Background: MicroRNA-146-deficient mice have developed some abnormal hematopoietic phenotypes.
Results: MicroRNA-146b-5p (miR-146b), transcriptionally activated by GATA-binding protein 1 (GATA-1), promotes human erythroid and megakaryocytic differentiation via regulating the PDGFRA signaling pathway.
Conclusion: A regulatory circuit comprising miR-146b, PDGFRA, and GATA-1 promotes erythroid and megakaryocytic differentiation.
Significance: This study provides new insights into miR-146b function and connects GATA-1 and miR-146b in human erythroid and megakaryocytic differentiation.
Keywords: Cell Differentiation, Erythropoiesis, GATA Transcription Factor, Hematopoiesis, MicroRNA (miRNA), GATA-1, PDGFRA, Megakaryocytopoiesis, MicroRNA-146b
Abstract
Emerging evidence has shown that microRNAs have key roles in regulating various normal physiological processes, whereas their deregulated expression is correlated with various diseases. The miR-146 family includes miR-146a and miR-146b, with a distinct expression spectrum in different hematopoietic cells. Recent work indicated that miR-146a has a close relationship with inflammation and autoimmune diseases. miR-146-deficient mice have developed some abnormal hematopoietic phenotypes, suggesting the potential functions of miR-146 in hematopoietic development. In this study, we found that miR-146b was consistently up-regulated in both K562 and CD34+ hematopoietic stem/progenitor cells (HSPCs) undergoing either erythroid or megakaryocytic differentiation. Remarkably, erythroid and megakaryocytic maturation of K562 cells was induced by excess miR-146b but inhibited by decreased miR-146b levels. More importantly, an mRNA encoding receptor tyrosine kinase, namely platelet-derived growth factor receptor α (PDGFRA), was identified and validated as a direct target of miR-146b in hematopoietic cells. Gain-of-function and loss-of-function assays showed that PDGFRA functioned as a negative regulator in erythroid and megakaryocytic differentiation. miR-146b could ultimately affect the expression of the GATA-1 gene, which is regulated by HEY1 (Hairy/enhancer-of-split related with YRPW motif protein 1), a transcriptional repressor, via inhibition of the PDGFRA/JNK/JUN/HEY1 pathway. Lentivirus-mediated gene transfer also demonstrated that the overexpression of miR-146b promoted erythropoiesis and megakaryocytopoiesis of HSPCs via its regulation on the PDGFRA gene and effects on GATA-1 expression. Moreover, we confirmed that the binding of GATA-1 to the miR-146b promoter and induction of miR-146b during hematopoietic maturation were dependent on GATA-1. Therefore, miR-146b, PDGFRA, and GATA-1 formed a regulatory circuit to promote erythroid and megakaryocytic differentiation.
Introduction
The discovery of microRNAs (miRNAs)3 has greatly expanded our understanding of the molecular mechanisms that regulate gene expression (1). As of this writing, thousands of miRNAs have been identified in the human genome, and up to 30% of the protein-coding genes are estimated to be regulated by them. The high conservation of miRNAs among species suggests a strong selective pressure and indicates important functions for them in animal development, disease, and evolution (2–4). The importance of miRNAs is further underscored by the observation that mice lacking Dicer, the enzyme essential for the generation of mature miRNAs, die at embryonic day 7.5 and lack multipotent stem cells (5, 6). Given that miRNAs appear to constitute one of the largest classes of gene regulators in animals, understanding their physiological roles and action modes is essential. Although many endogenous miRNAs have been identified in human, specific functions have remained largely undefined until now.
The miR-146 family includes miR-146a and miR-146b, with a distinct expression spectrum in different hematopoietic cells (7). Recent studies implicated that miR-146 has a close relationship with inflammation and autoimmune diseases (8, 9). miR-146a negatively regulates the acute innate immune response following activation by pattern-recognition receptors and pro-inflammatory cytokines (10). Furthermore, miR-146a is important for innate immune tolerance in the neonatal intestine because it targets IRAK1, thereby preventing apoptosis of intestinal epithelial cells following bacterial exposure (11, 12). In addition to its function in innate immunity, miR-146a also has an important role in the adaptive immune response (13). miR-146a expression is higher in Th1 cells and lower in Th2 cells compared with that in naive T cells (14). Furthermore, miR-146a can inhibit LPS-induced production of IFN-γ and inducible NOS in T cells (15). miR-146a is also highly expressed in both effector and central memory T cells and is induced in naive T cells upon TCR stimulation (14). An miR-146 knock-out mouse was also used to validate its function in the molecular control of immune functions. Interestingly, miR-146-deficient mice develop many of the same abnormal hematopoietic phenotypes described in a subset of myelodysplastic syndrome patients, suggesting the potential functions of miR-146 in hematopoietic development and malignancies (16). In this study, we found that miR-146b (miR-146b-5p) was consistently up-regulated in both K562 and CD34+ HSPCs undergoing either erythroid or megakaryocytic differentiation. Remarkably, ectopic expression of miR-146b promoted the accumulation of mature erythroid cells and formation of megakaryocytes in K562 cells treated with hemin and PMA, respectively. By contrast, decreased miR-146b levels inhibited erythroid and megakaryocytic maturation of K562 cells. More importantly, an mRNA that encodes platelet-derived growth factor receptor α (PDGFRA), a receptor tyrosine kinase, was revealed to be the direct target of miR-146b in hematopoietic cells. PDGF receptors are shown to be crucial for the proper development of several organs in the embryo, including kidneys, lungs, and the cardiovascular system (17–20). However, the function of PDGF ligands and their receptors in hematopoiesis remains unclear. In this study, we demonstrated that PDGFRA could function as a negative regulator in erythroid and megakaryocytic differentiation. The down-regulation of PDGFRA with hematopoietic maturation was achieved by miR-146b at the post-transcriptional level. Furthermore, miR-146b could ultimately affect GATA-1 expression via the inhibition of PDGFRA. The miR-146b promoter contained several binding sites for GATA-1, and induction of miR-146b during hematopoietic maturation was dependent on GATA-1. Therefore, miR-146b, PDGFRA, and GATA-1 formed a regulatory circuit to promote erythroid and megakaryocytic differentiation.
