS
FA protein identification
Homologues and putative homologues of FA-associated genes were identified using literature searches and reciprocal BLASTp and PSI-BLAST (http://www.ncbi.nlm.nih.gov/, http://www.arabidopsis.org/ and http://bioinformatics.psb.ugent.be/plaza).
Genetic material
The lines used in this study were Atmhf1–3 (N576310), Atfanci (N555483), Atfancd2 (N613293), Atfance (N553587), Atfancl (37079— identified from the Max-Planck Institute für Züchtungsforschung collection from Köln, Germany (12)), zip4-1 (EJD21) (13), zip4-2 (N568052) (13), shoc1-1 (N557589) (14), msh5-2 (N526553) (15), mus81-2 (N607515) (16), spo11-1-3 (N646172) (17), fancm-1 (6), hei10-2 (N514624) (18), fluorescent-tagged lines (FTLs) I2ab (FTL1506/FTL1524/FTL965/qrt1-2) (19). Genotyping by polymerase chain reaction was performed with two primer pairs. The first pair is specific to the wild-type allele, and the second pair is specific to the left border of the inserted sequence as follows: Atmhf1-3 (N576310U 5′-CCTAAACC-ATCCTCCAGCTTC-3′ and N576310L 5′-CAATTTAAAGACGCAGGATCG-3′, N576310L and LBSalk2 5′-GCTTTCTTCCCTTCCTTTCTC-3′); Atfanci (N555483U 5′-AGTCCAACACATGTCCTCCAC-3′ and N555483L 5′-TGAGTTTGGTGATTCGAAAGG-3′, N555483L and LBSalk2); Atfancd2 (N613293U 5′-AATTCACCGGAATGTCACAAC-3′ and N613293L 5′-AATTCACCGGAATGTCACAAC-3′, and N613293L and LBSalk2); Atfance (N553587U 5′- TCAGCTGATGAAGACAGCATG-3′ and N553587L 5′-ATGTCAACCCACAGAGGATTG-3′, and N553587L and LBSalk2); Atfancl (FANCL-U 5′-ACAGAGATAAGAAGGGAAGAG-3′ and FANCL-L ATTATCATTAACCCGTCATTC, and FANCL-L and LB Gabi o8409 5′-ATATTGACCATCATACTCATTGC-3′). mhf2 alleles were genotyped by dCAPS as follows: mhf2-1 locus amplification with 5′-ATCTGCGAGCTTTTTTATTCGATTGCGATGAA-3′ and 5′-AGGAGTTACGATACCAAATGA-3′, subsequent digestion by MboII (104+33 bp for the wild-type amplicon and 137 bp for the mutant); mhf2-2 locus amplification with 5′-AAGCGTTTATGTATTTTTAGA-3′ and 5′-CTTCTGGTTCGTTTATACACT-3′, subsequent digestion with BseNI (350 bp for the wild-type and 330+20 for the mutant).
Atzip4(s)2 (Atmhf2-1) was sequenced using Illumina technology. Mutations were identified through MutDetect pipeline developed by Bioinformatics and Informatics IJPB team (Supplementary Methods).
Cytology
Meiotic chromosome spreads have been performed as described previously (20). Immuno-localizations were performed as described in (21). Observations were made using a ZEISS AxioObserver microscope.
RESULTS AND DISCUSSION
zmm suppressor screens identified MHF2 as an anti-CO factor
We sought to find Arabidopsis mutants with increased CO-formation. However, increased meiotic CO formation does not confer any obvious macroscopic phenotype preventing easy genetic screening (6). In contrast, reduction in CO formation is easily detectable because without the physical connection provided by CO, pairs of homologous chromosomes do not associate as bivalents at metaphase I and appear cytologically as univalents that segregate randomly at anaphase I. At the macroscopic level, this lack of CO is reflected by reduced fertility easily noticed by shorter fruit. For instance, zmm mutants show a 75% reduction in bivalent formation and are almost sterile (2). Here we continue a previously described genetic screen, based on the idea that mutations increasing CO frequency will restore the fidelity of chromosome segregation and subsequently restore the fertility of zmm mutants (6). We continued the Atzip4 (13) suppressor screen that previously revealed AtFANCM as an anti-CO gene. Among 2000 lines screened, eight recessive suppressors were found, falling into three complementation groups, the first of which corresponding to FANCM (6). The second complementation group contained one line, zip4 suppressor 2 (zip4(s)2), and is the focus of this study. Map-based cloning defined a region between 27.15 Mb and 30.29 Mb on chromosome 1 as containing the causal mutation. Following whole genome sequencing, we identified a candidate mutation in the splice donor site of exon 2 in the gene At1g78790. In parallel, we ran a second screen looking for suppressors of another zmm mutant, Athei10 (18). Among 2000 lines screened, 19 suppressors were found. Systematic sequencing of At1g78790 in the suppressors revealed that two lines (hei10(s)174 and hei10(s)170) also contained a mutation in this gene: one non-sense mutation deleting the last five amino acids of the protein and one in the splice donor site of exon 4 (Supplementary Table S1, Supplementary Figure S1). The hei10(s)174 and hei10(s)170 mutations were shown to be allelic, confirming that mutations in At1g78790 cause the fertility restoration of zmm mutants.
