Abstract
Phospholipase D (PLD) regulates downstream effectors by generating phosphatidic acid. Growing links of dysregulation of PLD to human disease have spurred interest in therapeutics that target its function. Aberrant PLD expression has been identified in multiple facets of complex pathological states, including cancer and inflammatory diseases. Thus, it is important to understand how the signaling network of PLD expression is regulated and contributes to progression of these diseases. Interestingly, small molecule PLD inhibitors can suppress PLD expression as well as enzymatic activity of PLD and have been shown to be effective in pathological mice models, suggesting the potential for use of PLD inhibitors as therapeutics against cancer and inflammation. Here, we summarize recent scientific developments regarding the regulation of PLD expression and its role in cancer and inflammatory processes.
Keywords: Cancer, Gene Expression, Gene Regulation, Inflammation, Phospholipase D, Phospholipase D Inhibitor, Therapeutic Drugs
Introduction
Phospholipase D produces phosphatidic acid (PA)2 via hydrolysis of phospholipids such as phosphatidylcholine and cardiolipin. As a lipid second messenger, PA has been implicated in a wide range of pathophysiological processes including proliferation, oncogenesis, inflammation, phagocytosis, membrane fusion, and spermatogenesis. Phosphatidylcholine-specific PLD1 and PLD2 are the classic mammalian isoforms of PLD (1, 2). Growth factor, mitogen, and inflammatory cytokines up-regulate expression and activity of PLD; however, aberrant dysregulation of PLD occurs in various cancers and inflammation-related diseases. Accordingly, it is important to understand how expression of PLD is regulated and contributes to these diseases.
An association between inflammation and cancer has been observed (3) and supported by recent epidemiological data that indicated ∼20% of cancer deaths are linked to chronic infections and persistent inflammation (4). The PLD signaling pathway provides a possible molecular link between inflammation and cancer; however, systematic investigation of PLD as a therapeutic target has only begun within the last few years with the development of small molecule PLD inhibitors and PLD knock-out mice (5–13). This review covers recent advances in the regulation of PLD expression and its role in cancer and inflammation.
Expression and Activity of PLD Are Dysregulated in Cancer
Role of PLD Up-regulated in Cancer
Elevated expression and activity of PLD have been detected in various human cancers, including colon, breast, gastric, thyroid, brain, kidney, and uterine smooth muscle when compared with adjacent non-neoplastic tissues. PLD contributes to various mitogenic or oncogenic signaling pathways, including G protein-coupled receptor (14), receptor tyrosine kinases (15, 16), integrin (7, 17), mammalian target of rapamycin (mTOR) (18–20), and Wnt signaling (21, 22).
PLD isozymes have been linked to protumorigenic and prometastatic phenotypes, although they play different roles in this context (23). Overexpression of PLD1 and its activity are required for mutant H-Ras-induced transformation and tumorigenesis (24). Moreover, PEA-15, a binding partner of PLD1, promotes H-Ras-mediated cell transformation via enhanced expression and activation of PLD1 (23). PLD2-generated PA in response to EGF activates Ras by recruiting its immediate activator, SOS, to translocate to the plasma membrane (16). Interestingly, PLD2 binds to Ras and acts as a guanine nucleotide exchange factor (GEF) of Ras.
The GEF action of PLD2 is due to the PLD2 protein itself and occurs independent of the lipase activity. Moreover, the GEF activity of PLD2 is greatly elevated in rapidly growing and highly metastatic cancer cells (25). Thus, PLD1 might be a critical downstream mediator of H-Ras-induced tumorigenesis, and PLD2-generated PA and/or PLD2 protein itself play crucial roles in the activation of Ras. PLD also activates mTOR and Wnt signaling and is linked to oncogenesis (18–22). In addition, PLD enhances secretion of matrix metalloproteinases (MMPs), which degrade surrounding extracellular matrix to facilitate cellular movement (26, 27). Thus, it seems that aberrant expression and activation of PLD in cancers accelerates these signaling pathways and contributes to cancerous phenotypes.
PLD2 expression level is correlated with tumor size and survival of patients with colorectal carcinoma, indicating that it might be a prognostic marker in colorectal cancers (28). Elevated expression of PLD2 in low invasive breast cancer cells has been shown to induce a highly aggressive phenotype, with primary tumors that formed following xenotransplantation being larger, growing faster, and developing lung metastases more readily (29). Silencing of PLD2 and PLD inhibitors (FIPI: 5-fluoro-2-indolyl des-chlorohalopemide; NOPT: N-[2-(4-oxo-1-phenyl-1,3,8-triazaspiro[4,5]dec-8-yl)ethyl]-2-naphthalenecarboxamide) in highly metastatic aggressive breast cancer cells decreases tumor size and metastases formation in vivo (29). Moreover, overexpression of wild-type PLD2 enhanced processes favorable to lymphoma cell metastasis in vivo, whereas catalytically inactive PLD2 reduced liver metastasis relative to control cells (30).
Simpler organisms, especially those containing a single gene, offer a useful setting to examine the role of PLD in cellular physiology. Deficiency of PLD in yeast is tolerated during vegetable growth, but the enzyme plays a critical role during sporulation (31). Inactivation of PLD in Caenorhabditis elegans results in viable progeny with no overt phenotype (32). Drosophila also has a single gene, but deficiency again results in a benign phenotype (33). Zebrafish have two PLD genes, and inhibition of PLD1 expression impairs blood vessel development in this organism (34). However, mice lacking PLD1 and PLD2 are viable, fertile, and have very benign phenotypes (5–8). Thus, PLD may play distinct roles in different species. Accordingly, this information points to a need for further discussion about the true function of the PLD mammalian system.
It has been suggested that PLD1 or PLD2 ablation might be compensated for by the other isoform or other signaling enzymes that increase the formation or decrease the catabolism of PA (7). Thus, it can be assumed that PLDs have dispensable functions during development and in normal mouse physiology. However, PLD1 and PLD2 knock-out mice are protected under pathological conditions (5–8). Although pharmacological inhibition of PLD1 and PLD2 would be well tolerated, it seems that these observations do not fit the plethora of functions ascribed to these genes well.
Because specific inhibitors for PLD were unavailable until recently, many PLD functional studies have employed primary alcohols to inhibit PLD-dependent generation of PA. However, more recent studies have raised concerns about off-target effects of primary alcohols, even when the tertiary alcohol is used as a control, and emphasized that the role of PLD in cell functions should be reevaluated (12, 35, 36).
It has been suggested that mice lacking PLD1, but not PLD2, incurred fewer lung metastases than wild-type mice, and thus PLD1 in the tumor microenvironment is critical for tumor growth and metastasis (8). These studies report complementary portions of the role of PLD1 and PLD2 in tumorigenesis and metastasis, indicating that a small molecule capable of inhibiting both PLD1 and PLD2 may be used in cancer therapeutics. Although small molecule PLD inhibitors appear to have some value in cell culture systems, their usefulness for PLD inhibition in vivo in animal models is less well established. Recent studies have shown that pharmacologically and genetically induced PLD inhibition had no obvious side effects (8, 11); thus, such a safe therapy could be particularly advantageous in clinical practice. Accordingly, highly selective PLD inhibitors with greater potency need to be developed and analyzed to enable optimized drug delivery and bioavailability.