EXPERIMENTAL PROCEDURES
Cell Culture
Human erythroleukemia cell line K562 was maintained in DMEM supplemented with 10% FBS (Hyclone). Erythroid and megakaryocytic differentiation of K562 cells was induced using 30 μm hemin and 50 pm PMA (Sigma), respectively. Erythroid differentiation was determined by benzidine staining for hemoglobin expression, and megakaryocytic differentiation was determined by propidium iodide (PI) staining. 293T cells were obtained from American Type Culture Collection and grown in DMEM with 10% FBS.
Isolation and Culture of CD34+ HSPCs
Human umbilical cord blood was obtained from normal full-term deliveries after informed consent was obtained. This procedure was approved by the Research Ethics Committee of Peking Union Hospital (Beijing, China). Mononuclear cell fractions were isolated from umbilical cord blood by Percoll density gradient (d = 1.077; Amersham Biosciences). CD34+ cells were enriched from mononuclear cells through positive immunomagnetic selection (CD34 MultiSort kit, Miltenyi Biotec, Bergisch-Gladbach, Germany). The isolation system yielded ∼90% CD34-positive cells. The isolated CD34+ cells were cultured in Iscove's modified Dulbecco's medium supplemented with 30% FBS (Hyclone), 1% BSA, 100 μm 2-mercaptoethanol, 2 ng/ml recombinant human IL-3, 100 ng/ml recombinant human stem cell factor (Stem Cell Technologies, Vancouver, British Columbia, Canada), 60 mg/ml penicillin, 100 mg/ml streptomycin, and 2 units/ml recombinant human erythropoietin (for erythroid culture) or 4 units/ml recombinant human thrombopoietin (for megakaryocytic culture) (R&D Systems, Minneapolis, MN). Cells were harvested every 3–5 days.
Northern and Western Blot Analysis
Northern blot analysis of miRNAs was performed as described (21). The oligonucleotide probe sequences for miR-146b and U6 snRNA were 5′-AGCCTATGGAATTCAGTTCTCA-3′ and 5′-CCATGCTAATCTTCTCTGTATCGTTCCAA-3′, respectively. Whole-cell lysate or nuclear extract was subjected to Western blot analysis as detailed elsewhere (22). The following antibodies were used for Western blot. GAPDH, p-PDGFRA (sc-12910), p-JUN (S73, sc-7981), and JUN (sc-1694) were purchased from Santa Cruz Biotechnology. PDGFRA (BS3759), JNK (BS6448), and p-JNK (BS4764) were purchased from Bioworld Co. GATA-1 (ab11963) and HEY1 (ab22614) were purchased from Abcam (Cambridge, UK). p-JUN (S63, 2361) was purchased from Cell Signaling Technology.
RNA Isolation and Real Time qPCR
Total RNA was extracted from the cell harvest using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA was quantified according to absorbance at 260 nm. cDNA was synthesized by Moloney murine leukemia virus reverse transcriptase (Invitrogen) from 2 μg of total RNA or 20 ng of small RNA. Oligo(dT)18 was used as the RT primer for reverse transcription (RT) of mRNAs. Stem-loop RT primers were used for RT of miRNAs. For mRNAs, qPCR was carried out in a IQ5 real time PCR System (Bio-Rad) using an SYBR Premix Ex Taq kit (Takara, Dalian, China) according to the manufacturer's instructions. For mRNAs, data were normalized using the endogenous GAPDH control. For miRNAs, U6 snRNA was used as the endogenous control. The primer sequences used for real time RT-PCR were as follows: 146b RT primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCCTATGG-3′; 146b up, 5′-ATGCGCTGCTGAGAACTGAATT-3′; 146b down, 5′-CAGTGCAGGGTCCGAGGT-3′; U6 snRNA RT primer, 5′-AAAATATGGAACGCTTCACGAATTTG-3′; U6 snRNA up, 5′-CTCGCTTCGGCAGCACATATACT-3′; and U6 snRNA down, 5′-ACGCTTCACGAATTTGCGTGTC-3′.