This gene encodes a protein with high similarity with mammalian MHF2, and reciprocal Basic Local Alignment Search Tool (BLAST) analyses showed that At1g78790 encodes the single MHF2 homologue in the Arabidopsis genome (Supplementary Figure S1). We then named At1g78790, AtMHF2, and the three mutations Atmhf2-1 (zip4(s)2), Atmhf2-2 (hei10(s)174) and Atmhf2-3 (hei10(s)170) (Supplementary Table S1). Human MHF1 and MHF2 were recently identified as a heterotetramer promoting FANCM activity and participating in somatic DNA damage repair and genome maintenance (10,11). Further, MHF1 and MHF2 have been shown to direct meiotic recombination outcome to NCOs in fission yeast (7).
In the three suppressors with mutations in AtMHF2, chromosome spreads were performed to assess the level of bivalent formation. This showed that the restored fertility was indeed associated with increased bivalent formation at metaphase I compared to their zmm counterpart (Figure 1), suggesting that MHF2 has an anti-CO activity at meiosis in Arabidopsis. The restoration of bivalent formation was not complete, the zmm mutants, zmm mhf2 double mutants and wild type having ∼1, ∼4 and 5 bivalent pairs, respectively (Figure 1). In contrast, Atfancm mutation almost completely restored bivalent formation of zmm mutants (4.9 bivalent pairs) suggesting that mutating AtMHF2 has a lesser anti-CO effect than mutating FANCM at meiosis (Figure 1).
Figure 1.
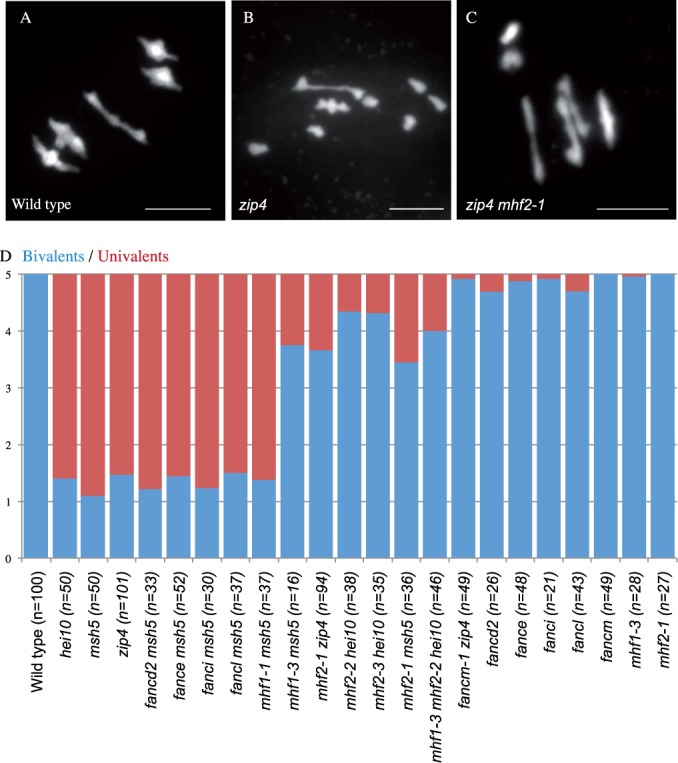
Bivalent formation analysis at metaphase I. (A–C) Metaphase I chromosome spreads of male meiocytes in three representative genotypes (A) wild type, (B) Atzip4 (C) Atzip4 Atmhf2-1. Scale bar = 5 μm. (D) Average number of bivalents (blue) and pairs of univalents (red) per male meiocyte at metaphase I. Number of cells analysed is indicated in parentheses. fancm zip4 and zip4 data are from (6).
In the single Atmhf2 mutants, metaphase I was indistinguishable from wild type with five bivalents (Figure 1, Supplementary Figure S2). Meiotic CO frequency in Atmhf2 was then measured genetically using pollen tetrad analysis (19,22,23) (Figure 2, Supplementary Table S2). In the single mutants Atmhf2-2 and Atmhf2-1 map distances increased by ∼60% compared to wild type on the two intervals tested (Z-test, P < 6×10−3), demonstrating that MHF2 is a CO-limiting factor. This increase, while significant, is lower that what is observed in fancm (P < 10−6), further supporting the conclusion that AtFANCM is a more effective barrier to CO formation than AtMHF2.
Figure 2.
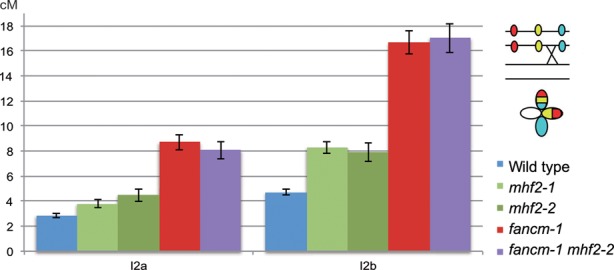
Genetic distances (cM) are increased in mhf2 mutants. Genetic distances in two adjacent intervals on chromosome 2 using FTLs (19) were calculated with the Perkins equation (23) and are given in centiMorgans (cM). Error bars indicate standard deviation (± SD). Raw data and calculation can be found in Supplementary Table S2. One tetrad example and its interpretation are shown on the top right corner.