Triple-negative breast cancers (TNBC) are difficult to treat due to their negative hormone receptor and ErbB2/HER2 status. In addition, TNBC are aggressive because of their frequent recurrence and high metastatic potential (37). Ceramide transfer protein (CERT) was recently reported to determine the signaling output of the EGF receptor (EGFR/ErbB1), which is up-regulated in TNBC (38). Reduced expression of CERT in TNBC is associated with alterations in plasma membrane organization and PLD2 activation (38). Heering et al. (38) suggested that the loss of CERT might trigger aberrant ligand-induced ErbB1 signaling through PLD2 activation, which may be relevant to the design of therapeutic interventions targeting TNBC. Choline kinase-α (ChK-α) is up-regulated in several cancers and a major contributor to increased phosphocholine, which is known as a metabolic hallmark in various cancers (39). Choline generated by PLD activation is used as a substrate of ChK-α. Recently, these two enzymes were found to be interactive, with depletion of ChK-α increasing PLD1 expression and vice versa in breast cancer cells and simultaneous depletion of both enzymes increasing apoptosis (40). Thus, ChK-α and PLD1 might be multiple target enzymes in choline phospholipid metabolism of breast cancer. Combined treatment with ChK-α inhibitor and PLD inhibitor will be more effective against breast cancer than individual treatments alone.
Genomic Alternation of PLD1 Gene in Cancer
Despite the dramatic advances in the field being driven by genome sequencing, somatic mutations or genomic alternations (rearrangements, copy number variations) at the PLD gene loci have not been widely observed in cancer. The Cancer Genome Atlas (TCGA) indicates that the PLD1 gene is mutated, amplified, and up-regulated in several types of cancers, including lung squamous cell carcinoma, ovarian serous cystadenocarcinoma, breast invasive carcinoma, uterine corpus endometrioid carcinoma, bladder urothelial carcinoma, and hepatocellular carcinoma. Accordingly, although changes in PLD expression may occur in some cancers, the mechanisms involved might be driven by environmental or genetic factors that constitute the primary cause of the disease.
Although PLD inhibition may have a place in cancer therapy, this approach might not be targeting the primary cause of the disease. A possible exception may be the 3q26 amplicon. Indeed, the 3q26 locus is frequently amplified in various types of cancer (ovarian, breast, and non-small cell lung cancers) and is correlated with poor prognosis and an invasive phenotype (41, 42). PLD1 was identified as a resident gene in the minimal common amplified region of 3q26 (42). However, the information relating copy number variation at this locus with PLD1 mRNA levels is not yet available in the TCGA. Although PLD1 has not been suggested as a transforming gene because overexpression of PLD1 did not show highly invasive capability in immortalized mammary epithelial cells (41), it is speculated that other genes at 3q26, including PLD1, may contribute to other cancer-related phenotypes. Overexpression of PLD1 may result in cancerous phenotypes via activation of various mitogenic or oncogenic signaling pathways. However, it is important to know how the very high expression levels needed to see these effects relate to changes in PLD1 expression as a result of 3q26 locus amplification.
PKCι and SOX2 were recently reported to be coamplified on chromosome 3q26, as well as to cooperate to activate hedgehog (Hh) signaling and drive a stem-like phenotype in lung squamous cell carcinoma (LSCC) (44). Aberrant Hh signaling has been implicated in the initiation and progression of various cancer subtypes. PKCι activates the Hh signaling pathway to stimulate oncosphere growth. Interestingly, RNA sequencing (RNA-Seq) data have revealed that knockdown of PKCι decreased expression of PLD1 and PLD2, which was increased in the oncosphere of lung squamous cell carcinoma (43). Furthermore, PLD inhibitors suppressed anchorage-independent growth of glioma stem cells (44). Thus, it is possible that PLD plays a role in stem-like cells and regulates the Hh signaling pathway.
MicroRNA Targeting PLD in Cancer
MicroRNA (miR), endogenous non-coding small strands of RNA 19–25 nucleotides in length, regulate gene expression through binding to the 3′-untranslated region (UTR) of target mRNAs, which eventually results in either mRNA degradation or translational repression. MiR is important in the development and progression of various types of cancer. Treatment of hypopharynx cancer cells with paclitaxel increased miR-29b-1*, which putatively mediated the decreased expression of PLD1 (45). miR-203 is reduced in various types of cancer, and acts as a tumor-suppressive microRNA by directly targeting certain oncogenes (46, 47). Expression of miR-203 is significantly lower in high World Health Organization (WHO) grade glioma tissues than low WHO grade glioma tissues and normal brain tissues (48). miR-203 inhibited the proliferation and invasion of glioma cells, at least in part, by targeting 3′-UTR of PLD2 and suppressing its expression (48). Moreover, PLD2 has been identified as a regulator of cell survival in glioblastoma through its regulation of the prosurvival kinase Akt (44). Thus, PLD2 as a new target of miR-203 may be a potential therapeutic candidate for the treatment of glioblastoma.
Dynamics of PLD Expression via Use of Different Transcription Factors in Cancer
Autoregulation of PLD Activity Is Coupled to Selective Induction of PLD1 Expression via NFκB
Despite the importance of the duration and amplitude of PLD signaling in cancer, mechanisms that regulate PLD expression remain poorly understood. Conversion of transient activation of signaling pathways to long term changes in cellular behavior is accomplished through changes in gene expression patterns. Despite the extensive information regarding the regulation of PLD activity, the long term effects of PLD activation on PLD expression have not yet been elucidated. It is well known that protein kinase C (PKC) stimulates PLD activity (49). Physiological activators of PLD (PDGF, EGF, or IL-1β) and phorbol-12-myristate-13-acetate, a PKC activator, enhanced the expression of PLD1, but not PLD2 (50). Phorbol-12-myristate-13-acetate and PDGF enhance binding of two functional NFκB sites to PLD1 promoter, suggesting that PLD1 is a downstream target of NFκB (50, 51). Depletion of PLD1 or PLD2 inhibited PDGF-induced PLD1 expression (51). Additionally, ectopic expression of wild-type PLD1 and PLD2, but not catalytically inactive PLD isozymes, increased the mRNA level and promoter activity of PLD1, but not PLD2. PA also increased PLD1 expression via the NFκB elements (52). Moreover, inhibitors selective for PLD1 and/or PLD2 (VU0155056, VU0155069, and VU0285655-1) suppressed the expression of PLD1 stimulated by the growth factor, but not that of PLD2 (52). Kang et al. (52) suggested that enzymatic activities originating from both PLD1 and PLD2 are required for selective induction of PLD1 via the positive feedback loop.
