Abstract
Human topoisomerase 1B has been simulated covalently bound to a negatively supercoiled DNA minicircle, and its behavior compared to the enzyme bound to a simple linear DNA duplex. The presence of the more realistic supercoiled substrate facilitates the formation of larger number of protein–DNA interactions when compared to a simple linear duplex fragment. The number of protein–DNA hydrogen bonds doubles in proximity to the active site, affecting all of the residues in the catalytic pentad. The clamp over the DNA, characterized by the salt bridge between Lys369 and Glu497, undergoes reduced fluctuations when bound to the supercoiled minicircle. The linker domain of the enzyme, which is implicated in the controlled relaxation of superhelical stress, also displays an increased number of contacts with the minicircle compared to linear DNA. Finally, the more complex topology of the supercoiled DNA minicircle gives rise to a secondary DNA binding site involving four residues located on subdomain III. The simulation trajectories reveal significant changes in the interactions between the enzyme and the DNA for the more complex DNA topology, which are consistent with the experimental observation that the protein has a preference for binding to supercoiled DNA.
INTRODUCTION
Topoisomerases are ubiquitous enzymes that control the topological state of DNA during fundamental cellular processes such as replication, transcription and chromosomal segregation (1–3). Human topoisomerase 1B (hTop1B) relaxes positive and negative supercoils by creating a nick on one strand of the DNA duplex and forming a transient phosphor-tyrosine bond (4). The enzyme mechanism is composed of five steps: (i) DNA binding and formation of a non-covalent complex; (ii) nucleophilic attack by a tyrosine residue on one DNA strand; (iii) rotation of the intact strand around the nick to resolve the supercoil; (iv) religation of the DNA strand and (v) enzyme release (4). The enzyme is highly active in cells undergoing a high degree of replication, such as cancer cells, and for this reason it is an important drug target. hTop1B is the unique molecular target of a class of anticancer compounds, camptothecins (CPTs) (5), which interact with the protein–DNA complex after cleavage and block the religation step (6,7).
The structure of the protein has been solved as a covalent and non-covalent complex with a 22 bp DNA substrate (8,9), and in the presence of different classes of inhibitors (6,10). The enzyme is composed of (i) an N-terminal domain, that due to its high degree of flexibility has never been crystallized, but which is dispensable for the catalytic activity and thought to be involved in nuclear localization and interaction with other proteins; (ii) a core domain further divided into subdomain I, II and III; (iii) a C-terminal domain owning the catalytic residue Tyr723 and (iv) a linker domain connecting subdomain III with the C-terminal (9,11). The protein first clamps the DNA forming a non-covalent complex that is stabilized by a salt bridge formed by residues Lys369 and Glu497 which are located on two loops defined as Lip1 and Lip2 (containing Lys369 and Glu497 respectively) and which stabilize the complex during the catalysis (1). The linker domain is formed by two long helices that are directly involved in the relaxation mechanism. The shape and positive charge of the linker enable it to interact with the DNA substrate downstream of the cleavage site; it is then thought to guide the relaxation of the supercoiled DNA, giving rise to the ‘controlled rotation’ mechanism (4). The direct correlation between the mobility of the linker and DNA relaxation has been demonstrated by showing that point mutations affecting the dynamics of the linker also alter the catalytic religation rate and the susceptibility of the enzyme to drugs (12–17). Reduced drug sensitivity and varied flexibility has also been shown in a chimeric enzyme in which the plasmodial linker has been substituted into the human protein (18).
One major limitation of both the X-ray structure and the simulations carried out to date is that the DNA substrates studied have all been linear. hTop1B in fact shows preferential binding to supercoiled DNA (19); however, mutational analysis of basic residues on the protein surface reduces this preference (20). Through improvements in computing technology, which now permit calculations on far larger systems, we have performed a series of atomistic molecular dynamics simulations of hTop1B in a covalent complex with a 240 bp negatively supercoiled plasmid with explicit water molecules, and we have compared the data obtained with a previous simulation of the protein in complex with a 22 bp linear substrate (21). Analysis of the dynamical properties of the protein bound to the more biologically relevant supercoiled substrate reveals an increased number of protein–DNA interactions and, most strikingly, the presence of a secondary protein–DNA binding site.
MATERIALS AND METHODS
In order to provide a negatively supercoiled substrate, a 5-ns GB/SA implicit solvent simulation of the supercoiled 240 bp DNA minicircle with linking number Lk = 20 and ΔLk = −1.5 was performed, using the simulation protocols and parameters described in our previous work (22). The twist of DNA is slightly underestimated by the AMBER force field (32.2 bp/turn), consequently, the relaxed linking (Lk0) number for a minicircle of this size is Lk0 = 21.5, which gives a superhelical density of σ = ΔLk/Lk0 = −1.5/21.5 = −0.07, which is marginally higher in magnitude than the σ = −0.06 found in bacterial DNA in physiological conditions. This simulation produced a DNA substrate with a writhe of −0.9, as calculated using the methodology previously described (22) and a global under-twist of ΔTw = −0.6 helical turns, as calculated using the CURVES+ program (23).