Mutations in MHF1, but not FANCL, FANCE, FANCI nor FANCD2 restore CO formation in zmm mutants
The identification of AtMHF2 (this study) and AtFANCM (6) as factors limiting CO formation in Arabidopsis, prompted us to test for a similar role of other FA proteins. First, we examined the conservation of FA proteins among a selection of eukaryotes through reciprocal BLAST analysis and literature survey (6–8,10,11,24-60) (Table 1). The FA proteins known in humans can be categorized into three classes, according to the cascade of events in the process of repairing blocked replication forks: the FA core complex, the FA-ID complex and the downstream factors (8,10,11). Many FA proteins are conserved in animals and plants, suggesting that the pathway may be conserved in these kingdoms, making Arabidopsis a suitable model to address the function of FA factors. In contrast, fungi seem to lack the majority of FA components. In Arabidopsis, unique homologues of components of the core complex (in addition to FANCM and MHF2) were identified (FANC -E, -L, MHF1), as well as all three FA-ID complex members (-D2, -I, FAN1). Among the downstream genes two unique homologues were identified (-D1, -O), and a third possesses two homologues organized as a tandem duplication (-J). No putative homologues were found for the other FA members (Table 1). For MHF1, two genes predictions corresponding to the same locus are present in the Arabidopsis databases: AT5E46180 (61) and AT5G50930.1 (62) but it has recently been shown that only the transcript corresponding to AT5E46180 exists in vivo (63).
Table 1. Conservation of FA proteins among a selection of eukaryotes.
| H. sapiens | A. thaliana | S. cerevisiae | S. pombe | D. melanogaster | C. elegans | |
|---|---|---|---|---|---|---|
| FA core complex | FANCA (8) | - | - | - | - | - |
| FANCB (24) | - | - | - | - | - | |
| FANCC (8) | - | - | - | - | - | |
| FANCE (8) | FANCE (At4g29560) | - | - | - | - | |
| FANCF (25) | - | - | - | - | - | |
| FANCG/XRCC9 (8) | - | - | - | - | - | |
| FANCL (8) | FANCL (At5g65740) (26) | - | - | FANCL (27)a | - | |
| FANCM/FAAP250 (8) | FANCM (At1g35530) (6,28)a | Mph1 (29,30)a | Fml1 (7,31) | FANCM (27) | FANCM-1/DRH3 (32,33)a | |
| MHF1/CENP-S/FAAP16 (10,11) | MHF1 (At5g50930) (34) | Mhf1 (10)a | Mhf1 (7)a | - | MHF1 (Y48E1C.1) | |
| MHF2/CENP-X/FAAP10 (10,11) | MHF2 (At1g78790) | Mhf2 (10)a | Mhf2 (7)a | - | MHF2 (F35H10.5) | |
| FAAP20 (35) | - | - | - | - | - | |
| FAAP24 (36) | - | - | - | - | - | |
| FAAP100 (37) | - | - | - | - | - | |
| - | ||||||
| Fml1 | ||||||
| Mhf1 | - | |||||
| Mhf2 | - | |||||
| FA-ID and FAN1 | FANCI (8) | FANCI (At5g49110) | - | - | FANCI | FANCI-1 (32,33)a |
| FANCD2 (8) | FANCD2 (At4g14970) (26) | - | - | FANCD2 (27)a | FACD-2 (32–33,38–39)a | |
| FAN1 (8) | FAN1 (At1g48360) | - | Fan1 (40)a | - | FAN-1 (41–43)a | |
| FA downstream partners | FANCD1/BRCA2 (8) | FANCD1 (At5g01630 & At4g00020) (64)a | - | - | BRCA2 (44)a | BRC-2 (33,45)a |
| FANCJ/BRIP1/ BACH1 (8) | FANCJ (At1g20720 & At1g20750) (46) | - | - | - | DOG-1 (47)a | |
| FANCN/PALB2 (8) | - | - | - | - | - | |
| FANCO/RAD51C (8) | FANCO/RAD51C (At2g45280) (48,65–66)a | - | - | Spindle D (49)a | RFS-1/RAD51C (50)a | |
| FANCP/SLX4/ BTBD12 (51) | - | Slx4 (52)a | Slx4 (53)a | MUS312 (54,55)a | HIM-18/SLX4 (56)a | |
| FANCQ/ERCC4/ XPF/RAD1 (57) | FANCQ/RAD1 (At5g41150) (58,67)a | |||||
| Rad1 (59)a | Rad16 (60)a | MEI9 (55)a | XPF (56)a |
Experimentally tested and putative homologues based on sequence similarity are shown.
The “-” symbol indicates no gene encoding protein with significant similarity was found.
aExperimental evidence of a role in DNA repair.
We analysed mutant lines in the three members of the core complex (AtMHF1, AtFANCE, AtFANCL) and two members of the FA-ID complex (AtFANCD2, AtFANCI) (see Materials and Methods).