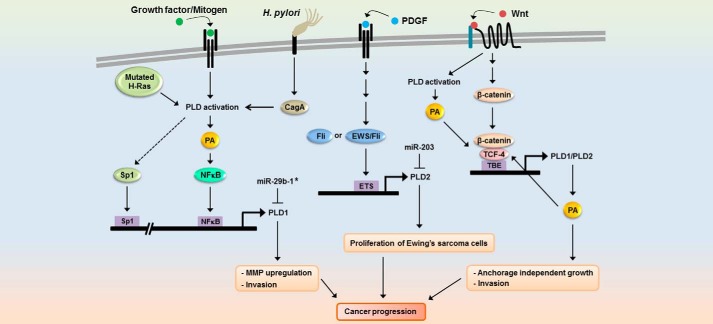
Helicobacter pylori induces chronic gastric inflammation and has been defined as a carcinogen leading to gastric cancer (53). H. pylori cagPAI (cag pathogenicity island) is a strain-specific locus that encodes a secretion system responsible for mediation of the translocation of bacterial virulence factor cagA protein into host epithelial cells (54). Infection of gastric cancer cells with cagA-positive H. pylori led to selective induction of PLD1 expression via cagA-dependent activation of NFκB (55), and H. pylori and expression of cagA increased the binding of NFκB to the PLD1 promoter. Additionally, expression of PLD1 was elevated in H. pylori-infected human gastric cancer tissues. Considering the direct link between cagA and the PLD1 signaling pathway, cagA-positive H. pylori-induced NFκB activation might lead to a risk of gastric cancer via up-regulation of PLD1 expression. Mutated H-Ras selectively increased PLD1, but not PLD2, via an interaction between Sp1 and its binding site in the 5′ promoter of PLD1 (56). Because mutated H-Ras is known to increase PLD activity, H-Ras can enhance PLD1 expression via NFκB. Autoregulation of PLD activity is coupled to selective induction of PLD1 expression via NFκB and contributes to cancer progression through increased MMP up-regulation and invasion (Fig. 1). Thus, PLD inhibitor may be useful for cancer therapy via inhibition of both PLD1 expression and PLD activity. However, it cannot exclude the possibility that such double inhibition would affect other key and necessary functions, such as membrane maintenance.
FIGURE 1.
Dynamics of differential regulation of PLD expression in cancer progression. Growth factor, mitogen, H. pylori infection, and Wnt signaling differentially express PLD isozymes via distinct signaling pathways or through different transcription factors depending on the physiological status of cells or cell type. TBE, TCF binding element.
Ewing Sarcoma Fusion Protein, EWS-Fli and Fli, Selectively Increase PLD2 Expression via the ETS Binding Motif
The reduced EWS-Fli-1 expression down-regulated PLD2 protein expression, resulting in repression of PDGF-mediated cell growth signaling in Ewing sarcoma (EWS) cells (57). EWS-Fli1, which is a fusion gene resulting from a chromosomal translocation t (11;22, q24;q12) found in EWS and primitive neuroectodermal tumors, encodes a transcriptional activator and promotes cellular transformation (58). Fli-1 genes are members of the erythroblast transformation-specific (ETS) family of transcription factors. Overexpression of EWS-Fli or Fli increases expression of PLD2 (60). PLD2, which is mainly involved in the increased PLD activity in EWS tumors (59), is a novel target of EWS-Fli-1 as well as Fli-1 transcription factor on the ETS binding site that functions as the principal promoter of the PLD2 gene. Other ETS transcription factors such as PU.1, ETS1 and ETS 2 could induce PLD2 gene expression, suggesting PLD2 as a general target of the ETS protein family. Thus, EWS-Fli1 might play a role in regulation of tumor proliferation-signaling enzymes via PLD2 expression in Ewing sarcoma cells. TC135 cells, an EWS cell line, showed predominant expression of PLD2, whereas PLD1 protein and gene expression were very weak. Although PLD1 promoter also contains six candidate ETS binding site motifs within the 5′ promoter region, PLD1 was not responsive to EWS-Fli-1 or Fli-1 transcription factors. The condition in histone acetylation around the PLD1 genome could be one possible mechanism of the insensitivity of PLD1 transcription to members of the ETS family of proteins. Kikuchi et al. (59) suggested that trichostatin A, an inhibitor of histone deacetylase, could increase PLD1 expression in EWS cells. However, expression of PLD1 gene by histone deacetylase inhibitor needs further analysis. EWS-Fli1 plays a role in PDGF-induced signaling by regulating PLD2 expression in EWS cells (57, 59). Based on our finding that PDGF-induced PLD activation is associated with PLD1 induction in breast cancer cells, regulation of the expression of each PLD isoform gene might be cell type-specific. Thus, differential expression of PLD isozymes might be mediated via a distinct signaling pathway or through different transcription factors dependent on the physiological status of cells or cell type (Fig. 1).
Wnt/β-Catenin Signaling Up-regulates Expression of Both PLD1 and PLD2 Isozymes via TCF
Aberrant activation of Wnt/β-catenin signaling followed by hyperactivation of T-cell transcription factor (TCF)-dependent transcription of target genes is linked to various cancers. Recently, both PLD1 and PLD2 were identified as target genes of Wnt/β-catenin signaling via binding of TCF4/β-catenin in their promoters (21, 22, 60). β-Catenin and TCF4 were shown to elevate expression and activity of PLD isozymes, whereas PLD isozymes were found to act as a positive feedback regulator of Wnt signaling, which subsequently promotes Wnt-driven anchorage-independent growth of colorectal cancer cells (Fig. 1). PLD enhances expression of the MMP-2 gene by increasing the DNA binding activity of NFκB and Sp1, and then promotes glioma cell invasion (27). Thus, it is believed that PLDs can promote tumor phenotype independence of TCF-induced genes. PLD activity is required for β-catenin/TCF-4 transcriptional activation, which occurs via increased formation of the β-catenin/TCF-4 complex (21). A PLD inhibitor that suppresses binding of TCF-4 to β-catenin may confer a clinical benefit during treatment of Wnt/β-catenin-driven malignancies. Further studies will enhance our understanding of the specific mechanisms by which increased expression of PLD contributes to the development and progression of cancer.