To position the protein on the supercoiled plasmid, a structural alignment between the 22 bp nicked DNA co-crystallized with the enzyme and the supercoiled DNA was performed with the SwissPdbViewer program (24). By definition, the base that is cleaved is referred to as −1; the bases upstream are then numbered starting from −2 and the bases downstream start from +1. Particular attention was paid to the alignment of the bases −5/+2, which display the largest number of interaction with the protein and comprise the cleavage site at the −1 position. The coordinates for the 22 bp covalent DNA were taken from the crystal structure 1K4S (10). Once the alignment identified the most suitable position for the protein, it was positioned on the supercoiled DNA (Figure 1) and the system was energy minimized using the Sybyl program version 6.0 (TRIPOS, http://www.tripos.com/) with the Powell method (25). The topology of the system was built using the Amber10 all atom force field, with the parmbsc0 forcefield corrections (26) using tleap, and at this stage the 3′-phosphotyrosyl bond between the Tyr723 and the thymine in position −1 was constructed. The octahedral simulation box consisted of 243288 water molecules. Four hundred and fifty-nine Na+ counterions were added to neutralize the system, giving a final number of 754972 atoms. Simulations were performed using the program NAMD 2.8b361 (27). The system was simulated for 50 ns in periodic boundary conditions, using a cutoff of 10 Å for the evaluation of short-range non bonded interactions and the Particle Mesh Ewald method for the long-range electrostatic interactions (28). The temperature was fixed at 300 K, using Langevin dynamics (29) whereas pressure was kept constant at 1 Atmosphere through the Langevin piston method (30). The SHAKE (31) and SETTLE (32) algorithms were used to restrain bond lengths, for the solute (DNA and protein) and water molecules, respectively. The atomic positions were saved every 250 steps (i.e. 0.5 ps) for the analysis with the Gromacs 4.5 package (33), or with code written in-house. The results were compared with a previous simulation in which the protein was covalently bound to a linear 22 bps DNA substrate (21). Clustering analysis was performed by fitting the structure of the protein during the simulation trajectories relative to the Cα atoms of the core and C-terminal domains (residues 201–624 and 713–765), and then clustering the structures of the linker Cα atoms (residues 625–712) using a root mean squared deviation (RMSD) cut-off of 1.6 Å.
Figure 1.
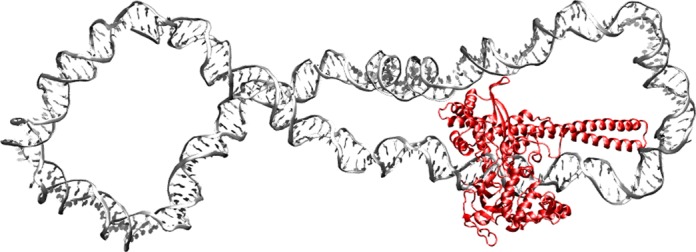
Molecular structure of the hTop1B in covalent complex with a 240 bp negatively supercoiled DNA after 20 ns of atomistic molecular dynamics, showing the formation of the secondary protein–DNA binding site.
In order to check the reproducibility of the results, four replicas of the simulation were performed. These replicas were generated by reassigning the atomic velocities for two configurations taken at 20 ns (replicas R1 and R2) and at 30 ns (replicas R3 and R4), respectively, in accordance with the required Maxwell-Boltzmann distribution. All quantitative comparisons of the behaviour of the primary and replica simulations considered only the last 20 ns of the trajectories to ensure that the trajectories are all of equivalent lengths.
To characterize the structure of the DNA during the MD trajectory, the each DNA base pair was firstly classified as either disrupted (e.g. containing a kink, bubble or wrinkle) (34) or non-disrupted. All DNA helical parameters were calculated using CURVES+ (23). Disrupted base pairs were identified using the following criteria: (i) if the time average displacement between the two complementary bases is >1 Å, the base pair is considered to contain a bubble; (ii) if the fluctuation in roll is >8°, the base pair step is classified as kinked and (iii) if the chi-dihedral angle is outside of the anti-region (between −75° and −150°), then the DNA is classified as wrinkled. Since DNA twist is highly sequence-dependent (for instance, MD simulations using parmbsc0 force field have shown that GC steps have an equilibrium twist of 35.6°, while AT steps have an equilibrium twist of only 30.5° (35)), to assess the change in DNA structure due to interactions with Top1B at its binding site, we have plotted the twist relative to the values reported for MD simulations of linear DNA (Δtw) rather than the absolute values.