Oligonucleotides and Transfection
miRNA-146b mimics, miRNA-146b inhibitors, and negative controls (scrambled control, mimic, and inhibitor) were obtained from Dharmacon (Austin, TX) and transfected with DharmFECT1 (Dharmacon, Austin, TX) in K562 cells at a final concentration of 60 nm. After 24 h of transfection, the K562 cells were washed with PBS and plated for hemin induction. siRNA smart pools (specifically for PDGFRA) and control siRNA pools were synthesized by Dharmacon and transfected into K562 cells (100 nm) using DharmFECT1. The medium was changed after 6 h, and cells were cultured for 48 h and harvested for Western blot analyses.
Constructs and Lentivirus
The reverse complementary sequence of miR-146b was inserted downstream of the firefly luciferase gene of pGL3 (Promega, WI) to generate a positive control (pGL3-miR-146b). The 3′-untranslated regions (3′UTR) of human PDGFRA mRNA containing the two predicted miR-146b sites were PCR-amplified and cloned into pGL3 to generate the corresponding reporter construct (PDGFRA). Primers for PDGFRA cloning were 5′-ATTGTATTAACTATCTTCTTTGGAC-3′ and 5′-TCATATGGTATCAGCAATTAAGCAG-3′. Three mutant constructs corresponding to PDGFRA_mut1 (the 994-nucleotide miR-146b site was mutant), PDGFRA_mut2 (the 1305-nucleotide miR-146b site was mutant), and PDGFRA_dmut (both sites were mutant) were created using a QuikChange site-directed mutagenesis kit (Stratagene, CA). Full-length cDNA of GATA-1 or PDGFRA gene was cloned into pcDNA3.1 vector to produce pcDNA3.1-GATA-1 or pcDNA-PDGFRA. The PDGFRA constructs were transfected with Lipofectamine® LTX reagent (Invitrogen) into K562 cells. These cells were washed the next day with PBS and plated for hemin induction. The self-inactivating transfer vector plasmid containing miR-146b (pMIR-lenti-146b) and packaging kit were purchased from System Biosciences (SBI) and operated according to the manufacturer's instructions. The harvested viral particles (Lenti-146b or Lenti-GFP) were added to the CD34+ cultured cells. Cells were washed the next day with PBS and plated for colony-forming experiments and liquid induction cultures.
Flow Cytometry
Cells were harvested at the indicated times and washed twice at 4 °C in PBS, 0.5% BSA to block Fc receptors. Transduced CD34+ HSPCs were assessed for CD235a and CD61 expression after staining with phycoerythrin-conjugated anti-CD235a or anti-CD61 antibody (BD Biosciences). Flow cytometry was carried out on a C6 Flow Cytometer® Instrument (BD Biosciences).
Luciferase Reporter Assay
For miRNA target analysis, 293T cells were co-transfected with 0.4 μg of the reporter construct, 0.02 μg of pRL-TK control vector, and 5 pmol of miRNA mimic or scrambled controls. Cells were harvested 48 h post-transfection and assayed by Dual-Luciferase assay (Promega, WI) according to the manufacturer's instructions. All transfection assays were carried out in triplicate.
Rescue Assay of miRNA Targets
To determine the effects of miRNAs and target genes on phenotypic changes, K562 cells in 6-well plates were first transfected with scrambled control miRNA inhibitor or miRNA-146b inhibitors (100 nm). After 24 h in culture, these cells were then co-transfected with a combination of scrambled control miRNA inhibitor (50 nm) and control siRNA (50 nm), miRNA inhibitors and control siRNA, or miRNA inhibitors and siRNA to PDGFRA. After co-transfection for 24 h, cells were induced by hemin or PMA. Cells were harvested at the indicated time points after hemin or PMA addition and assayed as required.
Chromatin Immunoprecipitation (ChIP) Assay
Antibodies anti-GATA-1 (ab11963) and rabbit isotype controls IgG (ab27472) were purchased from Abcam and used for ChIP studies. K562 cells induced by hemin for 48 h were collected and cross-linked with 1% formaldehyde for 10 min, washed in cold PBS buffer, resuspended in lysis buffer (0.1% SDS, 0.5% Triton X-100, 20 mm Tris-HCl (pH 8.1), 150 mm NaCl, protease inhibitor; Roche Applied Science), and sonicated to obtain chromatin fragments between 200 and 1000 bp. The sonicated chromatin fragments were resuspended in IP buffer and incubated overnight at 4 °C with magnetic bead-conjugated antibodies (Santa Cruz Biotechnology). IP was washed with lysis buffer, LiCl buffer (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris-HCl (pH 8.1)), and TE buffer and eluted in elution buffer (1% SDS, 0.1 m NaHCO3). DNA was then recovered by reversing the cross-links and purified by a Qiagen purification kit. An unenriched sample of DNA was treated similarly to serve as input. Subsequently, 0.1% of the input and 20% of the IPs were used for ChIP-PCR analysis with 25 cycles.