Regulation of PLD Expression in Inflammation
Selective Induction of PLD1 by Inflammatory Cytokines
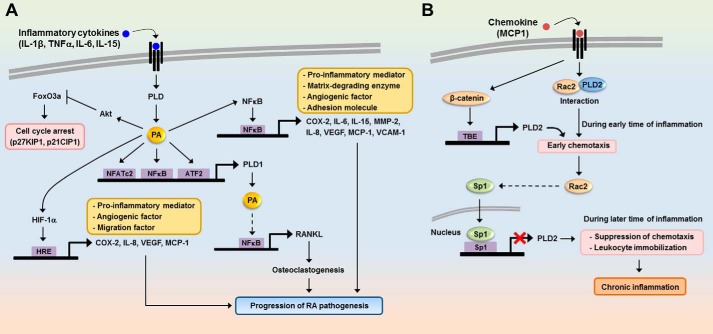
Expression of PLD1 has been shown to be up-regulated in a variety of inflammatory and autoimmune diseases, including acute pancreatitis (61), peritonitis (62), brain ischemia (63), rheumatoid arthritis (64), and Alzheimer disease (65). Thus, PLD may be involved in various inflammatory diseases. It has been reported that PLD2 expression is not affected in activated monocytes, and it appears to be a constitutive enzyme in circulating monocytes (66). Conversely, PLD1 is an inducible protein selectively induced during cell activation (52), suggesting different roles of PLD isoforms in mononuclear phagocytes. Therefore, selective induction of PLD1 by pathogen activation might play a relevant role in the late phase of immune response and in chronic inflammation. Microarray analysis during the transcriptional profiling of peripheral blood mononuclear cells associated with acute pancreatitis demonstrated that PLD1 was the top up-regulated gene (61), suggesting that it is a suitable molecular marker of acute pancreatitis. Genetic silencing of PLD1 effectively blocked the cytokine/chemokine production, vascular permeability, and leukocyte recruitment triggered by tumor necrosis factor α (TNFα) in an in vivo peritonitis model (62). Based on these findings, blockade of PLD1 has potential clinical implications for improving acute inflammatory conditions. PLD1 also plays a critical role in IL-1β-induced synoviocyte activation and the progression of chronic inflammatory arthritis (64). PLD1 expression and activity in the synovium or synoviocytes were found to be higher in rheumatoid arthritis (RA) patients than in osteoarthritis patients. RA is a chronic inflammatory disease characterized by the progressive destruction of articular cartilage and bone in the chronic phase. PLD1 expression correlates well with the severity of RA; thus, abnormal up-regulation of PLD1 may contribute to the pathogenesis of chronic arthritis. Increased expression of PLD1, but not PLD2, was triggered by proinflammatory cytokines such as IL-1β, TNFα, and IL-6 in RA synoviocytes, but not in osteoarthritis synoviocytes. IL-1β induced PLD1 expression via the NFκB and ATF-2 pathways. IL-1β enhances proinflammatory mediators, angiogenic factors, and proliferation and migration via PLD1-mediated NFκB or HIF-1α activation and the FoxO3a inactivation signaling pathway in RA synoviocytes (Fig. 2A). Osteoclastogenesis plays an important role in joint destruction in RA rheumatoid arthritis. It was recently suggested that osteoclastogenesis may occur through the expression of PLD1-induced receptor activator of nuclear factor-κB ligand (RANKL) in rheumatoid synoviocytes stimulated by IL-15 (67). Moreover, the PLD signaling pathway is involved in the lung cancer-derived IL-8-induced increase in osteoclastogenesis (68). Thus, regulation of PLD expression in RA patients may provide a new treatment strategy for inhibition of the bone destruction caused by this disease, as well as RA. IL-1β or TNFα has been reported to induce PLD1 expression by increasing the expression of calcium/calcineurin-regulated NFATc2/NFAT1 (69). It has also been suggested that PLD1 promoter contains putative NFAT binding sites. Taken together, these results indicate that proinflammatory cytokines selectively induce PLD1 expression via NFκB, ATF2, or NFATc2 transcription factors (Fig. 2A).
FIGURE 2.
Role of proinflammatory cytokine-induced PLD expression in chronic inflammation. A, proinflammatory cytokines selectively induce PLD1 expression via NFκB and ATF2. PLD1-derived PA promotes the binding of NFκB or HIF-1α to the promoters of its target genes and then increases the expression of proinflammatory mediators, migration, and angiogenic factors. However, PA suppresses the expression of cell cycle arrest genes via inhibition of FoxO3a transactivation. Moreover, PLD1 promotes osteoclastogenesis via up-regulation of RANKL expression in rheumatoid synovial fibroblasts stimulated by IL-15. Ultimately, up-regulation of PLD1 by proinflammatory cytokines can induce the pathogenesis of chronic inflammation. HRE, hypoxia-responsive element. B, during the early stages of inflammation, activation of leukocytes by MCP-1 induces the binding of Rac2 to PLD2 in the cytosol and then stimulates chemotaxis. During sustained inflammation, PLD2 expression is repressed by unknown mechanisms involving Rac and Sp1, and migration of the immune cells is suppressed. Prolonged immobilization of leukocytes by PLD2 repression can induce pathological stages of chronic inflammation. TBE, TCF binding element.
Role of PLD2 Expression in Inflammatory Cytokine-induced Migration of Immune Cells
IL-8, a potent chemoattractant, increases activity and expression of PLD2 and enables PLD2 to function as a facilitator for migration of immune cells (70). Leukocytes infiltrate into tissues at the site of inflammation and stay for prolonged periods of time until the insult is cleared, or remain longer in the case of chronic inflammation. Speranza et al. (71) recently demonstrated that, in the early stages of inflammation, activation of leukocytes by MCP-1/CCL2 induces the binding of Rac2 to PLD2 in the cytosol, which leads to stimulation of chemotaxis. Later in the process, Rac2 translocates into the nucleus and activates Sp1 via an unknown mechanism and then represses PLD2 expression. The subsequently reduced PLD2 activity inhibits the ability of cells to undergo chemotaxis. Although MCP-1 can activate β-catenin and increase PLD2 expression at earlier times in the process, Rac2 dominates at later times, exerting a negative effect on PLD2 expression. These findings suggest that if this negative effect is sustained, it could induce prolonged immobilization of leukocytes from normal functionality to the pathological stages of chronic inflammation (Fig. 2B).
Compounds Down-regulating PLD Expression
PLD isozymes are downstream transcriptional target molecules of NFκB, Sp1, TCF4, EWS-Fli or Fli, ATF-2, and NFATc2, which contribute to carcinogenesis or inflammation. As a result, compounds suppressing PLD expression might be useful in sensitizing resistant cancers and reducing inflammation. PLD inhibition abolishes growth factors and inflammatory cytokine-induced PLD1 expression, which induces proliferation, migration, invasion, angiogenesis, and chronic autoimmune inflammatory arthritis; therefore, selective PLD inhibitors may have therapeutic potential as anti-cancer and anti-inflammatory agents. Several compounds are known to suppress inflammation and proliferation of cancer cells via down-regulation of PLD. Triptolide, a natural product used in traditional Chinese medicine, has a myriad of therapeutic uses against inflammation and autoimmune disease (72). Rebamipide, a mucosal protective agent, has been used clinically for treatment of gastritis and peptic ulcers (73). Triptolide and rebamipide suppressed the expression and activity of both PLD1 and PLD2 isozymes in various cancer cells, which was followed by inhibition of the proliferation of cancer cells (74, 75). Triptolide- or rebamipide-mediated PLD1 repression was due to inhibition of NFκB activity. However, it is likely that the compounds inhibit PLD2 expression via a pathway distinct from that of PLD1 because NFκB is not responsible for PLD2 expression. PLD isozymes might be a potential therapeutic target of triptolide and rebamipide. Triptolide has been shown to interfere with a number of transcription factors at the molecular level, including NFκB, p53, NFAT, HSF-1, and xeroderma pigmentosum group B protein (XPB) (76–79), whereas rebamipide might contribute to the anti-tumorigenic effect of gastric cancer cells via inhibition of the H. pylori cagA-NFκB-PLD1 signaling pathway (55). Quercetin and caffeic acid phenethyl ester are natural compounds that have also been shown to exert a broad range of pharmacological effects, including anti-oxidant and anti-inflammatory activities. Quercetin and caffeic acid phenethyl ester abolished PLD1 expression via inhibition of NFκB transactivation, leading to inhibition of the invasion and proliferation of glioma cells (80, 81). These compounds mentioned above are known to indirectly inhibit PLD activity, but are inadequate as chemical probes because they have many molecular targets in various signaling pathways. Novartis Pharmaceuticals identified a psychotropic agent, halopemide, by high throughput screening, which was used as a starting point for PLD inhibitor development (10). Brown and colleagues (9) developed highly selective PLD1 and PLD2 inhibitors based on this halopemide scaffold. PLD-selective inhibitors will undoubtedly lead to new opportunities in investigations of PLDs and enable development of potential and therapeutic drugs for treatment of diseases related to PLD dysregulation such as cancer and inflammation that function by suppressing both the activity and the expression of PLD.