RESULTS AND DISCUSSION
Analysis of protein dynamics
The simulations of the hTop1B in covalent complex with a 240 bp plasmid DNA negatively supercoiled (hTop1Bsc) enable us to compare the dynamical behavior of the protein bound to a supercoiled DNA substrate with a simple linear 22 bp DNA fragment (hTop1Blin) (21). In both the linear and supercoiled DNA, the RMSD of the protein Cα atoms does not converge (Supplementary Figure S1A, full lines). However, when the contribution of the linker domain is eliminated, the RMSD of protein reaches a plateau with an average RMSD value lower than 0.15 nm (Supplementary Figure S1A, dashed lines), indicating that in both systems the linker is the most dynamic protein domain. This was also the case for each of the four replica simulations (Supplementary Figure S1B).
The highly mobile nature of the linker is confirmed by calculating the per-residue root mean square fluctuation (RMSF), as shown in Figure 2. The profile of the fluctuations is similar in the two systems, with the linker reaching values of 0.8 and 0.7 nm in the hTop1Bsc and hTop1Blin, respectively (Figure 2). A second peak is found for residues 635–644, which are located in part of the loop that connects subdomain III to the linker, and which have already been reported to have a role in modulating the linker mobility (36). The per residue profiles of the protein fluctuations are similar for all replica simulations (Supplementary Figure S2). Clustering of the Cα atoms of residues Ala625-Lys712, involving the linker and the loop preceding it, shows that the linker explores a different region of conformational space in the two systems. We identified 15 and 4 clusters for hTop1Blin and hTop1Bsc, respectively; the most occupied clusters are compared in red in Figure 3, where the less occupied clusters are shown in gray. For the linear DNA substrate, the first four clusters account for 96% of the total structures (cl1 = 62%, cl2 = 18%, cl3 = 9%, cl4 = 7%), whereas for the supercoiled substrate more than 98% of the total conformations sampled belong to the first cluster, as shown in Supplementary Figure S3. In the supercoiled system, the linker undergoes a conformational change from the starting structure after around 100 ps (Supplementary Figure S3) which allows it to participate in additional interactions with the longer DNA substrate. Supplementary Figure S4 shows molecular configurations extracted from the trajectory every 10 ns. These structures show that the complex topological state of the supercoiled DNA substrate permits additional interactions of the N- and C-terminal regions of the linker domain with the double helix, which is accompanied by a rotation of the coiled–coiled helices around their central axis (Supplementary Figure S4).
Figure 2.
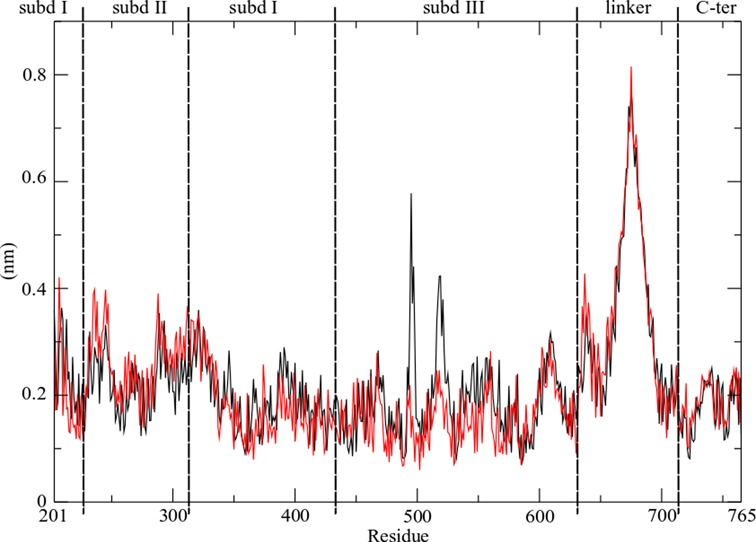
Per-residue RMSF of the protein in hTop1Blin (black line) and hTop1Bsc (red line) complexes. The different protein domains are defined by vertical lines.
Figure 3.