Statistics
Student's t test (two-tailed) was performed to analyze the data. Statistical significance was set at p < 0.05, as indicated by an asterisk (*, p < 0.05; **, p < 0.01).
RESULTS
miR-146b Expression Gradually Increases during Erythroid Differentiation and Megakaryocytic Differentiation of K562 Cells
Recent work has implicated miR-146 in the regulation of innate and adaptive immunity, inflammatory response, autoimmunity, and cancer. However, its involvement in normal hematopoiesis is undefined. Given the significant associations of miR-146b in lymphoid cell proliferation and differentiation, we examined the expression level of miR-146b in hematopoietic myeloid lineages, especially in K562 cells undergoing either erythroid or megakaryocytic differentiation. Northern blot showed that miR-146b continuously increased during hemin-induced erythroid differentiation (Fig. 1A) and PMA-induced megakaryocytic differentiation (Fig. 2A) of K562 cells. The expression spectrum of miR-146b suggests its possibly positive regulatory function in erythroid and megakaryocytic differentiation.
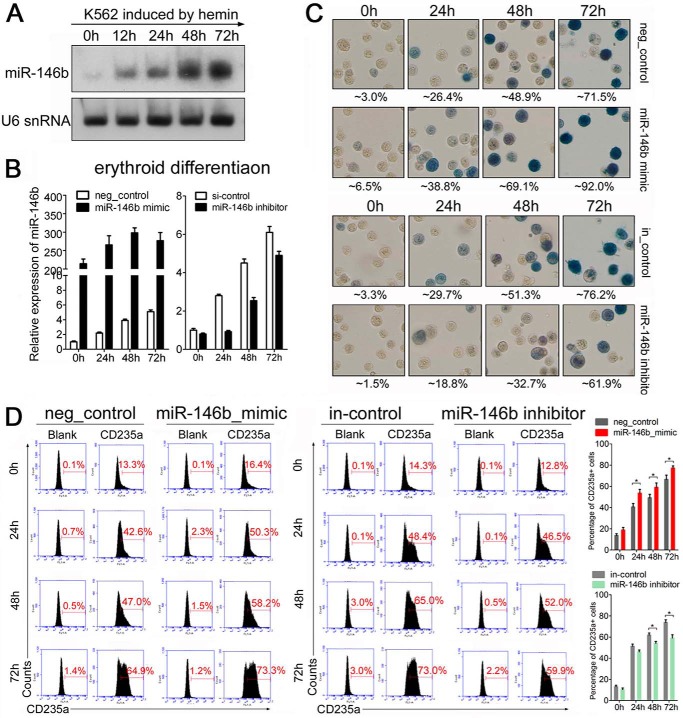
FIGURE 1.
miR-146b promotes erythroid differentiation of K562 cells. A, Northern blot analysis of miR-146b during hemin-induced erythroid differentiation of K562 cells. U6 snRNA was detected to check equal RNA loading. B, qRT-PCR analysis of miR-146b in K562 cells transfected with miR-146b mimic or inhibitor. K562 cells were transfected with scrambled control (neg_control or in_control), miR-146b mimic, or miR-146b inhibitor. After 24 h of transfection, the cells were induced by hemin for 0, 24, 48, or 72 h. C, benzidine staining of hemoglobin-containing cells. The hemoglobin-containing cells were stained dark blue/brown. The percentage of benzidine-positive cells is indicated below the panel. D, flow cytometry analysis of erythroid marker CD235a-positive cells in K562 cells treated as described above. A representative experiment is presented at left, and statistical analysis from three independent experiments is shown at right. Asterisk indicates significant changes in the indicated groups compared with the control (*, p < 0.05).