Conclusion and Future Perspectives
In the last few years, the function of mammalian PLD has been investigated in greater detail because of the development of selective PLD inhibitors and PLD knock-out mice. As a result, we now have a much better molecular understanding of the multiple roles of PLD. Increasingly, PLD has emerged as a critical modulator of cancer and inflammatory responses, and its expression and activities have been linked to multiple diseases, including cancer, inflammation, neurodegeneration, hypertension, thrombosis, and ischemic stroke. However, key questions still remain. For example, we still have a poor understanding of the molecular mechanisms differentially regulating PLD isozyme expression and of the precise role of each isoform. Additionally, it is not clear whether PLD isozyme is regulated in a coordinated fashion or separately in cancer and inflammation. However, future design of the next generation of isoform-specific therapeutics for the treatment of cancer and other diseases looks promising.
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (Grant 2012002009), a Translational Research Center for Protein Function Control Grant (NSF 2009-0083522), and National R&D program for Cancer Control, Ministry for Health, Welfare, and Family Affairs, Republic of Korea Grant 0920050. This is the third article in the Thematic Minireview Series “Phospholipase D and Cancer.”
- PA
- phosphatidic acid
- PLD
- phospholipase D
- GEF
- guanine nucleotide exchange factor
- TNBC
- triple-negative breast cancers
- CERT
- ceramide transfer protein
- ChK-α
- choline kinase-α
- EWS
- Ewing sarcoma
- RA
- rheumatoid arthritis
- MMP
- matrix metalloproteinase
- ETS
- erythroblast transformation-specific
- miR
- microRNA
- NFAT
- nuclear factor of activated T-cells
- Hh
- hedgehog
- TCF
- T-cell transcription factor.
REFERENCES
- 1. Hammond S. M., Altshuller Y. M., Sung T. C., Rudge S. A., Rose K., Engebrecht J., Morris A. J., Frohman M. A. (1995) Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J. Biol. Chem. 270, 29640–29643 [DOI] [PubMed] [Google Scholar]
- 2. Colley W. C., Sung T. C., Roll R., Jenco J., Hammond S. M., Altshuller Y., Bar-Sagi D., Morris A. J., Frohman M. A. (1997) Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr. Biol. 7, 191–201 [DOI] [PubMed] [Google Scholar]
- 3. Balkwill F., Mantovani A. (2001) Inflammation and cancer: back to Virchow? Lancet 357, 539–545 [DOI] [PubMed] [Google Scholar]
- 4. Kuper H., Adami H. O., Trichopoulos D. (2000) Infections as a major preventable cause of human cancer. J. Intern. Med. 248, 171–183 [DOI] [PubMed] [Google Scholar]
- 5. Oliveira T. G., Chan R. B., Tian H., Laredo M., Shui G., Staniszewski A., Zhang H., Wang L., Kim T. W., Duff K. E., Wenk M. R., Arancio O., Di Paolo G. (2010) Phospholipase D2 ablation ameliorates Alzheimer's disease-linked synaptic dysfunction and cognitive deficits. J. Neurosci. 30, 16419–16428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dall'Armi C., Hurtado-Lorenzo A., Tian H., Morel E., Nezu A., Chan R. B., Yu W. H., Robinson K. S., Yeku O., Small S. A., Duff K., Frohman M. A., Wenk M. R., Yamamoto A., Di Paolo G. (2010) The phospholipase D1 pathway modulates macroautophagy. Nat. Commun. 1, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elvers M., Stegner D., Hagedorn I., Kleinschnitz C., Braun A., Kuijpers M. E., Boesl M., Chen Q., Heemskerk J. W., Stoll G., Frohman M. A., Nieswandt B. (2010) Impaired αIIbβ3 integrin activation and shear-dependent thrombus formation in mice lacking phospholipase D1. Sci. Signal. 3, ra1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Q., Hongu T., Sato T., Zhang Y., Ali W., Cavallo J. A., van der Velden A., Tian H., Di Paolo G., Nieswandt B., Kanaho Y., Frohman M. A. (2012) Key roles for the lipid signaling enzyme phospholipase D1 in the tumor microenvironment during tumor angiogenesis and metastasis. Sci. Signal. 5, ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scott S. A., Selvy P. E., Buck J. R., Cho H. P., Criswell T. L., Thomas A. L., Armstrong M. D., Arteaga C. L., Lindsley C. W., Brown H. A. (2009) Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat. Chem. Biol. 5, 108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Monovich L., Mugrage B., Quadros E., Toscano K., Tommasi R., LaVoie S., Liu E., Du Z., LaSala D., Boyar W., Steed P. (2007) Optimization of halopemide for phospholipase D2 inhibition. Bioorg. Med. Chem. Lett. 17, 2310–2311 [DOI] [PubMed] [Google Scholar]
- 11. Stegner D., Thielmann I., Kraft P., Frohman M. A., Stoll G., Nieswandt B. (2013) Pharmacological inhibition of phospholipase D protects mice from occlusive thrombus formation and ischemic stroke: brief report. Arterioscler. Thromb. Vasc. Biol. 33, 2212–2217 [DOI] [PubMed] [Google Scholar]
- 12. Su W., Yeku O., Olepu S., Genna A., Park J. S., Ren H., Du G., Gelb M. H., Morris A. J., Frohman M. A. (2009) 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol. Pharmacol. 75, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Reilly M. C., Scott S. A., Brown K. A., Oguin T. H., 3rd, Thomas P. G., Daniels J. S., Morrison R., Brown H. A., Lindsley C. W. (2013) Development of dual PLD1/2 and PLD2 selective inhibitors from a common 1,3,8-triazaspiro[4.5]decane core: discovery of Ml298 and Ml299 that decrease invasive migration in U87-MG glioblastoma cells. J. Med. Chem. 56, 2695–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. López De Jesús M., Stope M. B., Oude Weernink P. A., Mahlke Y., Börgermann C., Ananaba V. N., Rimmbach C., Rosskopf D., Michel M. C., Jakobs K. H., Schmidt M. (2006) Cyclic AMP-dependent and Epac-mediated activation of R-Ras by G protein-coupled receptors leads to phospholipase D stimulation. J. Biol. Chem. 281, 21837–21847 [DOI] [PubMed] [Google Scholar]