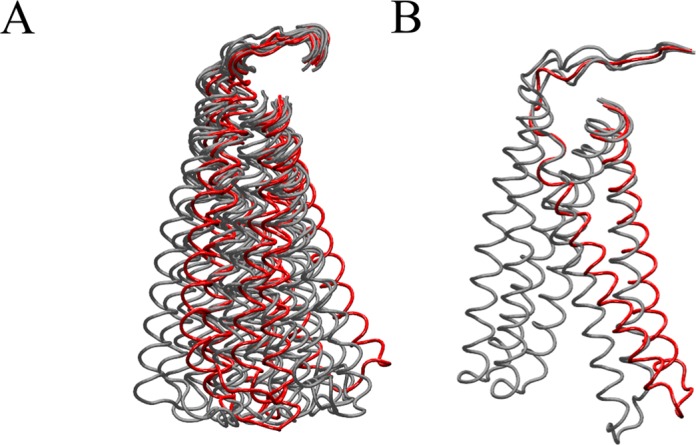
Clustering of the structures of the linker domain, comprising residues Ala625-Lys712, in the simulation of hTop1Blin (A) and hTop1Bsc complexes (B). The centroids of the four families representing 96% of total structures in hTop1Blinear and of the only one representing 98% of total structures in hTop1Bsc are reported in red.
Similarly, analysis of the persistent hydrogen bonds (e.g. those present for more than 60% of total simulation time) between the DNA and residues Ala625-Lys712 (which form the linker and the loop preceding it) shows that there are seven hydrogen bonds in the hTop1Bsc system, while only a single bond is found in for hTop1Blin (Table 1). Our simulations therefore show that the three-dimensional arrangement of the supercoiled double helix facilitates a larger number of hydrogen bonding interactions with the linker. Moreover, comparing the per-residue RMSF in the hTop1Blin system with that for the supercoiled DNA indicates that Glu497 (which is located on Lip2) undergoes larger fluctuations when the complex is formed with linear DNA. This residue is significant because in the X-ray crystal structures of both the protein–DNA covalent and non-covalent complexes it forms an electrostatic interaction with Lys369, (located on Lip1) and is implicated in the stabilization of the protein clamp around the DNA (1). In the hTop1Bsc simulation, as well as in the four replicas, this fluctuation is not observed, suggesting the presence of a tighter interaction between the Lip1 and Lip2, and a reduced conformational diversity of the protein–DNA complex than is observed for the linear substrate.
Table 1. Hydrogen bonds between residues Ala625-Lys712 and DNA which are present for more than 60% of total simulation time.
| hTop1Blin | hTop1Bsc |
|---|---|
| Arg634-Gua12 71% | His632-Gua436 64% |
| Lys654-Ade447 69% | |
| Arg693-Ade446 76% | |
| Gln697-Ade446 76% | |
| Lys700-Ade446 68% | |
| Gln704-Gua39O 81% | |
| Arg708-Cyt38O 73% |
Protein–DNA interactions and evidence of a secondary DNA binding site
The static X-ray structures of both the covalent and non-covalent complex of hTopoI with a 22 bp linear DNA fragment show that the protein establishes a large number of hydrogen bonds with DNA, in particular with the segment of the DNA corresponding to the −5/+2 bases of both strands. In the simulation of the enzyme bound to the linear substrate, 25 hydrogen bonds between the protein and DNA bases −10 to +12 DNA are present for more than 60% of the total simulation time. When we consider the same region in hTop1Bsc, then the number of hydrogen bonds increases to 54. Among these bonds, 19 are conserved between the linear and supercoiled DNA substrates, demonstrating that the network of interactions is similar in the simulations of the two systems, but that additional bonds are occupied in presence of the supercoiled DNA relative to the linear fragment (21). Among the new interactions appearing in hTop1Bsc, six involve the five residues of the catalytic pentad with both bases of the intact (i) or cleaved (c) strands, Arg488-Gua+1(c), Lys532-Ade-2(i), Lys532-Thy-1(c), Hys632-Gua+1(c), His632-Gua+2(c) and Gua+1(c)-Tyr723, and seven interactions involve the linker domain residues, as reported in Table 1. These six specific hydrogen bonding interactions, and indeed additional contacts not observed in the primary MD simulation, are also present in all of the replica simulations (see Supplementary Table S1).