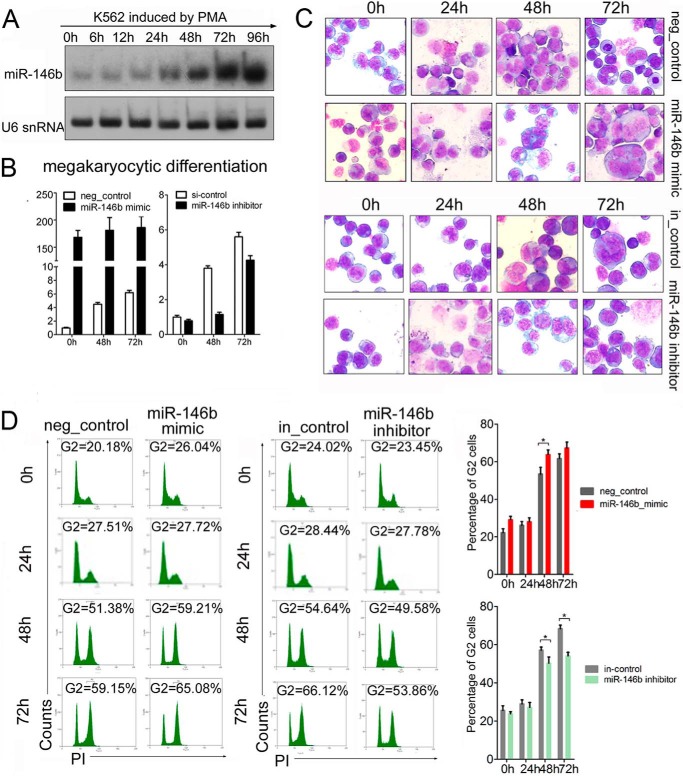
FIGURE 2.
miR-146b promotes megakaryocytic differentiation of K562 cells. A, Northern blot analysis of miR-146b during PMA-induced megakaryocytic differentiation of K562 cells. U6 snRNA was detected to check equal RNA loading. B, qRT-PCR analysis of miR-146b in K562 cells transfected with miR-146b mimic or inhibitor. K562 cells were transfected with scrambled control (neg_control or in_control), miR-146b mimic, or miR-146b inhibitor. After 24 h of transfection, the cells were induced by PMA for 0, 24, 48, or 72 h. C, morphological analysis of megakaryocytic differentiation by May-Grunwald Giemsa staining in the K562 cells that were transfected as described on the right. After 24 h of transfection, the cells were induced by PMA at the indicated times. D, flow cytometry analysis of megakaryocytic DNA content by PI staining. The left peak arose from 2N and the right peak from 4N (G2) cells. A representative experiment is presented in the left, and statistical analysis from three independent experiments is shown in the right. (*) Significant changes in the indicated groups compared with the control (*, p < 0.05).
miR-146b Promotes Erythroid Differentiation and Megakaryocytic Differentiation of K562 Cells
To examine the effect of miR-146b on erythroid and megakaryocytic differentiation, an miR-146b mimic was transfected into K562 cells, and qPCR was performed to confirm the transfection efficiency (Figs. 1B and 2B). We then evaluated hemin-driven differentiation in miR-146b mimic-transfected K562 cells. Benzidine staining demonstrated that ectopic miR-146b increased the proportion of hemoglobin-containing cells after hemin treatment in K562 cells (Fig. 1C). Fluorescence-activated cell sorting (FACS) analysis using a major erythroid cell surface marker (CD235a) showed that excess miR-146b increased the percentage of CD235a+ cells (∼8% at 24 h, 11% at 48 h, and 9% at 72 h after hemin induction) compared with cells transfected with the scrambled control (Fig. 1D). By contrast, the introduction of miRNA inhibitor of miR-146b (anti-146b), which resulted in a significantly reduced miR-146b level (Fig. 1B), inhibited hemin-induced erythroid differentiation of K562 cells. This inhibition was indicated by the decrease in benzidine-positive cells (Fig. 1C) and reduced percentage of CD235a+ cells (∼13% at 48 h and 14% at 72 h after hemin induction) (Fig. 1D) compared with scrambled control transfection. These results indicate that miR-146b promotes erythroid differentiation in K562 cells.
miR-146b mimic-transfected K562 cells were induced to megakaryocytic differentiation by PMA to assess its effect on megakaryocytic maturation. Similar to the aforementioned observations, the ectopic expression of miR-146b enhanced the megakaryocytic morphological features as follows: larger cells with polylobulated or polysegmented nuclei and a basophilic cytoplasm (Fig. 2C). These cells were then stained with PI. Ploidy analysis by flow cytometry showed that the peak corresponding to 4N (G2) cells increased in both miR-146b mimic-transfected and scrambled control-transfected K562 cells after PMA induction. Moreover, the percentage of 4N (G2) cells in K562 cells overexpressing miR-146b at both 48 and 72 h were higher (∼8% at 48 h and 6% at 72 h after PMA induction) than that in cells transfected with the scrambled control (Fig. 2D). By contrast, miR-146b inhibitor transfection delayed megakaryocytic maturation in K562 cells as indicated by the decrease in megakaryocytic morphological cells (Fig. 2C) and reduced percentage of 4N (G2) cells (∼5% at 48 h and 13% at 72 h after PMA induction) compared with scrambled control transfection (Fig. 2D). These results indicate the positively regulatory functions of miR-146b in megakaryocytic differentiation of K562 cells.