- 15. Lu Z., Hornia A., Joseph T., Sukezane T., Frankel P., Zhong M., Bychenok S., Xu L., Feig L. A., Foster D. A. (2000) Phospholipase D and RalA cooperate with the epidermal growth factor receptor to transform 3Y1 rat fibroblasts. Mol. Cell Biol. 20, 462–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao C., Du G., Skowronek K., Frohman M. A., Bar-Sagi D. (2007) Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol. 9, 706–712 [DOI] [PubMed] [Google Scholar]
- 17. Powner D. J., Payne R. M., Pettitt T. R., Giudici M. L., Irvine R. F., Wakelam M. J. (2005) Phospholipase D2 stimulates integrin-mediated adhesion via phosphatidylinositol 4-phosphate 5-kinase Iγb. J. Cell Sci. 118, 2975–2986 [DOI] [PubMed] [Google Scholar]
- 18. Chen Y., Rodrik V., Foster D. A. (2005) Alternative phospholipase D/mTOR survival signal in human breast cancer cells. Oncogene 24, 672–679 [DOI] [PubMed] [Google Scholar]
- 19. Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., Chen J. (2001) Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294, 1942–1945 [DOI] [PubMed] [Google Scholar]
- 20. Xu L., Salloum D., Medlin P. S., Saqcena M., Yellen P., Perrella B., Foster D. A. (2011) Phospholipase D mediates nutrient input to mammalian target of rapamycin complex 1 (mTORC1). J. Biol. Chem. 286, 25477–25486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kang D. W., Lee S. H., Yoon J. W., Park W. S., Choi K. Y., Min D. S. (2010) Phospholipase D1 drives a positive feedback loop to reinforce the Wnt/β-catenin/TCF signaling axis. Cancer Res. 70, 4233–4242 [DOI] [PubMed] [Google Scholar]
- 22. Kang D. W., Min D. S. (2010) Positive feedback regulation between phospholipase D and Wnt signaling promotes Wnt-driven anchorage-independent growth of colorectal cancer cells. PLoS One 5, e12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sulzmaier F. J., Valmiki M. K., Nelson D. A., Caliva M. J., Geerts D., Matter M. L., White E. P., Ramos J. W. (2012) PEA-15 potentiates H-Ras-mediated epithelial cell transformation through phospholipase D. Oncogene 31, 3547–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buchanan F. G., McReynolds M., Couvillon A., Kam Y., Holla V. R., Dubois R. N., Exton J. H. (2005) Requirement of phospholipase D1 activity in H-RasV12-induced transformation. Proc. Natl. Acad. Sci. U.S.A. 102, 1638–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henkels K. M., Mahankali M., Gomez-Cambronero J. (2013) Increased cell growth due to a new lipase-GEF (Phospholipase D2) fastly acting on Ras. Cell Signal. 25, 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kato Y., Lambert C. A., Colige A. C., Mineur P., Noël A., Frankenne F., Foidart J. M., Baba M., Hata R., Miyazaki K., Tsukuda M. (2005) Acidic extracellular pH induces matrix metalloproteinase-9 expression in mouse metastatic melanoma cells through the phospholipase D-mitogen-activated protein kinase signaling. J. Biol. Chem. 280, 10938–10944 [DOI] [PubMed] [Google Scholar]
- 27. Park M. H., Ahn B. H., Hong Y. K., Min D. S. (2009) Overexpression of phospholipase D enhances matrix metalloproteinase-2 expression and glioma cell invasion via protein kinase C and protein kinase A/NFκB/Sp1-mediated signaling pathways. Carcinogenesis 30, 356–365 [DOI] [PubMed] [Google Scholar]
- 28. Saito M., Iwadate M., Higashimoto M., Ono K., Takebayashi Y., Takenoshita S. (2007) Expression of phospholipase D2 in human colorectal carcinoma. Oncol. Rep. 18, 1329–1334 [PubMed] [Google Scholar]
- 29. Henkels K. M., Boivin G. P., Dudley E. S., Berberich S. J., Gomez-Cambronero J. (2013) Phospholipase D (PLD) drives cell invasion, tumor growth and metastasis in a human breast cancer xenograph model. Oncogene 32, 5551–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Knoepp S. M., Chahal M. S., Xie Y., Zhang Z., Brauner D. J., Hallman M. A., Robinson S. A., Han S., Imai M., Tomlinson S., Meier K. E. (2008) Effects of active and inactive phospholipase D2 on signal transduction, adhesion, migration, invasion, and metastasis in EL4 lymphoma cells. Mol. Pharmacol. 74, 574–584 [DOI] [PubMed] [Google Scholar]
- 31. Waksman M., Eli Y., Liscovitch M., Gerst J. E. (1996) Identification and characterization of a gene encoding phospholipase D activity in yeast. J. Biol. Chem. 271, 2361–2364 [DOI] [PubMed] [Google Scholar]
- 32. Matthies D. S., Fleming P. A., Wilkes D. M., Blakely R. D. (2006) The Caenorhabditis elegans choline transporter CHO-1 sustains acetylcholine synthesis and motor function in an activity-dependent manner. J. Neurosci. 26, 6200–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. LaLonde M., Janssens H., Yun S., Crosby J., Redina O., Olive V., Altshuller Y. M., Choi S. Y., Du G., Gergen J. P., Frohman M. A. (2006) A role for Phospholipase D in Drosophila embryonic cellularization. BMC Dev. Biol. 6, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zeng X. X., Zheng X., Xiang Y., Cho H. P., Jessen J. R., Zhong T. P., Solnica-Krezel L., Brown H. A. (2009) Phospholipase D1 is required for angiogenesis of intersegmental blood vessels in zebrafish. Dev. Biol. 328, 363–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Norton L. J., Zhang Q., Saqib K. M., Schrewe H., Macura K., Anderson K. E., Lindsley C. W., Brown H. A., Rudge S. A., Wakelam M. J. (2011) PLD1 rather than PLD2 regulates phorbol-ester-, adhesion-dependent and Fcγ-receptor-stimulated ROS production in neutrophils. J. Cell Sci. 124, 1973–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yanase Y., Carvou N., Frohman M. A., Cockcroft S. (2010) Reversible bleb formation in mast cells stimulated with antigen is Ca2+/calmodulin-dependent and bleb size is regulated by ARF6. Biochem. J. 425, 179–193 [DOI] [PubMed] [Google Scholar]
