Abstract
Phospholipase D (PLD) enzymes play a double vital role in cells: they maintain the integrity of cellular membranes and they participate in cell signaling including intracellular protein trafficking, cytoskeletal dynamics, cell migration, and cell proliferation. The particular involvement of PLD in cell migration is accomplished: (a) through the actions of its enzymatic product of reaction, phosphatidic acid, and its unique shape-binding role on membrane geometry; (b) through a particular guanine nucleotide exchange factor (GEF) activity (the first of its class assigned to a phospholipase) in the case of the mammalian isoform PLD2; and (c) through protein-protein interactions with a wide network of molecules: Wiskott–Aldrich syndrome protein (WASp), Grb2, ribosomal S6 kinase (S6K), and Rac2. Further, PLD interacts with a variety of kinases (PKC, FES, EGF receptor (EGFR), and JAK3) that are activated by it, or PLD becomes the target substrate. Out of these myriads of functions, PLD is becoming recognized as a major player in cell migration, cell invasion, and cancer metastasis. This is the story of the evolution of PLD from being involved in a large number of seemingly unrelated cellular functions to its most recent role in cancer signaling, a subfield that is expected to grow exponentially.
Keywords: Cancer Biology, Cell Growth, Cell Invasion, Cell Signaling, Lipid, Lipid Metabolism, Metastasis, Phosphatidic Acid
Introduction
Phospholipase D (PLD)2 hydrolyzes phosphatidylcholine (PC) to yield phosphatidic acid (PA) and free choline (1). PLD is necessary for normal maintenance of cellular or intracellular membranes (2, 3), and it also participates in several physiological cellular functions, such as intracellular protein trafficking, cytoskeletal dynamics, membrane remodeling, and cell proliferation in mammalian cells and meiotic division and sporulation in yeast (4).
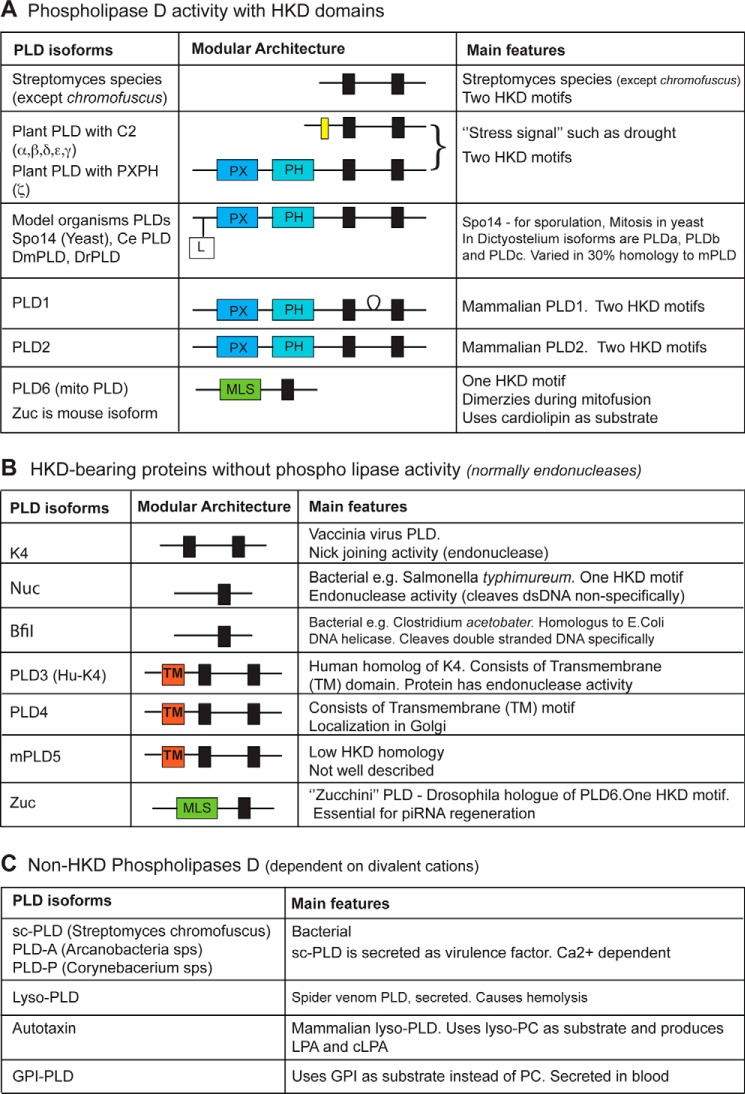
An important characteristic feature of members of the phospholipase D superfamily is the presence of two HKD motifs with the consensus amino acid sequence HXKX4DX6GSXN. However, there are exceptions where some of the PLDs lack these motifs and some have only one HKD motif. In Fig. 1, phospholipases are classified as follows: (a) active phospholipases with HKD motifs; (b) phospholipases with HKD motifs that lack lipase activity; and (c) non-HKD phospholipases.
FIGURE 1.
Classifications of PLDs. A, phospholipase D enzymes with HKD domains and intact lipase activity. B, phospholipase D enzymes with HKD domains that lack lipase activity. C, phospholipase D enzymes that lack HKD domains but are dependent on divalent cations for action. Parts of this panel are based on Ref. 99. Abbreviations: Phospholipase D activity, lipid phosphodiesterase toward PC (also phosphatidylethanolamine, phosphatidylserine, and cardiolipin) releasing PA and polar head; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Dr, Danio rerio; MLS, mitochondrial localization signal; mPLD5, mouse PLD5; sc, Streptomyces chromofuscus; GPI, glycosyl phosphatidyl inositol. Red boxes marked with TM, transmembrane domains; black boxes, HKD motifs (HXKX4DX6GSXN); yellow box, C2 domain.
Mammalian PLDs
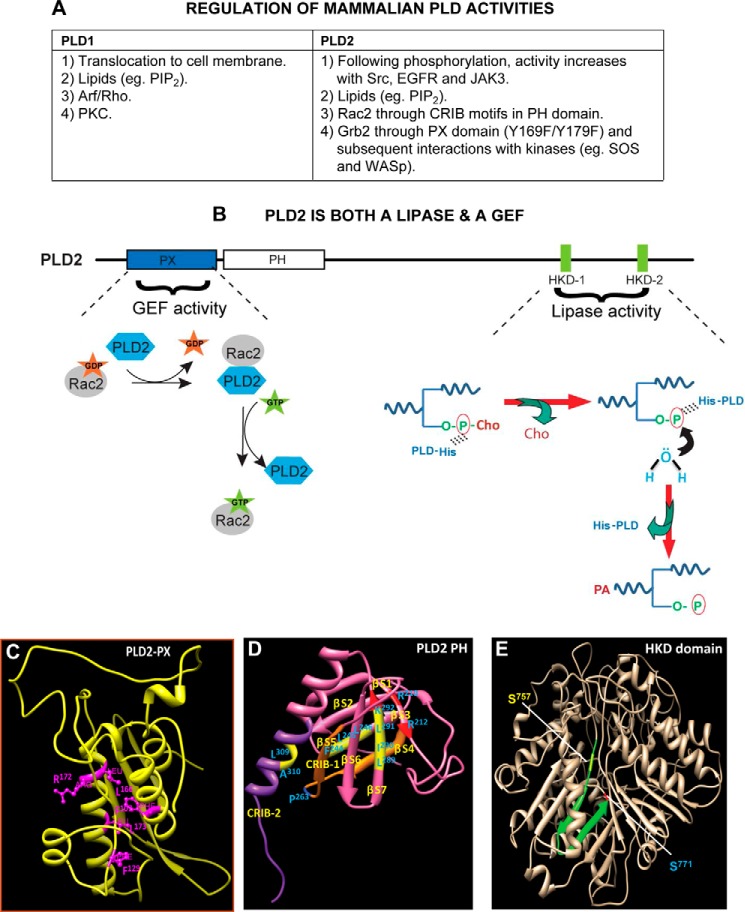
The two best characterized mammalian isoforms are PLD1 and PLD2 (5–8). Their genes share about 50% homology including two highly conserved phosphatidyltransferase HKD catalytic motifs that are requisite for catalytic activity. PLD1 and PLD2 also have phox homology (PX) and pleckstrin homology (PH) domains (2). A unique characteristic of PLD2 is that it possesses guanine nucleotide exchange factor (GEF) activity for the small GTPases Rac2 and Rho (9–11) (Fig. 2).