PDGFRA mRNA Is Identified as a Direct Target of miR-146b
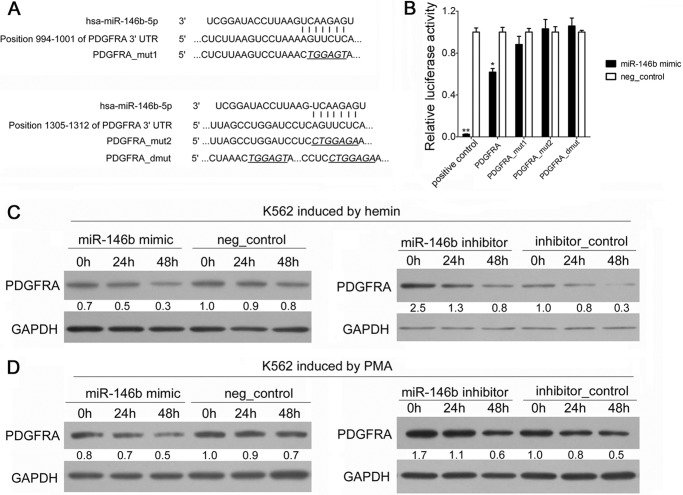
Given that miRNAs function by negatively regulating expression of their target genes, at post-transcriptional level through binding to the complementary sequence in mRNA 3′UTRs, we used TargetScan algorithm to predict potential mRNAs with miR-146b-binding sites. PDGFRA mRNA was predicted as a potential target of miR-146b based on the presence of miRNA-binding sites in its 3′UTR (Fig. 3A). We cloned the 3′UTR of PDGFRA into a luciferase reporter construct (pGL3). Reporter assays in 293T cells revealed miRNA-dependent repression of this 3′UTR, and introduction of mutations to either one or both of the two miR-146b-binding sites abrogated this reduction in luciferase activity (Fig. 3B). Consistent with the reporter assay, we observed a decrease in PDGFRA protein expression in the K562 cells that were transfected with miR-146b mimic and induced to either erythroid or megakaryocytic differentiation (Fig. 3, C and D). By contrast, PDGFRA levels increased after endogenous miR-146b was blocked by the transfection of miRNA inhibitor in both hemin- and PMA-treated K562 cells (Fig. 3, C and D). In addition, we observed the decreased expression of PDGFRA during either erythroid or megakaryocytic differentiation of K562 cells. This finding was reciprocal to the enhanced accumulation of miR-146b, verifying the functional significance of miR-146b and its target. These results demonstrate PDGFRA as a direct target of miR-146b during erythroid and megakaryocytic differentiation.
FIGURE 3.
PDGFRA mRNA was identified and validated as a direct target of miR-146b. A, computer prediction of conserved binding sites within the 3′UTR of PDGFRA mRNA for miR-146b. The mutated base sequences in the luciferase reporter assays are italicized and underlined. B, relative luciferase activity of the indicated PDGFRA reporter constructs. Error bars represent the standard deviation obtained from three independent experiments. *, p < 0.05; **, p < 0.01. C, immunoblot analysis of PDGFRA in the K562 cells transfected with scramble or miR-146b mimics or inhibitors, followed by hemin induction for 0, 24, or 48 h. D, immunoblot analysis of PDGFRA in K562 cells transfected with scrambled or miR-146b mimics or inhibitors, and PMA induction for 0, 24, or 48 h. For all Western blots, GAPDH antibody was used to assess equal protein loading. The signal in each lane was quantified using Gelpro32 software, and the ratio of PDGFRA to GAPDH was determined. PDGFRA expression was calculated as the relative fold with respect to its expression in the negative (neg)-control-transfected cells before differentiation induction.
PDGFRA Acts as a Negative Regulator of Erythroid and Megakaryocytic Differentiation
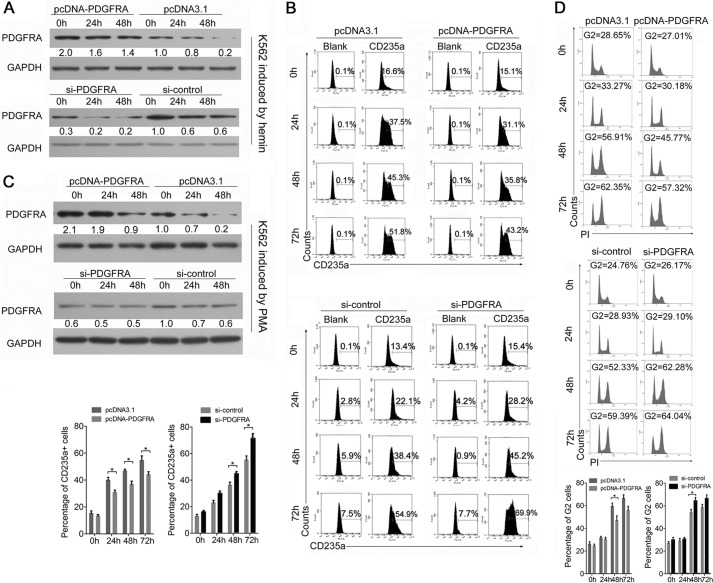
As of this writing, the function of PDGFRA in erythropoiesis and megakaryocytopoiesis is not well known. Given that PDGFRA was down-regulated in K562 cells undergoing erythroid differentiation and megakaryocytic differentiation, PDGFRA was hypothesized to be a negative regulator for lineage maturation. To investigate its biological function in erythroid differentiation, we performed either a loss-of-function or a gain-of-function experiment using PDGFRA siRNAs or pcDNA construct carrying PDGFRA ORF in K562 cells (Fig. 4A). As expected, PDGFRA knockdown increased the percentage of CD235a+ cells (∼7% at 48 h and 15% at 72 h after hemin induction), whereas PDGFRA overexpression decreased the percentage of CD235a+ K562 cells compared with the control (Fig. 4B). The same experiments were performed in PMA-treated K562 cells to examine the regulatory function of PDGFRA in megakaryocytic differentiation (Fig. 4C). Similarly, the loss-of-function study showed enhanced megakaryocytic maturation, and the gain-of-function study showed impaired megakaryocytic differentiation; these findings were revealed by changes in the percentage of 4N (G2) cells (Fig. 4D).