For each targeted gene, we identified a T-DNA insertion that disrupts the genomic coding sequence (Supplementary Figure S3). We did not analyse the downstream partners AtFANCD1/BRCA2 and AtFANCO/RAD51C because they are essential for the repair of meiotic recombination intermediates in Arabidopsis and CO formation (64–66), and are therefore unlikely candidates for a role in limiting meiotic COs that could be detected. We did not analyse FANCJ either, because it is present as a tandem duplication, making its mutation unrealistic to obtain. None of the tested lines showed any obvious somatic defects. Meiosis was indistinguishable from wild type on chromosome spreads in Atmhf1, as shown above for Atmhf2 (Supplementary Figure S2). This contrasts with the situation in fission yeast where MHF1/CENP-S and MHF2/CENP-X are required for balanced segregation of chromosomes at meiosis, through the establishment of proper kinetochore function, independently of FANCM and recombination (68). However a low frequency of univalents was detected in Atfancd2, Atfance, Atfanci and Atfancl, suggesting that these genes may have a minor role in promoting CO formation (Figure 1D and Supplementary Figure S2).
To assess the putative anti-CO activity of these genes we tested if their mutation could suppress the zmm lack of bivalents, as do the mutations in the genes FANCM and MHF2. For each T-DNA line, we obtained a double mutant with either Atmsh5 or Athei10 (15,18). None of the mutations of AtFANCE, AtFANCL, AtFANCD2, AtFANCI (Figure 1) nor AtFANCQ/RAD1 (67) increased the number of bivalent in a zmm background. Even if we cannot formally exclude that some activity could be retained in the T-DNA mutants (although it appears unlikely in view of the positions of the 4.5 kb T-DNA insertions; Supplementary Figure S3), these results suggest that these FA genes do not have any anti-CO activity like FANCM and MHF2. In contrast, the double mutant Atmhf1-3 Atmsh5 showed a large increase of bivalent formation compared to Atmsh5 (Figure 1), showing that AtMHF1 possesses a meiotic anti-CO function. The Salk_119435 insertion (mhf1-1 in (63)), which is inserted 41 base pairs in 3′ of the ATG was unable to restore bivalent formation of Atmsh5. This suggests that a functional MHF1 protein is produced at meiosis in this line.
The effect of AtMHF1 depletion at meiosis was similar to that of AtMHF2, and thus weaker than that of FANCM, suggesting that like MHF2, MHF1 is a less efficient barrier to CO formation than FANCM. In summary, these data suggest that only a subset of the FA associated proteins, namely FANCM, MHF1 and MHF2, are involved in limiting meiotic COs.
MHF1, MHF2 and FANCM act in the same pathway to limit meiotic COs
We then tested whether AtMHF1, AtMHF2 and AtFANCM act in the same pathway to limit meiotic COs. First, a triple mutant hei10 mhf1 mhf2 showed the same level of bivalent formation compared to msh5 mhf1, msh5 mhf2, or hei10 mhf2 (Figure 1D), suggesting that AtMHF1 and AtMHF2 act in the same pathway. As fancm mutation restores bivalent formation of zmm mutants to near wild-type levels, it cannot be tested if bivalent formation can be restored further in combination with mhf1or mhf2. This limitation can be overcome by measuring genetic CO formation. We therefore tested the effect of mutating both AtFANCM and AtMHF2, using tetrad analysis on one pair of adjacent intervals (I2a/I2b) (Figure 3). The genetic distances in Atfancm Atmhf2-2 double mutant was higher than wild type (Z-test, P < 10−5) and Atmhf2-2 (Z-test, P < 10−4), but not different from Atfancm (Z-test, P > 0.05), demonstrating that AtFANCM and AtMHF2 limit COs in the same genetic pathway. This predicts that the extra COs in an Atmhf2 mutant would arise from the class II pathway, as described for Atfancm (6). Consistently, the number of MLH1 foci per cell, a marker of class I, ZMM-dependent COs, are unchanged in Atmhf2-1 compared to wild type [9.2±1.7 (n = 21) and 8.9±1.4 (n = 16), T-test P = 0.54] (Supplementary Figure S4). Further, as class II COs do not display interference, interference should be impaired in Atmhf2 mutants. We used the tetrad data set to analyse interference through the calculation