In addition to the increased hydrogen bonding interactions within the enzyme active site and the tighter complex with the linker domain, the simulations of hTop1Bsc show the presence of a secondary DNA binding site. Within the core subdomain III, the four residues Lys466, Lys468, Lys545 and Lys549 form direct contacts with the DNA in spite of the fact that they are far from the canonical DNA binding site. The list of the interactions and their percentage occurrences are provided in Table 2 and in Figure 4, and the formation and persistence of the secondary binding site is illustrated as a function of simulation time in Supplementary Figure S5. The replica simulations indicate that the same amino acid residues are involved in formation of this additional protein–DNA interaction (see Table 2). This secondary DNA binding site involves positively charged and polar surface exposed residues located on subdomain III. A secondary binding site for hTopoIB was postulated by Champoux and co-workers, who demonstrated that reversing the charge of residues Lys466/Lys468 or Lys545/Lys549 affects the preferential binding of hTop1B to supercoiled DNA (36). In our simulations, the most persistent interactions involve these residues; in particular Lys545 and Lys549 are engaged in Lys545-Gua218 and Lys549-Cyt217 hydrogen bonding interactions for 64 and 41% of the total simulation time, respectively (Table 2, column 1). The simulations suggest that Lys545 is the residue that plays the primary role in the formation of this second binding site; the interaction observed between this positively charge residue and the DNA in the replica simulations reaches values of up to 95% (Table 2, columns 2–6). A secondary DNA binding site has been also described in the minimal functional enzyme from vaccinia virus (37,38), and was observed in the crystal structure of the Deinococcus radiorurans topoisomerase in complex with DNA, where it involved equivalent residues to those that are implicated in our simulations (e.g. residues 114, 116, 181 and 185 (39) which are equivalent to the human 466, 468, 545 and 549) (19). Moreover, the ability of the human enzyme to promote the formation of intra- and inter-molecular synapses between distal DNA segments has been demonstrated by atomic force microscopy (40). It is possible that this secondary binding site plays an important biological role; most interestingly, a mutation close to Lys545, namely Arg546Gln, has been recently reported in an individual with a rare form of autism (41).
Table 2. Hydrogen bonds between residues in the secondary binding site and supercoiled DNA along the total 50 ns and last 20 ns of the main (M) simulation (columns 1 and 2) and along the last 20 ns of replica (R1–4) simulations (columns 3–6).
| M (50 ns) | M (last 20 ns) | R1 (last 20 ns) | R2 (last 20 ns) | R3 (last 20 ns) | R4 (last 20 ns) | |
|---|---|---|---|---|---|---|
| Lys466-Ade267 | 28% | 44% | ||||
| Lys466-Thy268 | 30% | 47% | 60% | 84% | 51% | 33% |
| Lys468-Gua269 | 18% | 36% | 25% | |||
| Lys545-Cyt217 | 30% | 29% | 20% | 26% | ||
| Lys545-Gua218 | 64% | 59% | 95% | 72% | 40% | 92% |
| Lys545-Ade219 | 27% | 65% | 25% | |||
| Lys549-Cyt217 | 41% | 46% | 42% | 20% | 14% | 56% |
| Lys549-Gua218 | 26% | 40% | 52% | 95% | 14% | 44% |
Figure 4.
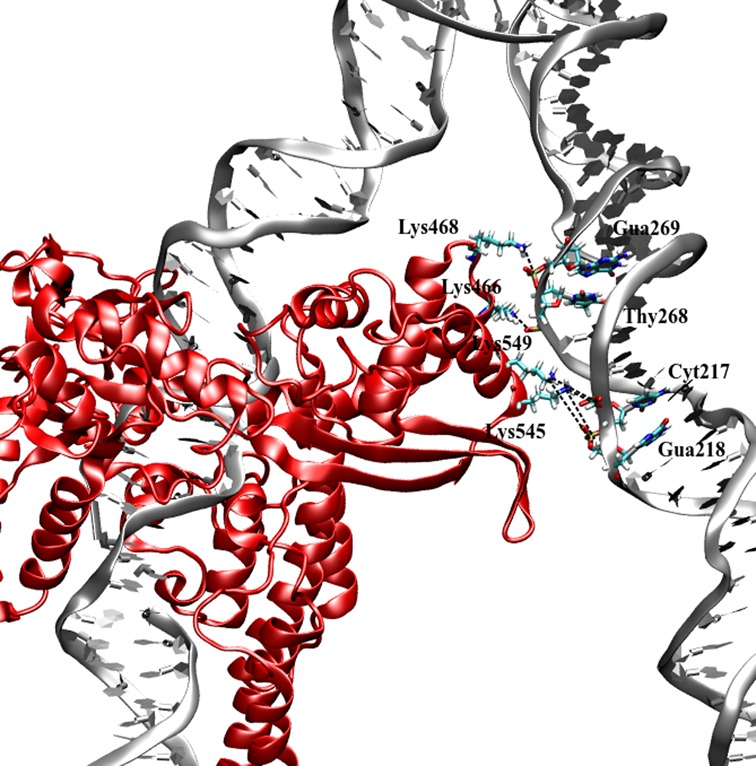
Protein–DNA hydrogen bonding network within the secondary binding site.
The presence of this secondary binding site, the tighter interaction we observe with the linker region of the protein and the increased number of hydrogen bonding interactions within the active site are all consistent with the experimental observation that hTop1B prefers supercoiled over linear DNA substrates.