FIGURE 4.
PDGFRA is a negative regulator of erythroid and megakaryocytic differentiation. A, immunoblot analysis of PDGFRA expression in transfected and hemin-induced K562 cells. K562 cells were transfected with a construct carrying PDGFRA ORF or an empty vector and with PDGFRA siRNAs or si-control. After transfection for 24 h, the cells were treated by hemin and harvested at the indicated times after hemin induction. GAPDH antibody was used to assess equal protein loading. PDGFRA expression was calculated as the relative fold change with respect to its expression in control-transfected cells before hemin induction. B, flow cytometry analysis of erythroid marker CD235a-positive cells in the K562 cells treated as described above. A representative experiment is presented in the upper panels, and statistical analysis is presented in the lower panels. Error bars represent the standard deviation obtained from three independent experiments. *, p < 0.05. C, immunoblot analysis of PDGFRA expression in transfected and PMA-induced K562 cells. The K562 cells were transfected with the above constructs and oligonucleotides. After transfection for 24 h, the cells were treated by PMA and harvested at the indicated times. D, flow cytometry analysis of megakaryocytic DNA content by PI staining. A representative experiment is presented in the upper panels, and statistical analysis from three independent experiments is presented in the lower panels. *, p < 0.05.
miR-146b Mediates Erythroid and Megakaryocytic Differentiation via Negative Regulation on Its Target PDGFRA
To clarify the biological link among miR-146b, PDGFRA, and the phenotypic changes, we designed a rescue experiment to assess their functional relevance in K562 cell differentiation. First, an increase in PDGFRA (Fig. 5A), which was accompanied by decreased percentages of differentiated erythroid (CD235a+ cells) (Fig. 5B) and megakaryocytic cells (G2 cells) (Fig. 5C), was observed after transfection of miR-146b inhibitor (miR-146b inhibitor + si_control versus inhibitor-control + si_control). Furthermore, the re-transfection of specific PDGFRA siRNAs led to an extra reduction upon the increase in PDGFRA protein resulting from miR-146b inhibitor transfection (Fig. 5A). This reduction was accompanied by restoration of the percentages of differentiated erythroid (Fig. 5B) and megakaryocytic cells (Fig. 5C) (miR-146b inhibitor + si_control versus miR-146b inhibitor + si-PDGFRA). These data confirm that miR-146b mediated both erythropoiesis and megakaryocytopoiesis via negative regulation on PDGFRA mRNA.
FIGURE 5.