FIGURE 2.
Regulation of PLD enzymatic activities. A, list of specific regulation(s) of PLD1 and PLD2, the most studied mammalian isoforms. PIP2, phosphatidylinositol 4,5-bisphosphate; CRIB motif, Cdc42/Rac interactive binding motif. B, phospholipase D2 is a dual enzyme that catalyzes a lipase activity, as well as a guanine nucleotide exchange. Shown are the N-terminal PLD2-PX (where part of the GEF activity resides) and the C-terminal HKD1/2 domains (where lipase activity resides). For the GEF reaction, PLD2 causes Rac2-based GDP dissociation upon interaction with Rac2-GDP. In a second step, PLD2 stabilizes nucleotide-free Rac2 until GTP binds, after which PLD2 is released from the complex, leading to the activation of Rac2. For the lipase reaction, the catalytic HKD motifs of PLD2 fold around the substrate phosphatidylcholine (P-Cho). In the first step, a phosphatidyl-histidine intermediate is generated due to a nucleophilic attack of the histidine of the lipases on the phosphate of phosphatidylcholine. In the next step, the hydroxyl group of water attacks the phosphatidyl-histidine intermediate, leading to the formation of phosphatidic acid, at which time the enzyme is regenerated for the next cycle of PC breakdown. C–E, schematic drawings of main domains in the PLD2 structure. C, ribbon model of PLD2-PX domain noting key amino acids needed for the GEF activity. D, ribbon model of PLD2-PH domain that includes CRIB-1 and CRIB-2 needed for interaction with small GTPases (e.g. Rac2). E, ribbon model of PLD2-HKD domain. Serine residues that are mutated for inhibitor studies but that retain lipase activity are highlighted. The structures in C–E were generated by using protein prediction servers such as I-TASSER and Phyre (52). Once the structures were obtained, they were validated using biochemistry data available from both my laboratory and those of published authors in the field.
The existence of isoforms PLD3, PLD4, PLD5, and PLD6 has been recently reported. All of these PLD isoforms lack PX and PH domains and, therefore, are termed as “non-classical PLDs” (Fig. 1). However, all but PLD6 do have two HKD motifs, whereas PLD6 has only one such motif. PLD3 was originally identified as viral K4L homologue and, hence, named as Hu-K4. Despite the presence of two HKD motifs, no similarity with PLD1 or PLD2 was been found. SAM9 is a murine orthologue of Hu-K4, which is expressed in brain and localized in the endoplasmic reticulum (12), as is PLD4. No activity has been assigned for the products of PLD3 or PLD4 so far (13). PLD6 (also termed mitoPLD) is localized in mitochondrial outer membranes. It is required for mitochondrial fusion during which PLD6 located on one mitochondrion dimerizes with PLD6 located on a second mitochondria and hydrolyzes cardiolipin to generate PA (14, 15).
PLD and Its Product of Reaction, PA, Affect Intracellular Signaling Dramatically
PLD enzymes are involved in a large variety of physiological cellular functions, and I will consider here three molecular mechanisms by which this occurs through their lipase action, through the GEF activity (in the case of PLD2), and through protein-protein interactions that initiate signaling independently of the enzymatic activities. PA is the catalytic product of the lipase reaction with phospholipids in the cell membrane, particularity PC. The biggest dilemma concerning the function of PLD is the lack of clarity over a PA binding site on target proteins and thus understanding of the mechanism of downstream action. This is particularly concerning given the plethora of PA-binding proteins that have been identified. Three studies can be cited where it is indicated that PA binds to the positively charged amino acid residues or surface-exposed hydrophobic residues or both in the target proteins (16–18), but clearly a specific PA binding site is lacking, and once found, the field should advance considerably. Related to this, the integration of PA and PLD has been addressed only in one review by Jang et al (19). The authors found that 9 out of 50 binding partners are common between PA and PLD. Based on this, the authors suggested a complex regulation patterns between PLD, PA, and their binding partners. This paucity of intersection indicates that indeed, PLD is an enzyme that cannot be confined to the sole actions derived from its enzymatic activity and, as I will discuss later, the protein-protein interactions involving the whole PLD or parts of the PLD molecular are central to PLD signaling, particularly in cell migration.
A further interest in this PA-PLD topic has become highlighted by the discovery of PLD2 as a GEF that makes more challenging a demarcation between lipase-mediated and/or GEF-mediated functions of PLD2 (9). However, the finding that PA regulates the GEF activity of PLD2 adds a further level of sophistication in the regulation of this enzyme that is necessary considering the key role it has in cellular functions (20). Further, with the discovery of the GEF catalytic site, it is now possible to use lipase-inactive or GEF-inactive mutants to determine lipase or GEF-mediated functions (11).
PLD Signaling as a Phosphoprotein and Its Interaction with Tyrosine Kinases
PLD is a phosphoprotein whose phosphorylation is regulated by kinases and phosphatases (Fig. 3). Protein kinase C (PKC) interacts with both PLD1 and PLD2 and enhances lipase activity (21, 22). PKCδ phosphorylates PLD2 by direct association, thereby aiding in the localization of PLD2 at lamellipodia and promoting integrin-mediated cell spreading (23). A physical association between PLD2 and PLCγ occurs in an EGF-dependent fashion and enhances PLD activity (24). Cdk5-mediated phosphorylation and activation of PLD2 is responsible for EGF-dependent insulin secretion (25). Phosphorylated PLD2 forms a ternary complex with both PTP1b and Grb2, a critical signal transducer of EGFR, which links PLD2 to cellular proliferation and the MAPK and Ras/Erk pathways (26).
FIGURE 3.
Multiple signaling pathways result in PLD2 contributing to many different steps in this process. As shown in the schematic drawing, PLD2 has many functions in the cell. The invasive phenotype of MDA-MB-231 cells is mediated by PLD2 under control of JAK3 and EGFR. Serum deprivation of cells results in an up-regulated EGFR/JAK3/PLD2-PA system, which is extremely sensitive to JAK3 and PLD2 inhibitors. JAK3 and FES greatly enhance PLD activity following protein-protein interaction through the SH2 domain and the Tyr-415 residue of PLD2. PA enhances FES activity in cancer cells, which provides a positive activation loop between FES and PLD2. PLD2 anchors WASp at the phagocytic cup through Grb2 following protein-protein interactions and also activates it, making key lipids available locally. The heterotrimer PLD2-Grb2-WASp then enables actin nucleation at the phagocytic cup and phagocytosis, which are at the center of the innate immune system function. PLD2 binds to the small GTPase Rac2, which results in a PLD2-GEF activity that switches Rac2 from the GDP-bound to the GTP-bound states, which impacts actin and cell motility. PLD-derived PA binds to ribosomal S6 kinase (S6K), whose enzymatic activity regulates the activation of actin nucleation, and to nuclear receptors (NR) in the nucleus, which contributes to synthesis of EGFR protein and increases cell proliferation.
Although PLD2 can be phosphorylated by the serine/threonine kinase AKT at residue Thr-175, which serves to up-regulate DNA synthesis, more typically PLD is known as a substrate for many receptor (EGFR and PDGFR) and non-receptor tyrosine kinases (Src and JAK3). Choi et al. (27) have found that PLD2 is specifically phosphorylated on residues Tyr-11, Tyr-14, Tyr-165, and Tyr-470. Phosphorylation targets within the PLD2 molecule have been mapped that are vital to its regulation as a lipase and thus correlated in vitro to at least three different tyrosine kinases, EGFR, Src, and Janus kinase 3 (JAK3) (28), that target Tyr-296, Tyr-511, and Tyr-415, respectively, and that yield either positive or negative effects on the lipase.