of the interference ratio (IR) (Supplementary Table S2). IR measures the effect of having recombination in one interval on the genetic distance of the adjacent interval. IR is close to 0 when having COs in one interval prevents CO formation in the adjacent interval, thus indicating positive interference; IR = 1 when interference is absent (22). Interference was detected in wild-type (IR I2b/I2a = 0.37; Z test P(IR = 1) = 1.2 10−6) but was undetectable in Atmhf2-1 (IR I2b/I2a = 0.89; P(IR = 1) = 0.6) and Atmhf2-2 (IR I2b/I2a = 1.02; P(IR = 1) = 0.9) (Supplementary Table S2C). Finally, as MUS81 promotes class II COs, we produced Atmhf1-3 Atmus81 and Atmhf2-1 Atmus81 double mutants (Figure 3). In these double mutants, chromosome fragmentation was observed at anaphase I, while this is not the case for the respective single mutants; Atmhf1, Atmhf2 and Atmus81 (Figure 3B). This shows that in absence of MHF1 or MHF2, MUS81 becomes necessary for efficient repair of DNA DSBs. This is reminiscent of the Atfancm Atmus81 meiotic defects (6). Altogether this confirms that MHF1 and MHF2 act in the same pathway of FANCM to restrain class II meiotic CO. However, based on the partial restoration of bivalent formation in a zmm context (Figure 1) and on the measurement of recombination levels (Figure 2), it appears that MHF1 and MHF2 have a less prominent role than FANCM in limiting COs. While Atfancm-1 Atmus81 plants are barely viable (6), growth and development of Atmhf1-3 Atmus81 and Atmhf2-1 Atmus81 plants did not have the same synthetic growth defect (Figure 3) until they enter into the reproductive phase and have reduced fertility. Similar results were reported by Dangel and colleagues (63). This suggests that MHF1 and MHF2 have a less important role than FANCM in the repair of somatic DNA damage, as they have a less important role in limiting meiotic COs. Similarly, in human HeLa cells, the absence of MHF1 or MHF2 leads to less severe genotoxic agent sensitivity than the absence of FANCM (10). Further, the MHF1 and MHF2 form a heterotetramer that enhances FANCM DNA binding and DNA branch migration activity in vitro but FANCM alone retains some activity independently of these two co-factors (10,11,69,70). We thus propose that during meiosis, MHF1 and MHF2 support the FANCM helicase anti-CO activity, but that FANCM is able to function partially in the absence of MHF1/MHF2. The other conserved members of the FA pathway, including the members of the core complex, do not seem to play a role in the FANCM-MHFs anti-CO activity. It has been previously suggested that FANCM, in addition to being a core component of the FA pathway, also has a function in somatic DNA repair independently of the FA pathway (discussed in (71)). Here we showed that the FANCM-MHF1-MHF2 module ensures a specific function as a barrier to CO formation in meiosis.
Figure 3.
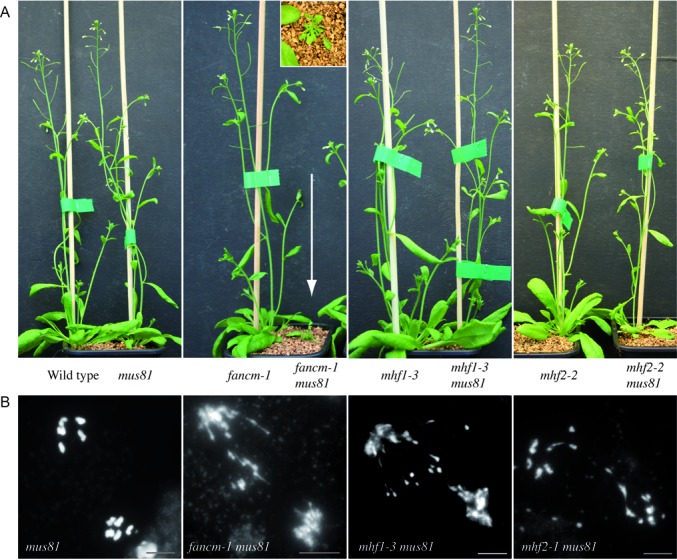
Genetic interaction of Atmhf1, Atmhf2 and Atfancm with Atmus81. (A) Six weeks old plants are shown with the corresponding genotype indicated below. The arrow points to the sick Atfancm Atmus81 double mutant for which an enlargement (top view) is shown. (B) Anaphase I chromosome spreads. Chromosome fragmentation can be observed in Atfancm-1 Atmus81, Atmhf1–3 Atmus81 and Atmhf2-1 Atmus81. Scale bar = 5 μm.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We wish to thank Mathilde Grelon and Christine Mezard for critical reading of the manuscript. We wish to thank Gregory Copenhaver for providing the FTL lines and Vincent Colot for access to sequencing facilities. We also thank Delphine Charif, Fabienne Garnier, Joseph Tran and Najla Ben Hassine for developing the MutDetect pipeline.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
Present address:
Wayne Crismani, DuPont-Pioneer, 8305 NW 62nd Avenue, PO Box 7060, Johnston, IA 50131, USA.
FUNDINGS
European Community's Seventh Framework Programme FP7/2007-2013 [KBBE-2009-222883 (MeioSys); ERC 2011 StG 281659 (MeioSight)]. Funding for open access charge: European Community's Seventh Framework Programme FP7/2007-2013 [KBBE-2009-222883 (MeioSys); ERC 2011 StG 281659 (MeioSight)].
Conflict of interest statement. None declared.