- 37. Hurvitz S. A., Finn R. S. (2009) What's positive about ‘triple-negative’ breast cancer? Future Oncol. 5, 1015–1025 [DOI] [PubMed] [Google Scholar]
- 38. Heering J., Weis N., Holeiter M., Neugart F., Staebler A., Fehm T. N., Bischoff A., Schiller J., Duss S., Schmid S., Korte T., Herrmann A., Olayioye M. A. (2012) Loss of the ceramide transfer protein augments EGF receptor signaling in breast cancer. Cancer Res. 72, 2855–2866 [DOI] [PubMed] [Google Scholar]
- 39. Janardhan S., Srivani P., Sastry G. N. (2006) Choline kinase: an important target for cancer. Curr. Med. Chem. 13, 1169–1186 [DOI] [PubMed] [Google Scholar]
- 40. Gadiya M., Mori N., Cao M. D., Mironchik Y., Kakkad S., Gribbestad I. S., Glunde K., Krishnamachary B., Bhujwalla Z. M. (2014) Phospholipase D1 and choline kinase-α are interactive targets in breast cancer. Cancer Biol. Ther. 15, 593–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Akagi I., Miyashita M., Makino H., Nomura T., Hagiwara N., Takahashi K., Cho K., Mishima T., Takizawa T., Tajiri T. (2008) SnoN overexpression is predictive of poor survival in patients with esophageal squamous cell carcinoma. Ann. Surg. Oncol. 15, 2965–2975 [DOI] [PubMed] [Google Scholar]
- 42. Hagerstrand D., Tong A., Schumacher S. E., Ilic N., Shen R. R., Cheung H. W., Vazquez F., Shrestha Y., Kim S. Y., Giacomelli A. O., Rosenbluh J., Schinzel A. C., Spardy N. A., Barbie D. A., Mermel C. H., Weir B. A., Garraway L. A., Tamayo P., Mesirov J. P., Beroukhim R., Hahn W. C. (2013) Systematic interrogation of 3q26 identifies TLOC1 and SKIL as cancer driver. Cancer Discov. 3, 1044–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Justilien V., Walsh M. P., Ali S. A., Thompson E. A., Murray N. R., Fields A. P. (2014) The PRCK1 and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell 25, 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bruntz R. C., Taylor H. E., Lindsley C. W., Brown H. A. (2014) Phospholipase D2 mediates survival signaling through direct regulation of Akt in glioblastoma cells. J. Biol. Chem. 289, 600–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu C. Z., Shi R. J., Chen D., Sun Y. Y., Wu Q. W., Wang T., Wang P. H. (2013) Potential biomarkers for paclitaxel sensitivity in hypopharynx cancer cell. Int. J. Clin. Exp. Pathol. 6, 2745–2756 [PMC free article] [PubMed] [Google Scholar]
- 46. Bian K., Fan J., Zhang X., Yang X. W., Zhu H. Y., Wang L., Sun J. Y., Meng Y. L., Cui P. C., Cheng S. Y., Zhang J., Zhao J., Yang A. G., Zhang R. (2012) MicroRNA-203 leads to G1 phase cell cycle arrest in laryngeal carcinoma cells by directly targeting survivin. FEBS Lett. 586, 804–809 [DOI] [PubMed] [Google Scholar]
- 47. Wang C., Zheng X., Shen C., Shi Y. (2012) MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. J. Exp. Clin. Cancer Res. 31, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen Z., Li D., Cheng Q., Ma Z., Jiang B., Peng R., Chen R., Cao Y., Wan X. (2014) MicroRNA-203 inhibits the proliferation and invasion of U251 glioblastoma cells by directly targeting PLD2. Mol. Med. Rep. 9, 503–508 [DOI] [PubMed] [Google Scholar]
- 49. Selvy P. E., Lavieri R. R., Lindsley C. W., Brown H. A. (2011) Phospholipase D-enzymology, functionality, and chemical modulation. Chem. Rev. 111, 6064–6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kang D. W., Park M. H., Lee Y. J., Kim H. S., Kwon T. K., Park W. S., Min D. S. (2008) Phorbol ester up-regulates phospholipase D1 but not phospholipase D2 expression through a PKC/Ras/ERK/NFκB-dependent pathway and enhances matrix metalloproteinase-9 secretion in colon cancer cells. J. Biol. Chem. 283, 4094–4104 [DOI] [PubMed] [Google Scholar]
- 51. Kang D. W., Min D. S. (2010) Platelet derived growth factor increases phospholipase D1 but not phospholipase D2 expression via NFκB signaling pathway and enhances invasion of breast cancer cells. Cancer Lett. 294, 125–133 [DOI] [PubMed] [Google Scholar]
- 52. Kang D. W., Park M. H., Lee Y. J., Kim H. S., Lindsley C. W., Alex Brown H., Min D. S. (2011) Autoregulation of phospholipase D activity is coupled to selective induction of phospholipase D1 expression to promote invasion of breast cancer cells. Int. J. Cancer 128, 805–816 [DOI] [PubMed] [Google Scholar]
- 53. Hatakeyama M. (2008) Linking epithelial polarity and caecinogenesis by multitasking Helicobacter pylori virulence factor CagA. Oncogene 27, 7047–7054 [DOI] [PubMed] [Google Scholar]
- 54. Hatakeyama M. (2004) Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat. Rev. Cancer 4, 688–694 [DOI] [PubMed] [Google Scholar]
- 55. Kang D. W., Hwang W. C., Park M. H., Ko G. H., Ha W. S., Kim K. S., Lee Y. C., Choi K. Y., Min D. S. (2013) Rebamipide abolishes Helicobacter pylori CagA-induced phospholipase D1 expression via inhibition of NFκB and suppresses invasion of gastric cancer cells. Oncogene 32, 3531–3542 [DOI] [PubMed] [Google Scholar]
- 56. Gao S., Murakami M., Ito H., Furuhata A., Yoshida K., Tagawa Y., Hagiwara K., Takagi A., Kojima T., Suzuki M., Banno Y., Nozawa Y., Murate T. (2009) Mutated RAS induced PLD1 gene expression through increased Sp1 transcription factor. Nagoya J. Med. Sci. 71, 127–136 [PMC free article] [PubMed] [Google Scholar]
- 57. Nozawa S., Ohno T., Banno Y., Dohjima T., Wakahara K., Fan D. G., Shimizu K. (2005) Inhibition of platelet-derived growth factor-induced cell growth signaling by a short interfering RNA for EWS-Fli-1 via down-regulation of phospholipase D2 in Ewing sarcoma cells. J. Biol. Chem. 280, 27544–27551 [DOI] [PubMed] [Google Scholar]
- 58. Arvand A., Denny C. T. (2001) Biology of EWS/ETS fusions in Ewing's family tumors. Oncogene 20, 5747–5754 [DOI] [PubMed] [Google Scholar]
- 59. Kikuchi R., Murakami M., Sobue S., Iwasaki T., Hagiwara K., Takagi A., Kojima T., Asano H., Suzuki M., Banno Y., Nozawa Y., Murate T. (2007) Ewing's sarcoma fusion protein, EWS/Fli-1 and Fli-1 protein induce PLD2 but not PLD1 gene expression by binding to an ETS domain of 5′ promoter. Oncogene 26, 1802–1810 [DOI] [PubMed] [Google Scholar]