Elevation of either PLD1 or PLD2 has the potential to transform rat fibroblasts and contribute to cancer progression of the malignant phenotype in cells that also have elevated levels of EGFR or Src tyrosine kinases (29). Contrarily, it has been hypothesized that PLD2 activity in certain breast cancer cell lines is comparatively low when compared with non-cancerous cells or other breast cancer cell lines because it is down-regulated by tyrosyl phosphorylation at Tyr-296 via EGFR (28). This low level of PLD activity can be increased by in vitro treatment with either JAK3 or Src. Src participates in the activation of PLD through the Ras pathway and the kinases Fyn and Fgr but not Lyn (27).
There are also protein-protein interactions between PLD2 and JAK3, as well as with another tyrosine kinase, FES, which is implicated in the proliferation of breast cancer cells (30). The PLD2-JAK-FES inter-regulation of this lipase and these kinases is implicated in the high proliferation rate of MDA-MB-231 breast cancer cells (30). Additionally, PLD interacts with type-Ia phosphatidylinositol-4-phosphate 5 (PI4P5) kinase. In turn, phosphatidylinositol 4,5-bisphosphate (PIP2) generated by phosphatidylinositol-4-phosphate 5 kinase is essential for PLD activity (31).
The Complex Interaction between Small GTPases with PLD
GTPases regulate PLD activity, and PLD in turn regulates GTPases (32). For GTPases regulating PLD, it was found initially that Arf1 and RalA directly interact with and activate PLD1 (33). Several other GTPases, such as RhoA, RhoB, Rac1, Rac2, and Cdc42, activate PLD. The Switch I region of Rho A directly interacts with the C-terminal region of PLD1 (34, 35). These GTPases must be GTP-bound to stimulate/activate PLD because mutation of the Rho binding site on PLD1 abrogates PLD1/Arf interaction.
There is a dual (positive and negative) effect of Rac2 on PLD2 activity that is implicated in regulation of chemotaxis. Rac2 localizes in vivo at the leading edge of leukocyte pseudopodia, with PLD2 being physically posterior to a wave of Rac2. This impedes the membrane association of PLD2 and thereby inhibits the lipase activity (36). Rac2 has a negative effect on PLD2 gene expression as well (37). Regulation of PLD2 activity by the small GTPase Sar1p is implicated in COPII-mediated endoplasmic reticulum export (38) (39). Further, PLD2 acts as a GTPase-activating factor (GAP) for dynamin (40).
PLD2 Is a GEF
Not only is PLD2 regulated by small GTPASES, as just discussed, but PLD2 also regulates GTPases; in fact, PLD2 is a GEF for small GTPases (Fig. 2). PLD2 but not PLD1 is upstream to small GTPases, such as Rac1, RhoA, and Rac2 via its GEF activity or via a PA-dependent manner (9, 10, 23). PLD2 possesses a GEF activity for the small GTPase Rac2 or RhoA (9, 10). After the discovery of the GEF activity of PLD2, PLD2-mediated functions are more challenging in terms of demarcating the lipase- or GEF-mediated functions of PLD2. By extensive mutational analysis, my laboratory discovered the essential amino acid residues for GEF catalysis: Phe-107, Phe-129, Leu-166, Arg-172, and Leu-173 (Fig. 2C) (11). This information is valuable in using either the mutant lipase-inactive PLD2 or the mutant GEF-inactive PLD2 to differentiate between varieties of PLD2-mediated functions. PLD2-GEF activity correlates with Ras activation in highly proliferative and metastatic breast cancer cells (41). This is a very important area to be pursued further, as not only mutations in Ras, but also hyperactivation of Ras, promote tumorigenesis (42).
PLD2 is a dual enzyme with GEF and lipase activities embedded in the N- and C-terminal regions, respectively. Very interestingly, for the dual GEF/lipase activity, the products of the lipase and the GEF reactions regulate the alternate activity. This involves the dual effect of PA on PLD2-GEF activity and a temporal switch in lipase and GEF activities (20).
WASp, Grb2, and Rac2: The Mechanism by Which PLD Acts on Cell Migration
PLD is an important player in the regulation of actin cytoskeletal regulation and, as such, a key element for cell migration (Fig. 4). A component of this effect is due to the product of its reaction, PA, and another is through protein-protein interaction with the intracellular motility machinery. PA regulates actin and leukocyte cell migration because lamellipodia structures and membrane ruffles can be hindered if PLD is inhibited (43). PLD activation plays a vital role in actin cytoskeleton formation (4). ARF6 activation by ARNO stimulates epithelial cell migration through Rac1 and PLD (44), and PLD is necessary for actin localization and actin-based motility in Dictyostelium via phosphatidylinositol 4,5-bisphosphate (45). PLD2 mediates adhesion via regulation of cell surface integrins (46) and is involved in cytoskeletal organization, macrophage phagocytosis and neutrophil recruitment (43, 47, 48). In leukocytes, PA is a chemoattractant that acts via ribosomal S6 kinase (S6K) (49) and Fer (17), and 5-fluoro-2-indolyl des-chlorohalopemide (FIPI) is a PLD inhibitor that alters cell spreading and inhibits chemotaxis (50). DOCK2 is controlled by PLD during neutrophil chemotaxis (51) and, conversely Rac2 controls PLD2 regulation during the onset and termination of chemotaxis (36).
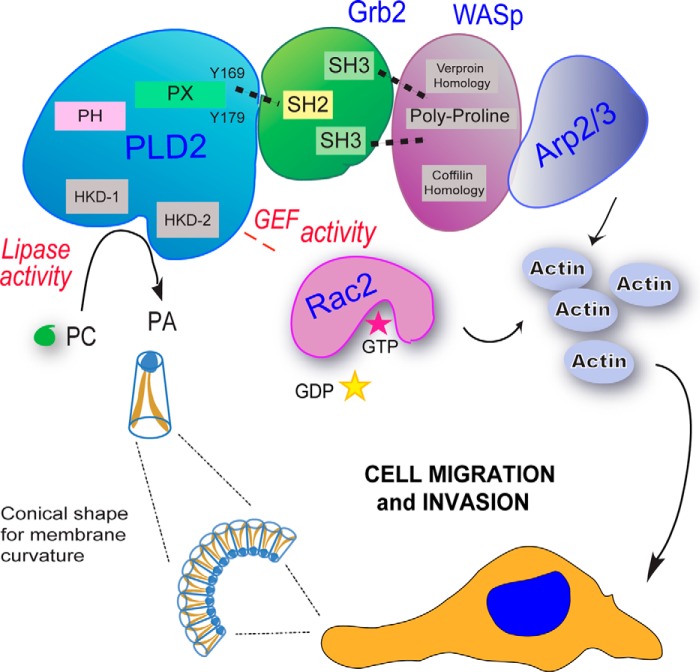
FIGURE 4.
Role of PLD in cell migration. PLD is a key component of cell migration of both cancer and inflammatory cells that includes a variety of different intracellular events (such as cytoskeletal organization, vesicle trafficking, endocytosis/exocytosis, etc.) using both PA-mediated mechanisms and protein-protein interactions. The figure shows that PA provides a curvature in the cell membrane that is conducive to formation of lamellipodia. On the other hand, PA is a second messenger on its own right, and carries the signal from the membrane to several proteins in the cytoplasm and in the nucleus. The GEF function of PLD2 targets small GTPases involved in cell migration, such as Rac2. Lastly, multiple protein-protein interactions have been described with PLD2 and motility proteins, such as Grb2-WASp.