REFERENCES
- 1.Youds J.L., Boulton S.J. The choice in meiosis—defining the factors that influence crossover or non-crossover formation. J. Cell Sci. 2011;124:501–513. doi: 10.1242/jcs.074427. [DOI] [PubMed] [Google Scholar]
- 2.Lynn A., Soucek R., Börner G.V. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 3.Osman K., Higgins J.D., Sanchez-Moran E., Armstrong S.J., Franklin F.C.H. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 2011;190:523–544. doi: 10.1111/j.1469-8137.2011.03665.x. [DOI] [PubMed] [Google Scholar]
- 4.Berchowitz L.E., Copenhaver G.P. Genetic interference: don't stand so close to me. Curr. Genomics. 2010;11:91–102. doi: 10.2174/138920210790886835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson I.R. Control of meiotic recombination frequency in plant genomes. Curr. Opin. Plant Biol. 2012;15:556–561. doi: 10.1016/j.pbi.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Crismani W., Girard C., Froger N., Pradillo M., Santos J.L., Chelysheva L., Copenhaver G.P., Horlow C., Mercier R. FANCM limits meiotic crossovers. Science. 2012;336:1588–1590. doi: 10.1126/science.1220381. [DOI] [PubMed] [Google Scholar]
- 7.Lorenz A., Osman F., Sun W., Nandi S., Steinacher R., Whitby M.C. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science. 2012;336:1585–1588. doi: 10.1126/science.1220111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deakyne J.S., Mazin A. V. Fanconi anemia: at the crossroads of DNA repair. Biochemistry. 2011;76:36–48. doi: 10.1134/s0006297911010068. [DOI] [PubMed] [Google Scholar]
- 9.Kottemann M.C., Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan Z., Delannoy M., Ling C., Daee D., Osman F., Muniandy P.A., Shen X., Oostra A.B., Du H., Steltenpool J., et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh T.R., Saro D., Ali A.M., Zheng X.-F., Du C., Killen M.W., Sachpatzidis A., Wahengbam K., Pierce A.J., Xiong Y., et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol. Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ríos G., Lossow A., Hertel B., Breuer F., Schaefer S., Broich M., Kleinow T., Jásik J., Winter J., Ferrando A., et al. Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. Plant J. 2002;32:243–253. doi: 10.1046/j.1365-313x.2002.01416.x. [DOI] [PubMed] [Google Scholar]
- 13.Chelysheva L., Gendrot G., Vezon D., Doutriaux M.-P., Mercier R., Grelon M. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 2007;3:e83. doi: 10.1371/journal.pgen.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macaisne N., Novatchkova M., Peirera L., Vezon D., Jolivet S., Froger N., Chelysheva L., Grelon M., Mercier R. SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr. Biol. 2008;18:1432–1437. doi: 10.1016/j.cub.2008.08.041. [DOI] [PubMed] [Google Scholar]
- 15.Higgins J.D., Vignard J., Mercier R., Pugh A.G., Franklin F.C.H., Jones G.H. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 2008;55:28–39. doi: 10.1111/j.1365-313X.2008.03470.x. [DOI] [PubMed] [Google Scholar]
- 16.Berchowitz L.E., Francis K.E., Bey A.L., Copenhaver G.P. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 2007;3:e132. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stacey N.J., Kuromori T., Azumi Y., Roberts G., Breuer C., Wada T., Maxwell A., Roberts K., Sugimoto-Shirasu K. Arabidopsis SPO11–2 functions with SPO11-1 in meiotic recombination. Plant J. 2006;48:206–216. doi: 10.1111/j.1365-313X.2006.02867.x. [DOI] [PubMed] [Google Scholar]
- 18.Chelysheva L., Vezon D., Chambon A., Gendrot G., Pereira L., Lemhemdi A., Vrielynck N., Le Guin S., Novatchkova M., Grelon M. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 2012;8:e1002799. doi: 10.1371/journal.pgen.1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berchowitz L.E., Copenhaver G.P. Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat. Protoc. 2008;3:41–50. doi: 10.1038/nprot.2007.491. [DOI] [PubMed] [Google Scholar]
- 20.Ross K.J., Fransz P., Jones G.H. A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosom. Res. 1996;4:507–516. doi: 10.1007/BF02261778. [DOI] [PubMed] [Google Scholar]
- 21.Chelysheva L., Grandont L., Vrielynck N., le Guin S., Mercier R., Grelon M. An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 2010;129:143–153. doi: 10.1159/000314096. [DOI] [PubMed] [Google Scholar]
- 22.Malkova A., Swanson J., German M., McCusker J.H., Housworth E.A., Stahl F.W., Haber J.E. Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics. 2004;168:49–63. doi: 10.1534/genetics.104.027961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perkins D.D. Biochemical mutants in the smut fungus ustilago maydis. Genetics. 1949;34:607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCauley J., Masand N., McGowan R., Rajagopalan S., Hunter A., Michaud J.L., Gibson K., Robertson J., Vaz F., Abbs S., et al. X-linked VACTERL with hydrocephalus syndrome: further delineation of the phenotype caused by FANCB mutations. Am. J. Med. Genet. A. 2011;155A:2370–2380. doi: 10.1002/ajmg.a.33913. [DOI] [PubMed] [Google Scholar]
- 25.De Winter J.P., Rooimans M.A., van Der Weel L., van Berkel C.G., Alon N., Bosnoyan-Collins L., de Groot J., Zhi Y., Waisfisz Q., Pronk J.C., et al. The Fanconi anaemia gene FANCF encodes a novel protein with homology to ROM. Nat. Genet. 2000;24:15–16. doi: 10.1038/71626. [DOI] [PubMed] [Google Scholar]