Structure and dynamics of the DNA in the hTop1B–DNA complex
The effect of the protein binding on the structural features of a negatively supercoiled DNA plasmid in complex with hTop1B have been monitored by following the twist (tw) between each base pair and plotting the Δtw as a function of time in Figure 5A. Δtw within undisrupted regions of the DNA was found to be slightly negative (−1.1°) due to the negative supercoiling, as would be expected. The plot also shows the presence of disrupted regions located throughout the supercoil (depicted in green). Interestingly, the regions showing the most evident disruption, with values lower than −20 to −30°, are those in proximity of the regions contacting the protein active site, the linker and the secondary binding site on subdomain III (Figure 5A), which we hypothesise is most likely due to the strong interaction between the DNA and the protein.
Figure 5.
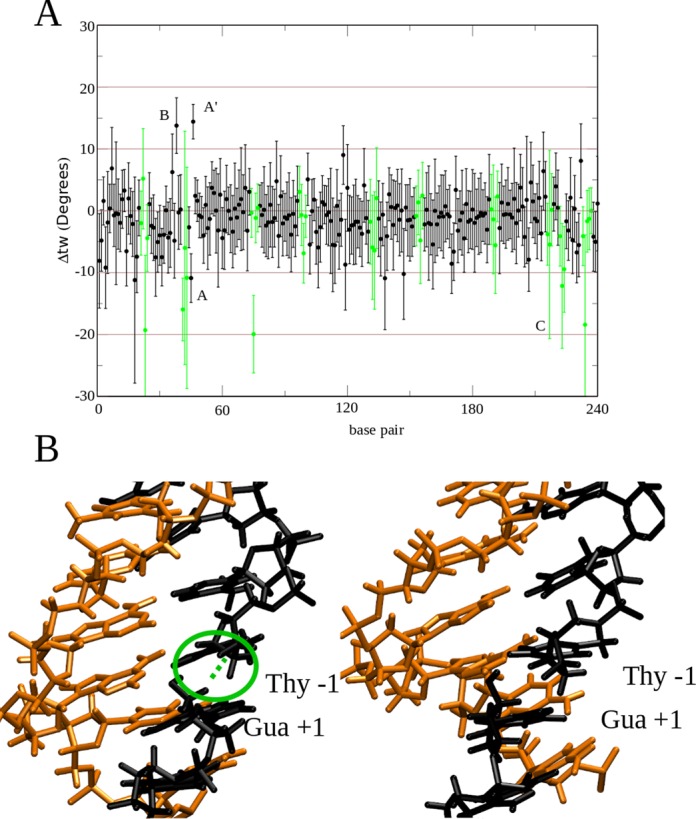
(A) Time average and S.D. error of Δtw for each base-pair step. Catalytic site (A and A′), linker binding site (B) and secondary (distal) binding site (C) are highlighted in the plot. ΔTw of the disrupted DNA bases are shown in green. (B) Cleavage site of the linear DNA (left panel) and of the supercoiled DNA plasmid (right panel). The presence of the hydrogen bond between the −1/+1 bases in the linear DNA is displayed in green.
The catalytic reaction of topoisomerase results in the cleavage of base pair step T−1s/G+1s, which allows the stressed negatively supercoiled DNA to relax. In the linear DNA, the hydrogen bond between the two ends of the cleavage site persists, in spite of the absence of the phosphate group (Figure 5B, left panel), while in supercoiled DNA, this hydrogen bond is lost (Figure 5B, right panel), most likely because the torsional stress has been partly relieved during the 50 ns timescale. The restoring torque within the supercoiled structure is reflected in the over-twisting of the twist at the cleavage site and the under-twisting of the next base pair step downstream, as shown in Figure 5B.
CONCLUSIONS
Molecular dynamics simulations of hTop1B in covalent complex with a negatively supercoiled plasmid containing 240 bp (Figure 1) have provided the first atomistic insight into the importance of supercoiling in mediating the interaction between the enzyme and its DNA substrate. While the 240 bp minicircle investigated in these calculations is certainly far smaller than either the plasmids generally employed in biochemical assays or the size of topological domains within the human genome (42), these tight loops may nevertheless be representative of the ends of plectonemically supercoiled loops and moreover are the largest DNA minicircles simulated atomistically in the presence of a bound protein in aqueous solution to date.
The simulations provide new insight into the preference for hTop1B for supercoiled over linear DNA substrates. Firstly, the calculations show that DNA supercoiling can influence the structure and the dynamical behavior of hTop1B linker, which is implicated in the controlled rotation mechanism associated with supercoil relaxation. In the presence of supercoiled DNA, the linker undergoes a rotation to a new conformation, which then has a tighter interaction with the DNA substrate. In addition, the protein active site has additional hydrogen bonding interactions with the supercoiled minicircle compared to the linear DNA fragment. Most striking, however, is the formation of a secondary DNA binding site which can only occur within complex DNA topologies, and which would not be detectable for a simple linear substrate. These simulations therefore emphasize the importance of DNA supercoiling in mediating in DNA recognition events, and consequently highlight the role of topology in genome regulation more generally.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We acknowledge PRACE for awarding us access to resource FERMI based in Italy at Cineca.