There are at least two ways by which PLD is connected to cell migration. The first involves Rho family GTPases (10, 53). The second way is through PLD-mediated cell migration, which is also regulated by specific protein-protein interactions, such as Grb2, which is a docking protein for PLD2 that is dependent on the SH2 domain of Grb2 and involves Tyr-169 and Tyr-179 of PLD2 (26). Upon interaction, Grb2 promotes lipase activity and regulates the localization of PLD2 (54). PLD2 recruits WASp to the plasma membrane and enhances phagocytic cup formation via Grb2 (55) (Fig. 4C). PLD activity and Rac2 cooperation are increased in macrophages following binding of PLD2 to Grb2, which stimulates actin polymerization and membrane ruffling (56). PLD2/Grb2-mediated chemotaxis and phagocytosis of RAW264.7/LR5 macrophages is dependent upon Grb2 interacting with other proteins, such as Rac2, PTP1b, and especially WASp (54, 57).
PLD Close Interaction with Other Lipid Enzymes
PLD-derived PA binds to and regulates sphingosine kinase 1 (SK1) (58). The product of SK1, sphingosine 1-phosphate, acts as a survival signal in cancer and also mediates tumorigenesis (59, 60). More importantly sphingosine-1-phosphate is also involved in transactivation of various growth factors (61) that are upstream of PLD activity. This suggests the possibility of cross-talk between SK1/sphingosine-1-phosphate and possibly PLD/PA pathways that might play a crucial role in cancer progression.
Lipid phosphate phosphatases (LPPs) hydrolyze a variety of phospholipids including PA (62). LPPs possess an inhibitory effect on lysophosphatidic acid-mediated PLD activity (63). Although LPP expression is low, PLD levels are high in a variety of cancers (64). In addition to the PLD inhibitors, the cross-signaling between LPPs and PLD/PA thus seems to be an area of interest in cancer perspective.
PLD in Tumor and Cancer Metastasis
PLD2 overexpression leads to elevated adhesion invasion and metastasis in a lymphoma cell line (65). Further, elevated PLD activity, as well as expression, has been reported in a wide variety of cancers, such as gastric, colorectal, renal, stomach, esophagus, lung, and breast. In addition, a PLD2 gene polymorphism was shown to be prevalent in colorectal cancer, where it was demonstrated that a C → T mutation resulting in Thr → Ile is associated with colorectal cancer. However, lipase activity was not affected with this mutation (66). A clear correlation was observed between PLD2 expression and the tumor size, as well as patient survival, and it has been proposed that PLD2 might be a prognostic indicator in colon cancers (67).
PLD also acts as a survival signal for cancers, such as renal cancer cells where PLD regulates hypoxia-inducible factor 1a (HIF-1a) at the translation level, in a von Hippel-Lindau (vHL)-independent fashion, and promotes cancer cell proliferation (68). In ovarian cancer cells, PLD is shown to be essential for agonist-induced lysophosphatidic acid production and promotes motility, growth, and proliferation (69). PLD2 enhances the expression of anti-apoptotic proteins such as Bcl-2 and Bcl-xL in lymphoma cells (70).
PLD signaling with other cancer regulators (Ras, PDGF, TGF, and kinases) provides survival signals, thereby promoting tumorigenesis (71). PLD2 is linked to the progression of EWS-Fli sarcoma due to its cross-talk with PDGF-mediated signaling (72). A transmodulation between PLD2 and the oncogenic kinase RET is evident in thyroid cancer cells where PLD2 enhances STAT3 phosphorylation and transcriptional activation (73). A role for kinase-mediated regulation of PLD2 was seen in cell proliferation (74).
Recent Developments in Cancer and PLD Research
Some important clues indicating a role for PLD in cancer were given by the fact that PLD was involved in cell proliferation and in cell invasion. Additionally, it has been demonstrated that active PLD enhances lymphoma cell metastasis, and inactive PLD2 inhibits metastasis (75), MMP-2 expression, and glioma cell invasion (76). PLD2, EGFR, and JAK3 are involved in common pathways that maximize cancer cell invasion (77, 78). Several PLD-specific inhibitors interfere with cancer cell invasion (79). Because of this role of PLD in cell migration, chemotaxis, and cell invasion, the role of PLD in cancer has been significantly expanded.
The last 5–6 years have witnessed an exponential growth in research in PLD and cancer. PLD inhibitors have a negative effect on tumor growth in mice (75, 80, 81). A PLD2-specific inhibitor (ML298) and a dual PLD1/PLD2 inhibitor (ML299) were both found to have a potential role in treating brain cancer (82). FES and JAK3 were found to elevate PLD2 expression, and this interaction was found to be a reason for the elevated proliferation rate of MDA-MB-231 cells (30).
Elevated levels of PA are observed in colorectal tumors, which are driven by the Wnt/β-catenin pathway. In the same study, it has been reported that PLD1 and PLD2 are targets of the Wnt/β-catenin pathway (83–85). A potential therapeutic target for osteolytic bone metastases in lung cancer patients has been proposed (86). PLD inhibitors inhibit the invasion of breast cancer cells in culture or their proliferation (87, 88).
Cell Invasion and Metastasis, Central to the Tumorigenic Potency of PA
As indicated earlier, PLD2 has a direct role in cell migration, and it is also key to cell invasion and metastasis (65, 75, 80). Knowledge of the particular molecular mechanisms of PLD in cancer tissues now enable us to take advantage of the many new biological tools, and these mechanisms are only now coming to light. A tumorigenic role for PLD2 was established by xenotransplantation of human breast cancer cells into SCID mice (80). Primary tumors from xenotransplanted mice were larger, grew faster, and developed more lung metastases. Micro-osmotic pumps that delivered PLD-specific small-molecule inhibitors were implanted into xenotransplanted SCID mice, which inhibited primary tumor growth and lung metastases. Ablation of PLD1 in the tumor environment compromised the neovascularization and growth of tumors (81). PLD1 deficiency reduced tumor angiogenesis in a xenograft model. In addition, mice lacking PLD1 or treatment with 5-fluoro-2-indolyl des-chlorohalopemide incurred fewer lung metastases than did wild-type mice.
Very recent studies have indicated that PLD1 specific inhibitors prefer (S)-configuration on the methyl carbon adjacent to the amide linkage, whereas PLD2 selective inhibitors prefer spiro ring fused with lactam. Based on these factors, 4-aminopyrazolopyrimidines (used as kinase inhibitors) have been developed, which have IC50 values of 5 and 15 nm for PLD1 and PLD2, respectively (89). Although targeting PLD isoforms is the main focus for abrogating the effects of PLD on cancer growth, using indirect inhibitors of upstream regulators of PLD is another approach. Rebamipide, an antiulcer drug, has been shown to inhibit Helicobacter pylori-induced PLD1 expression and activity in gastric cancer cells (90).
Inhibition of PLD2 but not PLD1 or diacylglycerol kinase (DGK) inhibited nuclear ERK activity in a variety of cancer cells, causing a reduction in ERK-targeted gene expression. This suggests that PLD2 is upstream of ERK and that targeting PLD2 will further suppress ERK-mediated cancer cell growth factor signaling (91). Breast cancer cells expressing an oncogene FAM83B have been shown to possess high PLD1 but not PLD2 activity. In addition, PLD1 activity is an essential factor required for the transformation mediated by Ras and FAM83B (92).
One of the major problems in cancer treatment is resistance of cancer cells to chemotherapy and radiation. Radiation in combination with PLD inhibition (PLD1 and PLD2) has been shown to be an efficient way to improve radiosensitivity of the human breast cancer cell line, MDA-MB-231 (93). In agreement with the involvement of PLD in inducing resistance of cancer cells, it has been shown in laryngeal cancer cells that membrane-associated estrogen receptor α36 (ERα36) activates PKC, which in turn enhances PLD activity via estradiol (E2) (94).
Unresponsiveness of cancer cells to upstream chemokines makes them more aggressive. In this context, PLD1/Arf signaling has been demonstrated as one of the key factors that contribute to this unresponsiveness of leukemia cells (95). The activation of PLD improves chemotherapeutic sensitivity via reducing the gene expression of multidrug resistance (96).