- 26.Meetei A.R., Yan Z., Wang W. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle. 2004;3:179–181. [PubMed] [Google Scholar]
- 27.Marek L.R., Bale A.E. Drosophila homologs of FANCD2 and FANCL function in DNA repair. DNA Repair (Amst). 2006;5:1317–1326. doi: 10.1016/j.dnarep.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 28.Knoll A., Higgins J.D., Seeliger K., Reha S.J., Dangel N.J., Bauknecht M., Schröpfer S., Franklin F.C.H., Puchta H. The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell. 2012;24:1448–1464. doi: 10.1105/tpc.112.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheller J., Schürer A., Rudolph C., Hettwer S., Kramer W. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection ofthe genome from spontaneous and chemically induced damage. Genetics. 2000;7:1069–1081. doi: 10.1093/genetics/155.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prakash R., Satory D., Dray E., Papusha A., Scheller J., Kramer W., Krejci L., Klein H., Haber J.E., Sung P., et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun W., Nandi S., Osman F., Ahn J.S., Jakovleska J., Lorenz A., Whitby M.C. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K.Y., Chung K.Y., Koo H.-S. The involvement of FANCM, FANCI, and checkpoint proteins in the interstrand DNA crosslink repair pathway is conserved in C. elegans. DNA Repair (Amst). 2010;9:374–382. doi: 10.1016/j.dnarep.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 33.Youds J.L., Barber L.J., Boulton S.J. C. elegans: a model of Fanconi anemia and ICL repair. Mutat. Res. 2009;668:103–116. doi: 10.1016/j.mrfmmm.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Dangel N.J., Knoll A., Puchta H. MHF1 plays FANCM-dependentand -independent roles in DNA repair and homologous recombination in plants. Plant J. 2014;78:822–833. doi: 10.1111/tpj.12507. [DOI] [PubMed] [Google Scholar]
- 35.Leung J.W.C., Wang Y., Fong K.W., Huen M.S.Y., Li L., Chen J. Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand cross-link repair. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4491–4496. doi: 10.1073/pnas.1118720109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciccia A., Ling C., Coulthard R., Yan Z., Xue Y., Meetei A.R., Laghmani E.H., Joenje H., McDonald N., de Winter J.P., et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Ling C., Ishiai M., Ali A.M., Medhurst A.L., Neveling K., Kalb R., Yan Z., Xue Y., Oostra A.B., Auerbach A.D., et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26:2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collis S.J., Barber L.J., Ward J.D., Martin J.S., Boulton S.J. C. elegans FANCD2 responds to replication stress and functions in interstrand cross-link repair. DNA Repair (Amst). 2006;5:1398–1406. doi: 10.1016/j.dnarep.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 39.Lee K.Y., Yang I., Park J.E., Baek O.R., Chung K.Y., Koo H.S. Developmental stage- and DNA damage-specific functions of C. elegans FANCD2. Biochem. Biophys. Res. Commun. 2007;352:479–485. doi: 10.1016/j.bbrc.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 40.Fontebasso Y., Etheridge T.J., Oliver A.W., Murray J.M., Carr A.M. The conserved Fanconi anemia nuclease Fan1 and the SUMO E3 ligase Pli1 act in two novel Pso2-independent pathways of DNA interstrand crosslink repair in yeast. DNA Repair (Amst). 2013;12:1011–1023. doi: 10.1016/j.dnarep.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kratz K., Schöpf B., Kaden S., Sendoel A., Eberhard R., Lademann C., Cannavó E., Sartori A.A., Hengartner M.O., Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 42.Smogorzewska A., Desetty R., Saito T.T., Schlabach M., Lach F.P., Sowa M.E., Clark A.B., Kunkel T.A., Harper J.W., Colaiácovo M.P., et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacKay C., Déclais A.-C., Lundin C., Agostinho A., Deans A.J., MacArtney T.J., Hofmann K., Gartner A., West S.C., Helleday T., et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klovstad M., Abdu U., Schüpbach T. Drosophila brca2 is required for mitotic and meiotic DNA repair and efficient activation of the meiotic recombination checkpoint. PLoS Genet. 2008;4:e31. doi: 10.1371/journal.pgen.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin J.S., Winkelmann N., Petalcorin M.I.R., McIlwraith M.J., Boulton S.J. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol. Cell. Biol. 2005;25:3127–3139. doi: 10.1128/MCB.25.8.3127-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knoll A., Puchta H. The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J. Exp. Bot. 2011;62:1565–1579. doi: 10.1093/jxb/erq357. [DOI] [PubMed] [Google Scholar]
- 47.Youds J.L., Barber L.J., Ward J.D., Collis S.J., O'Neil N.J., Boulton S.J., Rose A.M. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol. Cell. Biol. 2008;28:1470–1479. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bleuyard J.-Y., Gallego M.E., Savigny F., White C.I. Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 2005;41:533–545. doi: 10.1111/j.1365-313X.2004.02318.x. [DOI] [PubMed] [Google Scholar]