FUNDING
Associazione Italiana Ricerca Cancro (AIRC) with the Grant No. 10121 to AD. Funding for open access charge: Associazione Italiana Ricerca Cancro (AIRC) with the Grant No. 10121 to AD and the Grant No. BB/I019472/1 to SH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Champoux J.J. DNA topoisomerase I-mediated nicking of circular duplex DNA. Methods Mol. Biol. 2001;95:81–87. doi: 10.1385/1-59259-057-8:81. [DOI] [PubMed] [Google Scholar]
- 2.Wang J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 3.Corbett K.D., Berger J.M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 4.Stewart L., Redinbo M.R., Qiu X., Hol W.G., Champoux J.J. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 5.Castelli S., Coletta A., D'Annessa I., Fiorani P., Tesauro C., Desideri A. Interaction between natural compounds and human topoisomerase I. Biol. Chem. 2012;393:1327–1340. doi: 10.1515/hsz-2012-0240. [DOI] [PubMed] [Google Scholar]
- 6.Staker B.L., Feese M.D., Cushman M., Pommier Y., Zembower D., Stewart L., Burgin A.B. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005;48:2336–2345. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]
- 7.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 8.Redinbo M.R., Stewart L., Champoux J.J., Hol W.G. Structural flexibility in human topoisomerase I revealed in multiple non-isomorphous crystal structures. J. Mol. Biol. 1999;292:685–696. doi: 10.1006/jmbi.1999.3065. [DOI] [PubMed] [Google Scholar]
- 9.Redinbo M.R., Stewart L., Kuhn P., Champoux J.J., Hol W.G. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 10.Staker B.L., Hjerrild K., Feese M.D., Behnke C.A., Burgin A.B., Jr., Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog; Proc. Natl. Acad. Sci. U.S.A.; 2002. pp. 15387–15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart L., Ireton G.C., Parker L.H., Madden K.R., Champoux J.J. Biochemical and biophysical analyses of recombinant forms of human topoisomerase I. J. Biol. Chem. 1996;271:7593–7601. doi: 10.1074/jbc.271.13.7593. [DOI] [PubMed] [Google Scholar]
- 12.Fiorani P., Bruselles A., Falconi M., Chillemi G., Desideri A., Benedetti P. Single mutation in the linker domain confers protein flexibility and camptothecin resistance to human topoisomerase I. J. Biol. Chem. 2003;278:43268–43275. doi: 10.1074/jbc.M303899200. [DOI] [PubMed] [Google Scholar]
- 13.Fiorani P., Tesauro C., Mancini G., Chillemi G., D'Annessa I., Graziani G., Tentori L., Muzi A., Desideri A. Evidence of the crucial role of the linker domain on the catalytic activity of human topoisomerase I by experimental and simulative characterization of the Lys681Ala mutant. Nucleic Acids Res. 2009;37:6849–6858. doi: 10.1093/nar/gkp669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chillemi G., D'Annessa I., Fiorani P., Losasso C., Benedetti P., Desideri A. Thr729 in human topoisomerase I modulates anti-cancer drug resistance by altering protein domain communications as suggested by molecular dynamics simulations. Nucleic Acids Res. 2008;36:5645–5651. doi: 10.1093/nar/gkn558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Annessa I., Tesauro C., Fiorani P., Chillemi G., Castelli S., Vassallo O., Capranico G., Desideri A. Role of flexibility in protein-DNA-drug recognition: the case of Asp677Gly-Val703Ile topoisomerase mutant hypersensitive to camptothecin. J. Amino Acids. 2012;206083:1–8. doi: 10.1155/2012/206083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tesauro C., Morozzo Della Rocca B., Ottaviani A., Coletta A., Zuccaro L., Arno B., D'Annessa I., Fiorani P., Desideri A. Molecular mechanism of the camptothecin resistance of Glu710Gly topoisomerase IB mutant analyzed in vitro and in silico. Mol. Cancer. 2013;12:100–113. doi: 10.1186/1476-4598-12-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Annessa I., Tesauro C., Wang Z., Arno B., Zuccaro L., Fiorani P., Desideri A. The human topoisomerase 1B Arg634Ala mutation results in camptothecin resistance and loss of inter-domain motion correlation. Biochim. Biophys. Acta. 2013;1834:2712–2721. doi: 10.1016/j.bbapap.