The involvement of PLD in inhibiting multidrug resistance (99) is in contradiction with other studies that support the role of PLD in making the cancer cells resistant (98). One possibility might be that this phenomenon of PLD might be cell/tissue- or cancer-dependent mechanism rather than a general mechanism. However, it is essential to confirm the chemotherapeutic sensitivity-promoting nature of the otherwise cancer-promoting PLD2. At any rate, a more conclusive explanation awaits. This is important because a compelling case will be needed for use of PLD inhibitors in the treatment of cancer, even if such information is used to determine which cancers are likely to respond to such inhibitors in a manner that has therapeutic utility, i.e. leading to a stratified approach.
Cancer, Autophagy, and PLD
Despite its role in promoting cancer, the mechanism behind PLD-mediated cancer is not clearly understood, and some subtopics are not entirely settled yet. Take for example the role of PLD in autophagy and cancer. On the one hand, PLD appears to inhibit autophagy (97) because PLD/PA has been shown to activate mammalian target of rapamycin (mTOR), which is an inhibitor of autophagy. Therefore, PLD inhibitors increase autophagy, which in this case leads to cell death. In contrast, another group of researchers (98) has indicated that PLD activates autophagy as inhibition of PLD reduces autophagy, leading to a decrease in cell viability, whereby autophagy might be a protective cell survival mechanism. In addition, these cancers might have different dependence on AKT or mTOR for regulating the cellular outcome of the autophagic response in a particular cancer. This discrepancy might be a result of dependence on cell or cancer type. Because the research on the effects of PLD on autophagy is novel, it is very important to investigate the same in various types of cancers and determine whether it is a general phenomenon or cancer type-dependent.
Remaining Challenges
At least four challenges remain for the immediate future. First, there is no crystal structure of mammalian PLD2 currently. To understand the mechanism underlying the multiple roles of PLD2 as a lipase, GEF, and as a signaling protein by itself via protein interactions, it is essential to obtain a three-dimensional structure of PLD2. This will further facilitate the investigation of PLD2-mediated biochemical functions and develop novel PLD molecule-specific inhibitors or modulators that can be developed to regulate PLD activities/protein interactions.
Second, although PLD2 activity is shown to be necessary for cellular processes like chemotaxis and phagocytosis, deregulated PLD2 levels were reported in several cancers such as breast, colorectal, and renal cancers. All this suggests increasing demand for the understanding of the in vivo mechanisms for which there is an abundant amount of information regarding in vitro and cultured cells, but it remains to be seen which of those are applicable to in vivo cancer studies.
Third, and as studies with autophagy and ARF have amply demonstrated, PLD might be cell/tissue- or cancer-dependent mechanism rather than a general mechanism. Genome sequencing of specific cancer cells derived from patients at several stages of the disease should clarify this, and this should provide a better understanding of which PLD inhibitor (or appropriate therapy) should be followed.
Fourth, it is becoming evident that several lipid enzymes are deregulated in cancer tissues. It will probably not come as a surprise that the effects of PLD are not alone, but rather, they are the result of cooperation with other lipid enzymes, particularly sphingosine kinase and/or lipid phosphate phosphatases.
In conclusion, further study of the pathways and mechanisms in which PLD is key to cancer will be developed from patients representing the different stages of breast cancer (0 to IV). In addition, more studies on PLD inhibitors will need to be conducted in order for PLD inhibitors to be possibly used clinically in cancer.
This work was supported, in whole or in part, by National Institutes of Health Grant by HL056653-14 and 13GRNT17230097 from the American Heart Association (to J. G.-C.). This is the first article in the Thematic Minireview Series “Phospholipase D and Cancer.”
- PLD
- phospholipase D
- PC
- phosphatidylcholine
- PA
- phosphatidic acid
- PH
- pleckstrin homology
- PX
- phox homology
- GEF
- guanine nucleotide exchange factor
- EGFR
- EGF receptor
- LPP
- lipid phosphate phosphatase
- WASp
- Wiskott–Aldrich syndrome protein
- SH
- Src homology domain
- Arf
- ADP ribosylation factor.
REFERENCES
- 1. Wang X., Xu L., Zheng L. (1994) Cloning and expression of phosphatidylcholine-hydrolyzing phospholipase D from Ricinus communis L. J. Biol. Chem. 269, 20312–20317 [PubMed] [Google Scholar]
- 2. Frohman M. A., Sung T. C., Morris A. J. (1999) Mammalian phospholipase D structure and regulation. Biochim. Biophys. Acta 1439, 175–186 [DOI] [PubMed] [Google Scholar]
- 3. Hammond S. M., Jenco J. M., Nakashima S., Cadwallader K., Gu Q., Cook S., Nozawa Y., Prestwich G. D., Frohman M. A., Morris A. J. (1997) Characterization of two alternately spliced forms of phospholipase D1: activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C-α. J. Biol. Chem. 272, 3860–3868 [DOI] [PubMed] [Google Scholar]
- 4. Powner D. J., Wakelam M. J. (2002) The regulation of phospholipase D by inositol phospholipids and small GTPases. FEBS Lett. 531, 62–64 [DOI] [PubMed] [Google Scholar]
- 5. Colley W. C., Altshuller Y. M., Sue-Ling C. K., Copeland N. G., Gilbert D. J., Jenkins N. A., Branch K. D., Tsirka S. E., Bollag R. J., Bollag W. B., Frohman M. A. (1997) Cloning and expression analysis of murine phospholipase D1. Biochem. J. 326, 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kodaki T., Yamashita S. (1997) Cloning, expression, and characterization of a novel phospholipase D complementary DNA from rat brain. J. Biol. Chem. 272, 11408–11413 [DOI] [PubMed] [Google Scholar]
- 7. Hammond S. M., Altshuller Y. M., Sung T. C., Rudge S. A., Rose K., Engebrecht J., Morris A. J., Frohman M. A. (1995) Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J. Biol. Chem. 270, 29640–29643 [DOI] [PubMed] [Google Scholar]
- 8. Lopez I., Arnold R. S., Lambeth J. D. (1998) Cloning and initial characterization of a human phospholipase D2 (hPLD2): ADP-ribosylation factor regulates hPLD2. J. Biol. Chem. 273, 12846–12852 [DOI] [PubMed] [Google Scholar]
- 9. Mahankali M., Peng H. J., Henkels K. M., Dinauer M. C., Gomez-Cambronero J. (2011) Phospholipase D2 (PLD2) is a guanine nucleotide exchange factor (GEF) for the GTPase Rac2. Proc. Natl. Acad. Sci. U.S.A. 108, 19617–19622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jeon H., Kwak D., Noh J., Lee M. N., Lee C. S., Suh P. G., Ryu S. H. (2011) Phospholipase D2 induces stress fiber formation through mediating nucleotide exchange for RhoA. Cell. Signal. 23, 1320–1326 [DOI] [PubMed] [Google Scholar]
- 11. Mahankali M., Henkels K. M., Alter G., Gomez-Cambronero J. (2012) Identification of the catalytic site of phospholipase D2 (PLD2) newly described guanine nucleotide exchange factor activity. J. Biol. Chem. 287, 41417–41431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Munck A., Böhm C., Seibel N. M., Hashemol Hosseini Z., Hampe W. (2005) Hu-K4 is a ubiquitously expressed type 2 transmembrane protein associated with the endoplasmic reticulum. FEBS J. 272, 1718–1726 [DOI] [PubMed] [Google Scholar]