- 49.Abdu U., González-Reyes A., Ghabrial A., Schüpbach T. The Drosophila spn-D gene encodes a RAD51C-like protein that is required exclusively during meiosis. Genetics. 2003;165:197–204. doi: 10.1093/genetics/165.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward J.D., Barber L.J., Petalcorin M.I., Yanowitz J., Boulton S.J. Replication blocking lesions present a unique substrate for homologous recombination. EMBO J. 2007;26:3384–3396. doi: 10.1038/sj.emboj.7601766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang M.H. Cancel all Hollidays for SLX4 mutations: identification of a new Fanconi anemia subtype, FANCP. Clin. Genet. 2011;80:28–30. doi: 10.1111/j.1399-0004.2011.01679.x. [DOI] [PubMed] [Google Scholar]
- 52.Mullen J.R., Kaliraman V., Ibrahim S.S., Brill S.J. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coulon S., Gaillard P.-H.L., Chahwan C., McDonald W.H., Yates J.R., Russell P. Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol. Biol. Cell. 2004;15:71–80. doi: 10.1091/mbc.E03-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersen S.L., Bergstralh D.T., Kohl K.P., LaRocque J.R., Moore C.B., Sekelsky J.J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol. Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majumder S., Kramer B., Sekelsky J.J., Carolina N., Hill C. Drosophila MUS312 Interacts with the Short Article MEI-9 to Generate Meiotic Crossovers. Mol. Cell. 2002;10:1503–1509. doi: 10.1016/s1097-2765(02)00782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saito T.T., Youds J.L., Boulton S.J., Colaiácovo M.P. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 2009;5:e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bogliolo M., Schuster B., Stoepker C., Derkunt B., Su Y., Raams A., Trujillo J.P., Minguillón J., Ramírez M.J., Pujol R., et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause fanconi anemia. Am. J. Hum. Genet. 2013;92:800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dubest S., Gallego M.E., White C.I. Role of the AtRad1p endonuclease in homologous recombination in plants. EMBO Rep. 2002;3:1049–1054. doi: 10.1093/embo-reports/kvf211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schiestl R.H., Prakash S. RAD1, an excision repair gene of Saccharomyces cerevisiae is also involved in recombination. Mol. Cell. Biol. 1988;8:3619–3626. doi: 10.1128/mcb.8.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Basmacioglu C.N., Subramani S., Clegg M., Nasim A., Carr A.M., Schmidt H., Kirchhoff S., Muriel W.J., Sheldrick K.S., Griffiths D.J., et al. he radl6 gene of Schizosaccharomyces pombe: a homolog of the RADI gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:2029–2040. doi: 10.1128/mcb.14.3.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dèrozier S., Samson F., Tamby J.-P., Guichard C., Brunaud V., Grevet P., Gagnot S., Label P., Leplé J.-C., Lecharny A., et al. Exploration of plant genomes in the FLAGdb++ environment. Plant Methods. 2011;7:8. doi: 10.1186/1746-4811-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M., et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dangel N.J., Knoll A., Puchta H. MHF1 plays Fanconi anaemia complementation group M protein (FANCM)-dependent and FANCM-independent roles in DNA repair and homologous recombination in plants. Plant J. 2014;78:822–833. doi: 10.1111/tpj.12507. [DOI] [PubMed] [Google Scholar]
- 64.Siaud N., Dray E., Gy I., Gérard E., Takvorian N., Doutriaux M.-P. Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 2004;23:1392–1401. doi: 10.1038/sj.emboj.7600146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abe K., Osakabe K., Nakayama S., Endo M., Tagiri A., Todoriki S., Ichikawa H., Toki S. Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol. 2005;139:896–908. doi: 10.1104/pp.105.065243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li W., Yang X., Lin Z., Timofejeva L., Xiao R., Makaroff C.A., Ma H. The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol. 2005;138:965–976. doi: 10.1104/pp.104.058347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crismani W., Portemer V., Froger N., Chelysheva L., Horlow C., Vrielynck N., Mercier R. MCM8 is required for a pathway of meiotic double-strand break repair independent of DMC1 in Arabidopsis thaliana. PLoS Genet. 2013;9:e1003165. doi: 10.1371/journal.pgen.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhattacharjee S., Osman F., Feeney L., Lorenz A., Bryer C., Whitby M.C. MHF1–2/CENP-S-X performs distinct roles in centromere metabolism and genetic recombination. Open Biol. 2013;3:130102. doi: 10.1098/rsob.130102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Tao Y., Jin C., Li X., Qi S., Chu L., Niu L., Yao X., Teng M. The structure of the FANCM–MHF complex reveals physical features for functional assembly. Nat. Commun. 2012;3:782. doi: 10.1038/ncomms1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang H., Zhang T., Tao Y., Wu L., Li H., Zhou J., Zhong C., Ding J. Saccharomyces cerevisiae MHF complex structurally resembles the histones (H3-H4)2 heterotetramer and functions as a heterotetramer. Structure. 2012;20:364–370. doi: 10.1016/j.str.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 71.Whitby M.C. The FANCM family of DNA helicases/translocases. DNA Repair (Amst). 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