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 18.Arno B., D'Annessa I., Tesauro C., Zuccaro L., Ottaviani A., Knudsen B., Fiorani P., Desideri A. Replacement of the human topoisomerase linker domain with the plasmodial counterpart renders the enzyme camptothecin resistant. PLoS One. 2013;8:e68404. doi: 10.1371/journal.pone.0068404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madden K.R., Stewart L., Champoux J.J. Preferential binding of human topoisomerase I to superhelical DNA. EMBO J. 1995;14:5399–5409. doi: 10.1002/j.1460-2075.1995.tb00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z., Carey J.F., Champoux J.J. Mutational analysis of the preferential binding of human topoisomerase I to supercoiled DNA. FEBS J. 2009;276:5906–5919. doi: 10.1111/j.1742-4658.2009.07270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mancini G., D'Annessa I., Coletta A., Chillemi G., Pommier Y., Cushman M., Desideri A. Binding of an Indenoisoquinoline to the topoisomerase-DNA complex induces reduction of linker mobility and strengthening of protein-DNA interaction. PLoS One. 2012;7:e51354. doi: 10.1371/journal.pone.0051354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell J., Harris S.A. The thermodynamics of DNA writhe from atomistic molecular dynamics simulations. Phys. Rev. Lett. 2013;110:148105–148110. doi: 10.1103/PhysRevLett.110.148105. [DOI] [PubMed] [Google Scholar]
- 23.Lavery R., Moakher M., Maddocks J.H., Petkeviciute D., Zakrzewska K. Conformational analysis of nucleic acids revisited: curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 25.Powell M.J.D. An efficient method for finding the minimum of a function of several variables without calculating derivatives. Comput. J. 1964;17:155–162. [Google Scholar]
- 26.Duan Y., Wu C., Chowdhury S., Lee M.C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 27.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheatham T.E., Miller J.L., Fox T., Darden T.A., Kollman P.A. Molecular-dynamics simulations on solvated biomolecular systems—the particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J. Am. Chem. Soc. 1995;117:4193–4194. [Google Scholar]
- 29.Ceriotti M., Bussi G., Parrinello M. Langevin equation with colored noise for constant-temperature molecular dynamics simulations. Phys. Rev. Lett. 2009;102:020601–020604. doi: 10.1103/PhysRevLett.102.020601. [DOI] [PubMed] [Google Scholar]
- 30.Feller S.E., Zhang Y., Pastor R.W., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 31.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 32.Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 33.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithm for highly Efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell J.S., Laughton C.A., Harris S.A. Atomistic simulations reveal bubbles, kinks and wrinkles in supercoiled DNA. Nucleic Acids Res. 2011;39:3928–3938. doi: 10.1093/nar/gkq1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavery R., Zakrzewska K., Beveridge D., Bishop T.C., Case D.A., Cheatham T., 3rd, Dixit S., Jayaram B., Lankas F., Laughton C., et al. A systematic molecular dynamics study of nearest-neighbor effects on base pair and base pair step conformations and fluctuations in B-DNA. Nucleic Acids Res. 2010;38:299–313. doi: 10.1093/nar/gkp834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mancini G., D'Annessa I., Coletta A., Sanna N., Chillemi G., Desideri A. Structural and dynamical effects induced by the anticancer drug topotecan on the human topoisomerase I - DNA complex. PLoS One. 2010;5:e10934. doi: 10.1371/journal.pone.0010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shuman S., Bear D.G., Sekiguchi J. Intramolecular synapsis of duplex DNA by vaccinia topoisomerase. EMBO J. 1997;16:6584–6589. doi: 10.1093/emboj/16.21.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreno-Herrero F., Holtzer L., Koster D.A., Shuman S., Dekker C., Dekker N.H. Atomic force microscopy shows that vaccinia topoisomerase IB generates filaments on DNA in a cooperative fashion. Nucleic acids Res. 2005;33:5945–5953. doi: 10.1093/nar/gki906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel A., Yakovleva L., Shuman S., Mondragon A. Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site. Structure. 2010;18:725–733. doi: 10.1016/j.str.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramani R., Juul S., Rotaru A., Andersen F.F., Gothelf K.V., Mamdouh W., Besenbacher F., Dong M., Knudsen B.R. A novel secondary DNA binding site in human topoisomerase I unravelled by using a 2D DNA origami platform. ACS Nano. 2010;4:5969–5977. doi: 10.1021/nn101662a. [DOI] [PubMed] [Google Scholar]
- 41.Neale B.M., Kou Y., Liu L., Ma'ayan A., Samocha K.E., Sabo A., Lin C.F., Stevens C., Wang L.S., Makarov V., et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dekker J., Marti-Renom M.A., Mirny L.A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