- 13. Otani Y., Yamaguchi Y., Sato Y., Furuichi T., Ikenaka K., Kitani H., Baba H. (2011) PLD$ is involved in phagocytosis of microglia: expression and localization changes of PLD4 are correlated with activation state of microglia. PLoS One 6, e27544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi S. Y., Huang P., Jenkins G. M., Chan D. C., Schiller J., Frohman M. A. (2006) A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 8, 1255–1262 [DOI] [PubMed] [Google Scholar]
- 15. Huang H., Frohman M. A. (2009) Lipid signaling on the mitochondrial surface. Biochim. Biophys. Acta 1791, 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stace C. L., Ktistakis N. T. (2006) Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta 1761, 913–926 [DOI] [PubMed] [Google Scholar]
- 17. Itoh T., Hasegawa J., Tsujita K., Kanaho Y., Takenawa T. (2009) The tyrosine kinase Fer is a downstream target of the PLD-PA pathway that regulates cell migration. Sci. Signal. 2, ra52. [DOI] [PubMed] [Google Scholar]
- 18. Kooijman E. E., Tieleman D. P., Testerink C., Munnik T., Rijkers D. T., Burger K. N., de Kruijff B. (2007) An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J. Biol. Chem. 282, 11356–11364 [DOI] [PubMed] [Google Scholar]
- 19. Jang J. H., Lee C. S., Hwang D., Ryu S. H. (2012) Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners. Prog. Lipid Res. 51, 71–81 [DOI] [PubMed] [Google Scholar]
- 20. Mahankali M., Henkels K. M., Gomez-Cambronero J. (2013) A GEF-to-phospholipase molecular switch caused by phosphatidic acid, Rac and JAK tyrosine kinase that explains leukocyte cell migration. J. Cell Sci. 126, 1416–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J. S., Exton J. H. (2004) Regulation of phospholipase D2 activity by protein kinase C α. J. Biol. Chem. 279, 22076–22083 [DOI] [PubMed] [Google Scholar]
- 22. Oishi K., Takahashi M., Mukai H., Banno Y., Nakashima S., Kanaho Y., Nozawa Y., Ono Y. (2001) PKN regulates phospholipase D1 through direct interaction. J. Biol. Chem. 276, 18096–18101 [DOI] [PubMed] [Google Scholar]
- 23. Chae Y. C., Kim K. L., Ha S. H., Kim J., Suh P. G., Ryu S. H. (2010) Protein kinase Cδ-mediated phosphorylation of phospholipase D controls integrin-mediated cell spreading. Mol. Cell. Biol. 30, 5086–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jang I. H., Lee S., Park J. B., Kim J. H., Lee C. S., Hur E. M., Kim I. S., Kim K. T., Yagisawa H., Suh P. G., Ryu S. H. (2003) The direct interaction of phospholipase C-γ1 with phospholipase D2 is important for epidermal growth factor signaling. J. Biol. Chem. 278, 18184–18190 [DOI] [PubMed] [Google Scholar]
- 25. Lee H. Y., Jung H., Jang I. H., Suh P. G., Ryu S. H. (2008) Cdk5 phosphorylates PLD2 to mediate EGF-dependent insulin secretion. Cell. Signal. 20, 1787–1794 [DOI] [PubMed] [Google Scholar]
- 26. Di Fulvio M., Lehman N., Lin X., Lopez I., Gomez-Cambronero J. (2006) The elucidation of novel SH2 binding sites on PLD2. Oncogene 25, 3032–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choi W. S., Hiragun T., Lee J. H., Kim Y. M., Kim H. P., Chahdi A., Her E., Han J. W., Beaven M. A. (2004) Activation of RBL-2H3 mast cells is dependent on tyrosine phosphorylation of phospholipase D2 by Fyn and Fgr. Mol. Cell. Biol. 24, 6980–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henkels K. M., Peng H. J., Frondorf K., Gomez-Cambronero J. (2010) A comprehensive model that explains the regulation of phospholipase D2 activity by phosphorylation-dephosphorylation. Mol. Cell. Biol. 30, 2251–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu L., Frankel P., Jackson D., Rotunda T., Boshans R. L., D'Souza-Schorey C., Foster D. A. (2003) Elevated phospholipase D activity in H-Ras- but not K-Ras-transformed cells by the synergistic action of RalA and ARF6. Mol. Cell. Biol. 23, 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ye Q., Kantonen S., Henkels K. M., Gomez-Cambronero J. (2013) A new signaling pathway (JAK-Fes-phospholipase D) that is enhanced in highly proliferative breast cancer cells. J. Biol Chem. 288, 9881–9891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Divecha N., Roefs M., Halstead J. R., D'Andrea S., Fernandez-Borga M., Oomen L., Saqib K. M., Wakelam M. J., D'Santos C. (2000) Interaction of the type Iα PIPkinase with phospholipase D: a role for the local generation of phosphatidylinositol 4, 5-bisphosphate in the regulation of PLD2 activity. EMBO J. 19, 5440–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peng H. J., Henkels K. M., Mahankali M., Dinauer M. C., Gomez-Cambronero J. (2011) Evidence for two CRIB domains in phospholipase D2 (PLD2) that the enzyme uses to specifically bind to the small GTPase Rac2. J. Biol. Chem. 286, 16308–16320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim J. H., Lee S. D., Han J. M., Lee T. G., Kim Y., Park J. B., Lambeth J. D., Suh P. G., Ryu S. H. (1998) Activation of phospholipase D1 by direct interaction with ADP-ribosylation factor 1 and RalA. FEBS Lett. 430, 231–235 [DOI] [PubMed] [Google Scholar]
- 34. Bae C. D., Min D. S., Fleming I. N., Exton J. H. (1998) Determination of interaction sites on the small G protein RhoA for phospholipase D. J. Biol. Chem. 273, 11596–11604 [DOI] [PubMed] [Google Scholar]
- 35. Yamazaki M., Zhang Y., Watanabe H., Yokozeki T., Ohno S., Kaibuchi K., Shibata H., Mukai H., Ono Y., Frohman M. A., Kanaho Y. (1999) Interaction of the small G protein RhoA with the C terminus of human phospholipase D1. J. Biol. Chem. 274, 6035–6038 [DOI] [PubMed] [Google Scholar]
- 36. Peng H. J., Henkels K. M., Mahankali M., Marchal C., Bubulya P., Dinauer M. C., Gomez-Cambronero J. (2011) The dual effect of Rac2 on phospholipase D2 regulation that explains both the onset and termination of chemotaxis. Mol. Cell. Biol. 31, 2227–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Speranza F. J., Mahankali M., Gomez-Cambronero J. (2013) Macrophage migration arrest due to a winning balance of Rac2/Sp1 repression over β-catenin-induced PLD expression. J. Leukoc. Biol. 94, 953–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pathre P., Shome K., Blumental-Perry A., Bielli A., Haney C. J., Alber S., Watkins S. C., Romero G., Aridor M. (2003) Activation of phospholipase D by the small GTPase Sar1p is required to support COPII assembly and ER export. EMBO J. 22, 4059–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Speranza F. J., Mahankali M., Gomez-Cambronero J. (2013) Macrophage migration arrest due to a winning balance of Rac2/Sp1 repression over β-catenin-induced PLD expression. J. Leukoc. Biol. 94, 953–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee C. S., Kim I. S., Park J. B., Lee M. N., Lee H. Y., Suh P. G., Ryu S. H. (2006) The phox homology domain of phospholipase D activates dynamin GTPase activity and accelerates EGFR endocytosis. Nat. Cell Biol. 8, 477–484 [DOI] [PubMed] [Google Scholar]
- 41. Henkels K. M., Mahankali M., Gomez-Cambronero J. (2013) Increased cell growth due to a new lipase-GEF (Phospholipase D2) fastly acting on Ras. Cell. Signal. 25, 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eckert L. B., Repasky G. A., Ulkü A. S., McFall A., Zhou H., Sartor C. I., Der C. J. (2004) Involvement of Ras activation in human breast cancer cell signaling, invasion, and anoikis. Cancer Res. 64, 4585–4592 [DOI] [PubMed] [Google Scholar]