- 60. Kang D. W., Choi K. Y., Min D. S. (2011) Phospholipase D meets Wnt signaling: a new target for cancer therapy. Cancer Res. 71, 293–297 [DOI] [PubMed] [Google Scholar]
- 61. Bluth M., Lin Y. Y., Zhang H., Viterbo D., Zenilman M. (2008) Use of gene expression profiles in cells of peripheral blood to identify new molecular markers of acute pancreatitis. Arch. Surg. 143, 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sethu S., Pushparaj P. N., Melendez A. J. (2010) Phospholipase D1 mediates TNFα-induced inflammation in a murine model of TNFα-induced peritonitis. PLoS ONE 5, e10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee M. Y., Kim S. Y., Min D. S., Choi Y. S., Shin S. L., Chun M. H., Lee S. B., Kim M. S., Jo Y. H. (2000) Upregulation of phospholipase D in astrocytes in response to transient forebrain ischemia. Glia 30, 311–317 [DOI] [PubMed] [Google Scholar]
- 64. Kang D. W., Park M. K., Oh H. J., Lee D. G., Park S. H., Choi K. Y., Cho M. L., Min D. S. (2013) Phospholipase D1 has a pivotal role in interleukin-1β-driven chronic autoimmune arthritis through regulation of NFκB, hypoxia-inducible factor 1α, and FoxO3a. Mol. Cell Biol. 33, 2760–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jin J. K., Ahn B. H., Na Y. J., Kim J. I., Kim Y. S., Choi E. K., Ko Y. G., Chung K. C., Kozlowski P. B., Min D. S. (2007) Phospholipase D1 is associated with amyloid precursor protein in Alzheimer's disease. Neurobiol. Aging 28, 1015–1027 [DOI] [PubMed] [Google Scholar]
- 66. Locati M., Riboldi E., Bonecchi R., Transidico P., Bernasconi S., Haribabu B., Morris A. J., Mantovani A., Sozzani S. (2001) Selective induction of phospholipase D1 in pathogen-activated human monocytes. Biochem. J. 358, 119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Park M. K., Her Y. M., Cho M. L., Oh H. J., Park E. M., Kwok S. K., Ju J. H., Park K. S., Min D. S., Kim H. Y., Park S. H. (2011) IL-15 promotes osteoclastogenesis via the PLD pathway in rheumatoid arthritis. Immunol. Lett. 139, 42–51 [DOI] [PubMed] [Google Scholar]
- 68. Hsu Y. L., Hung J. Y., Ko Y. C., Hung C. H., Huang M. S., Kuo P. L. (2010) Phospholipase D signaling pathway is involved in lung cancer-derived IL-8 increased osteoclastogenesis. Carcinogenesis 31, 587–596 [DOI] [PubMed] [Google Scholar]
- 69. Yang T. T., Ung P. M., Rincón M., Chow C. W. (2006) Role of the CCAAT/enhancer-binding protein NFATc2 transcription factor cascade in the induction of secretory phospholipase A2. J. Biol. Chem. 281, 11541–11552 [DOI] [PubMed] [Google Scholar]
- 70. Tabatabaian F., Dougherty K., Di Fulvio M., Gomez-Cambronero J. (2010) Mammalian target of rapamycin (mTOR) and S6 kinase down-regulate phospholipase D2 basal expression and function. J. Biol. Chem. 285, 18991–19001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Speranza F. J., Mahankali M., Gomez-Cambronero J. (2013) Macrophage migration arrest due to a winning balance of Rac2/Sp1 repression over β-catenin-induced PLD expression. J. Leukocyte Biol. 94, 953–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Qiu D., Kao P. N. (2003) Review Immunosuppressive and anti-inflammatory mechanisms of triptolide, the principal active diterpenoid from the Chinese medicinal herb Tripterygium wilfordii Hook. f. Drugs. R. D. 4, 1–18 [DOI] [PubMed] [Google Scholar]
- 73. Arakawa T., Kobayashi K., Yoshikawa T., Tarnawski A. (1998) Rebamipide: overview of its mechanisms of action and efficacy in mucosal protection and ulcer healing. Dig. Dis. Sci. 43, 5S–13S [PubMed] [Google Scholar]
- 74. Kang D. W., Lee J. Y., Oh D. H., Park S. Y., Woo T. M., Kim M. K., Park M. H., Jang Y. H., Min D. S. (2009) Triptolide-induced suppression of phospholipase D expression inhibits proliferation of MDA-MB-231 breast cancer cells. Exp. Mol. Med. 41, 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kang D. W., Min G., Park do. Y., Hong K. W., Min D. S. (2010) Rebamipide-induced downregulation of phospholipase D inhibits inflammation and proliferation in gastric cancer cells. Exp. Mol. Med. 42, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Qiu D., Zhao G., Aoki Y., Shi L., Uyei A., Nazarian S., Ng J. C., Kao P. N. (1999) Immunosuppressant PG490 (triptolide) inhibits T-cell interleukin-2 expression at the level of purine-box/nuclear factor of activated T-cells and NFκB transcriptional activation. J. Biol. Chem. 274, 13443–13450 [DOI] [PubMed] [Google Scholar]
- 77. Chang W. T., Kang J. J., Lee K. Y., Wei K., Anderson E., Gotmare S., Ross J. A., Rosen G. D. (2001) Triptolide and chemotherapy cooperate in tumor cell apoptosis: a role for the p53 pathway. J. Biol. Chem. 276, 2221–2227 [DOI] [PubMed] [Google Scholar]
- 78. Westerheide S. D., Kawahara T. L., Orton K., Morimoto R. I. (2006) Triptolide, an inhibitor of the human heat shock response that enhances stress-induced cell death. J. Biol. Chem. 281, 9616–9622 [DOI] [PubMed] [Google Scholar]
- 79. Titov D. V., Gilman B., He Q. L., Bhat S., Low W. K., Dang Y., Smeaton M., Demain A. L., Miller P. S., Kugel J. F., Goodrich J. A., Liu J. O. (2011) XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat. Chem. Biol. 7, 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Park M. H., Min D. S. (2011) Quercetin-induced downregulation of phospholipase D1 inhibits proliferation and invasion in U87 glioma cells. Biochem. Biophys. Res. Commun. 412, 710–715 [DOI] [PubMed] [Google Scholar]
- 81. Park M. H., Kang D. W., Jung Y., Choi K. Y., Min D. S. (2013) Caffeic acid phenethyl ester downregulates phospholipase D1 via direct binding and inhibition of NFκB transactivation. Biochem. Biophys. Res. Commun. 442, 1–7 [DOI] [PubMed] [Google Scholar]