- 43. Lehman N., Di Fulvio M., McCray N., Campos I., Tabatabaian F., Gomez-Cambronero J. (2006) Phagocyte cell migration is mediated by phospholipases PLD1 and PLD2. Blood 108, 3564–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Santy L. C., Casanova J. E. (2001) Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 154, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zouwail S., Pettitt T. R., Dove S. K., Chibalina M. V., Powner D. J., Haynes L., Wakelam M. J., Insall R. H. (2005) Phospholipase D activity is essential for actin localization and actin-based motility in Dictyostelium. Biochem. J. 389, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Powner D. J., Payne R. M., Pettitt T. R., Giudici M. L., Irvine R. F., Wakelam M. J. (2005) Phospholipase D2 stimulates integrin-mediated adhesion via phosphatidylinositol 4-phosphate 5-kinase Iγb. J. Cell Sci. 118, 2975–2986 [DOI] [PubMed] [Google Scholar]
- 47. Corrotte M., Chasserot-Golaz S., Huang P., Du G., Ktistakis N. T., Frohman M. A., Vitale N., Bader M. F., Grant N. J. (2006) Dynamics and function of phospholipase D and phosphatidic acid during phagocytosis. Traffic 7, 365–377 [DOI] [PubMed] [Google Scholar]
- 48. Ali W. H., Chen Q., Delgiorno K. E., Su W., Hall J. C., Hongu T., Tian H., Kanaho Y., Di Paolo G., Crawford H. C., Frohman M. A. (2013) Deficiencies of the lipid-signaling enzymes phospholipase D1 and D2 alter cytoskeletal organization, macrophage phagocytosis, and cytokine-stimulated neutrophil recruitment. PLoS One 8, e55325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Frondorf K., Henkels K. M., Frohman M. A., Gomez-Cambronero J. (2010) Phosphatidic acid (PA) is a leukocyte chemoattractant that acts through S6 kinase signaling. J. Biol. Chem. 285, 15837–15847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Su W., Yeku O., Olepu S., Genna A., Park J. S., Ren H., Du G., Gelb M. H., Morris A. J., Frohman M. A. (2009) 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol. Pharmacol. 75, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nishikimi A., Fukuhara H., Su W., Hongu T., Takasuga S., Mihara H., Cao Q., Sanematsu F., Kanai M., Hasegawa H., Tanaka Y., Shibasaki M., Kanaho Y., Sasaki T., Frohman M. A., Fukui Y. (2009) Sequential regulation of DOCK2 dynamics by two phospholipids during neutrophil chemotaxis. Science 324, 384–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gomez-Cambronero J. (2012) Structure analysis between the SWAP-70 RHO-GEF and the newly described PLD2-GEF. Small GTPases 3, 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kam Y., Exton J. H. (2001) Phospholipase D activity is required for actin stress fiber formation in fibroblasts. Mol. Cell. Biol. 21, 4055–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Di Fulvio M., Frondorf K., Henkels K. M., Lehman N., Gomez-Cambronero J. (2007) The Grb2/PLD2 interaction is essential for lipase activity, intracellular localization and signaling in response to EGF. J. Mol. Biol. 367, 814–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kantonen S., Hatton N., Mahankali M., Henkels K. M., Park H., Cox D., Gomez-Cambronero J. (2011) A novel phospholipase D2-Grb2-WASp heterotrimer regulates leukocyte phagocytosis in a two-step mechanism. Mol. Cell. Biol. 31, 4524–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mahankali M., Peng H. J., Cox D., Gomez-Cambronero J. (2011) The mechanism of cell membrane ruffling relies on a phospholipase D2 (PLD2), Grb2 and Rac2 association. Cell. Signal. 23, 1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Knapek K., Frondorf K., Post J., Short S., Cox D., Gomez-Cambronero J. (2010) The molecular basis of phospholipase D2-induced chemotaxis: elucidation of differential pathways in macrophages and fibroblasts. Mol. Cell. Biol. 30, 4492–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Delon C., Manifava M., Wood E., Thompson D., Krugmann S., Pyne S., Ktistakis N. T. (2004) Sphingosine kinase 1 is an intracellular effector of phosphatidic acid. J. Biol. Chem. 279, 44763–44774 [DOI] [PubMed] [Google Scholar]
- 59. Pchejetski D., Golzio M., Bonhoure E., Calvet C., Doumerc N., Garcia V., Mazerolles C., Rischmann P., Teissié J., Malavaud B., Cuvillier O. (2005) Sphingosine kinase-1 as a chemotherapy sensor in prostate adenocarcinoma cell and mouse models. Cancer Res. 65, 11667–11675 [DOI] [PubMed] [Google Scholar]
- 60. Nava V. E., Hobson J. P., Murthy S., Milstien S., Spiegel S. (2002) Sphingosine kinase type 1 promotes estrogen-dependent tumorigenesis of breast cancer MCF-7 cells. Exp. Cell Res. 281, 115–127 [DOI] [PubMed] [Google Scholar]
- 61. Pyne N. J., Pyne S. (2010) Sphingosine 1-phosphate and cancer. Nat. Rev. Cancer 10, 489–503 [DOI] [PubMed] [Google Scholar]
- 62. Brindley D. N., Waggoner D. W. (1998) Mammalian lipid phosphate phosphohydrolases. J. Biol. Chem. 273, 24281–24284 [DOI] [PubMed] [Google Scholar]
- 63. Jasinska R., Zhang Q. X., Pilquil C., Singh I., Xu J., Dewald J., Dillon D. A., Berthiaume L. G., Carman G. M., Waggoner D. W., Brindley D. N. (1999) Lipid phosphate phosphohydrolase-1 degrades exogenous glycerolipid and sphingolipid phosphate esters. Biochem. J. 340, 677–686 [PMC free article] [PubMed] [Google Scholar]
- 64. Brindley D. N., Pilquil C. (2009) Lipid phosphate phosphatases and signaling. J. Lipid Res. 50, (suppl.) S225–S230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zheng Y., Rodrik V., Toschi A., Shi M., Hui L., Shen Y., Foster D. A. (2006) Phospholipase D couples survival and migration signals in stress response of human cancer cells. J. Biol. Chem. 281, 15862–15868 [DOI] [PubMed] [Google Scholar]
- 66. Yamada Y., Hamajima N., Kato T., Iwata H., Yamamura Y., Shinoda M., Suyama M., Mitsudomi T., Tajima K., Kusakabe S., Yoshida H., Banno Y., Akao Y., Tanaka M., Nozawa Y. (2003) Association of a polymorphism of the phospholipase D2 gene with the prevalence of colorectal cancer. J. Mol. Med. 81, 126–131 [DOI] [PubMed] [Google Scholar]
- 67. Saito M., Iwadate M., Higashimoto M., Ono K., Takebayashi Y., Takenoshita S. (2007) Expression of phospholipase D2 in human colorectal carcinoma. Oncol. Rep. 18, 1329–1334 [PubMed] [Google Scholar]
- 68. Toschi A., Edelstein J., Rockwell P., Ohh M., Foster D. A. (2008) HIFα expression in VHL-deficient renal cancer cells is dependent on phospholipase D. Oncogene 27, 2746–2753 [DOI] [PubMed] [Google Scholar]
- 69. Luquain C., Singh A., Wang L., Natarajan V., Morris A. J. (2003) Role of phospholipase D in agonist-stimulated lysophosphatidic acid synthesis by ovarian cancer cells. J. Lipid Res. 44, 1963–1975 [DOI] [PubMed] [Google Scholar]
- 70. Oh K. J., Lee S. C., Choi H. J., Oh D. Y., Kim S. C., Min do S., Kim J. M., Lee K. S., Han J. S. (2007) Role of phospholipase D2 in anti-apoptotic signaling through increased expressions of Bcl-2 and Bcl-xL. J. Cell. Biochem. 101, 1409–1422 [DOI] [PubMed] [Google Scholar]
- 71. Shi M., Zheng Y., Garcia A., Xu L., Foster D. A. (2007) Phospholipase D provides a survival signal in human cancer cells with activated H-Ras or K-Ras. Cancer Lett. 258, 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nozawa S., Ohno T., Banno Y., Dohjima T., Wakahara K., Fan D. G., Shimizu K. (2005) Inhibition of platelet-derived growth factor-induced cell growth signaling by a short interfering RNA for EWS-Fli1 via down-regulation of phospholipase D2 in Ewing sarcoma cells. J. Biol. Chem. 280, 27544–27551 [DOI] [PubMed] [Google Scholar]
- 73. Kim Y. R., Byun H. S., Won M., Park K. A., Kim J. M., Choi B. L., Lee H., Hong J. H., Park J., Seok J. H., Kim D. W., Shong M., Park S. K., Hur G. M. (2008) Modulatory role of phospholipase D in the activation of signal transducer and activator of transcription (STAT)-3 by thyroid oncogenic kinase RET/PTC. BMC Cancer 8, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Di Fulvio M., Frondorf K., Gomez-Cambronero J. (2008) Mutation of Y179 on phospholipase D2 (PLD2) upregulates DNA synthesis in a PI3K-and Akt-dependent manner. Cell. Signal. 20, 176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Knoepp S. M., Chahal M. S., Xie Y., Zhang Z., Brauner D. J., Hallman M. A., Robinson S. A., Han S., Imai M., Tomlinson S., Meier K. E. (2008) Effects of active and inactive phospholipase D2 on signal transduction, adhesion, migration, invasion, and metastasis in EL4 lymphoma cells. Mol. Pharmacol. 74, 574–584 [DOI] [PubMed] [Google Scholar]
- 76. Park M. H., Ahn B. H., Hong Y. K., Min do S. (2009) Overexpression of phospholipase D enhances matrix metalloproteinase-2 expression and glioma cell invasion via protein kinase C and protein kinase A/NF-κB/Sp1-mediated signaling pathways. Carcinogenesis 30, 356–365 [DOI] [PubMed] [Google Scholar]
- 77. Henkels K. M., Farkaly T., Mahankali M., Segall J. E., Gomez-Cambronero J. (2011) Cell invasion of highly metastatic MTLn3 cancer cells is dependent on phospholipase D2 (PLD2) and Janus kinase 3 (JAK3). J. Mol. Biol. 408, 850–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ye Q., Kantonen S., Gomez-Cambronero J. (2013) Serum deprivation confers the MDA-MB-231 breast cancer line with an EGFR/JAK3/PLD2 system that maximizes cancer cell invasion. J. Mol. Biol. 425, 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lavieri R. R., Scott S. A., Selvy P. E., Kim K., Jadhav S., Morrison R. D., Daniels J. S., Brown H. A., Lindsley C. W. (2010) Design, synthesis, and biological evaluation of halogenated N-(2-(4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-8-yl)ethyl)benzamides: discovery of an isoform-selective small molecule phospholipase D2 inhibitor. J. Med. Chem. 53, 6706–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Henkels K. M., Boivin G. P., Dudley E. S., Berberich S. J., Gomez-Cambronero J. (2013) Phospholipase D (PLD) drives cell invasion, tumor growth and metastasis in a human breast cancer xenograph model. Oncogene 32, 5551–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen Q., Hongu T., Sato T., Zhang Y., Ali W., Cavallo J. A., van der Velden A., Tian H., Di Paolo G., Nieswandt B., Kanaho Y., Frohman M. A. (2012) Key roles for the lipid signaling enzyme phospholipase D1 in the tumor microenvironment during tumor angiogenesis and metastasis. Sci. Signal. 5, ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. O'Reilly M. C., Scott S. A., Brown K. A., Oguin T. H., 3rd, Thomas P. G., Daniels J. S., Morrison R., Brown H. A., Lindsley C. W. (2013) Development of dual PLD1/2 and PLD2 selective inhibitors from a common 1,3,8-triazaspiro[4.5]decane core: discovery of Ml298 and Ml299 that decrease invasive migration in U87-MG glioblastoma cells. J. Med. Chem. 56, 2695–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kang D. W., Lee S. H., Yoon J. W., Park W. S., Choi K. Y., Min do S. (2010) Phospholipase D1 drives a positive feedback loop to reinforce the Wnt/β-catenin/TCF signaling axis. Cancer Res. 70, 4233–4242 [DOI] [PubMed] [Google Scholar]
- 84. Kang D. W., Min do S. (2010) Positive feedback regulation between phospholipase D and Wnt signaling promotes Wnt-driven anchorage-independent growth of colorectal cancer cells. PLoS One 5, e12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kang D. W., Min G., Park do Y., Hong K. W., Min do S. (2010) Rebamipide-induced downregulation of phospholipase D inhibits inflammation and proliferation in gastric cancer cells. Exp. Mol. Med. 42, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hsu Y. L., Hung J. Y., Ko Y. C., Hung C. H., Huang M. S., Kuo P. L. (2010) Phospholipase D signaling pathway is involved in lung cancer-derived IL-8 increased osteoclastogenesis. Carcinogenesis 31, 587–596 [DOI] [PubMed] [Google Scholar]
- 87. Su W., Chen Q., Frohman M. A. (2009) Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis. Future Oncol. 5, 1477–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kang D. W., Lee J. Y., Oh D. H., Park S. Y., Woo T. M., Kim M. K., Park M. H., Jang Y. H., Min do S. (2009) Triptolide-induced suppression of phospholipase D expression inhibits proliferation of MDA-MB-231 breast cancer cells. Exp. Mol. Med. 41, 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kulkarni A., Quang P., Curry V., Keyes R., Zhou W., Cho H., Baffoe J., Török B., Stieglitz K. (2014) 1,3-Disubstituted-4-aminopyrazolo [3, 4-d] pyrimidines, a new class of potent inhibitors for Phospholipase D. Chem. Biol. Drug Des. 10.1111/cbdd.12319 [DOI] [PubMed] [Google Scholar]
- 90. Kang D. W., Hwang W. C., Park M. H., Ko G. H., Ha W. S., Kim K. S., Lee Y. C., Choi K. Y., Min D. S. (2013) Rebamipide abolishes Helicobacter pylori CagA-induced phospholipase D1 expression via inhibition of NFκB and suppresses invasion of gastric cancer cells. Oncogene 32, 3531–3542 [DOI] [PubMed] [Google Scholar]
- 91. Zhang F., Wang Z., Lu M., Yonekubo Y., Liang X., Zhang Y., Wu P., Zhou Y., Grinstein S., Hancock J. F., Du G. (2014) Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Mol. Cell. Biol. 34, 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cipriano R., Bryson B. L., Miskimen K. L., Bartel C. A., Hernandez-Sanchez W., Bruntz R. C., Scott S. A., Lindsley C. W., Brown H. A., Jackson M. W. (2014) Hyperactivation of EGFR and downstream effector phospholipase D1 by oncogenic FAM83B. Oncogene 33, 3298–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cheol Son J., Woo Kang D., Mo Yang K., Choi K. Y., Gen Son T., Min Do S. (2013) Phospholipase D inhibitor enhances radiosensitivity of breast cancer cells. Exp. Mol. Med. 45, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Schwartz N., Chaudhri R. A., Hadadi A., Schwartz Z., Boyan B. D. (2014) 17β-Estradiol promotes aggressive laryngeal cancer through membrane-associated estrogen receptor-α 36. Horm. Cancer 5, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pye D. S., Rubio I., Pusch R., Lin K., Pettitt A. R., Till K. J. (2013) Chemokine unresponsiveness of chronic lymphocytic leukemia cells results from impaired endosomal recycling of Rap1 and is associated with a distinctive type of immunological anergy. J. Immunol. 191, 1496–1504 [DOI] [PubMed] [Google Scholar]
- 96. Marguerite V., Gkikopoulou E., Alberto J. M., Guéant J. L., Merten M. (2013) Phospholipase D activation mediates cobalamin-induced downregulation of Multidrug Resistance-1 gene and increase in sensitivity to vinblastine in HepG2 cells. Int. J. Biochem. Cell Biol. 45, 213–220 [DOI] [PubMed] [Google Scholar]
- 97. Jang Y. H., Choi K. Y., Min D. S. (2014) Phospholipase D-mediated autophagic regulation is a potential target for cancer therapy. Cell Death Differ. 21, 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bruntz R. C., Taylor H. E., Lindsley C. W., Brown H. A. (2014) Phospholipase D2 mediates survival signaling through direct regulation of Akt in glioblastoma cells. J. Biol. Chem. 289, 600–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Selvy P. E., Lavieri R. R., Lindsley C. W., Brown H. A. (2011) Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev. 111, 6064–6119 [DOI] [PMC free article] [PubMed] [Google Scholar]