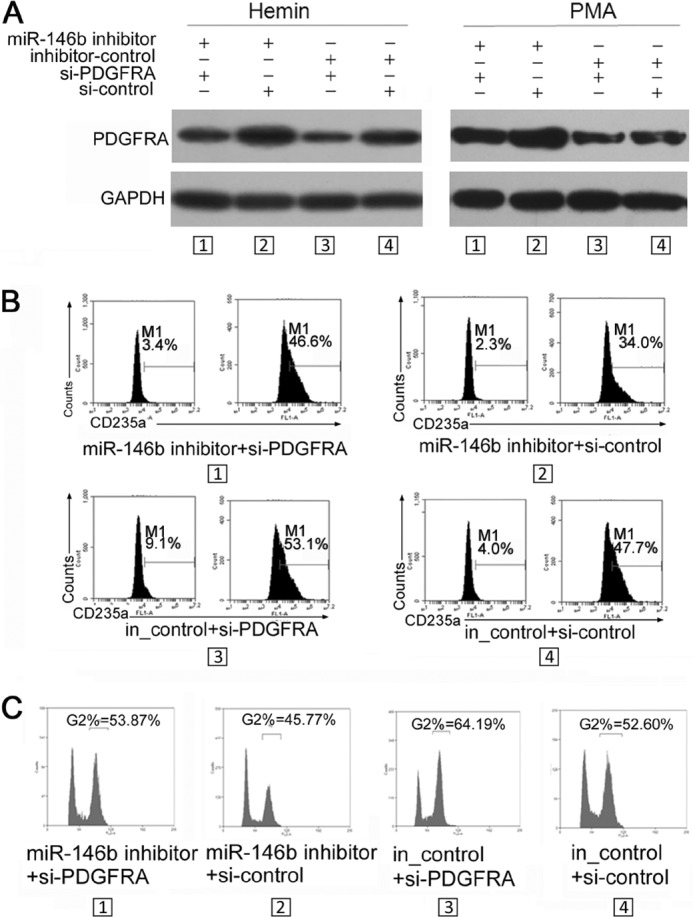
Rescue assays demonstrated that miR-146b promotes erythroid and megakaryocytic differentiation via directly targeting and down-regulating PDGFRA. A, immunoblot analysis of PDGFRA in K562 cells with different treatments. K562 cells were transfected with miR-146b inhibitor or scrambled inhibitor controls. After 24 h of transfection, the cells were subsequently re-transfected with control siRNAs or siRNAs specific to PDGFRA and treated with hemin or PMA for 48 h. B, flow cytometry analysis of erythroid differentiation of K562 cells treated as described above. C, PI staining analysis of megakaryocytic differentiation of K562 cells treated as described above.
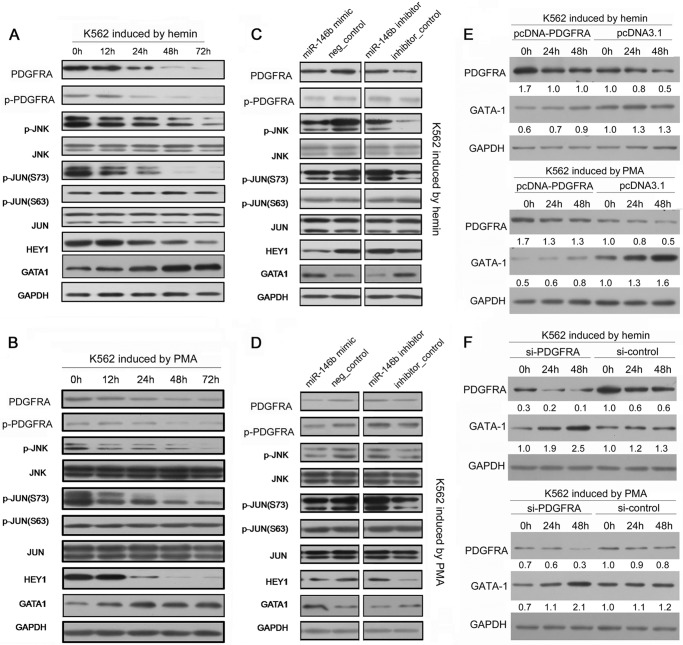
miR-146b Regulates GATA-1 Expression via the JNK/JUN/HEY1 Pathway
PDGFR signals can mediate the synergistic expansion of primary erythroid/megakaryocytic precursors by activating the JNK-JUN pathway (23, 24). Another report indicated that JUN can block erythropoiesis via transcriptional inhibition of GATA-1 mediated by HEY1 (25). These results suggested the possible cascade downstream of miR-146b/PDGFRA. To verify this hypothesis, we first assessed the existence of this pathway in erythroid and megakaryocytic differentiation of K562 cells. As shown in Fig. 6, A and B, the levels of PDGFRA, p-JNK, p-JUN, and HEY1 continuously decreased, whereas GATA-1 levels increased after hemin or PMA induction. We then further determined the effects of miR-146b overexpression or inhibition on the activity of the PDGFRA/JNK/JUN/HEY1 pathway in K562 cells treated by hemin and PMA, respectively. miR-146b overexpression significantly silenced this pathway as indicated by the decreased levels of PDGFRA, p-JNK, p-JUN, and the negative regulator of GATA-1 gene, HEY1, resulting in increased GATA-1 expression (Fig. 6, C and D). By contrast, anti-miR-146b activated this pathway and inhibited the GATA-1 expression (Fig. 6, C and D).
FIGURE 6.
miR-146b regulates the PDGFRA-JNK-JUN-GATA-1 pathways in erythroid and megakaryocytic differentiation. A, immunoblot analysis of PDGFRA, JNK, and JUN levels; phosphorylated PDGFRA, JNK, and JUN levels; and HEY1 and GATA-1 levels during hemin-induced erythroid differentiation of K562 cells. B, immunoblot analysis of PDGFRA, JNK, and JUN levels; phosphorylated PDGFRA, JNK, and JUN levels; and HEY1 and GATA-1 levels during PMA-induced megakaryocytic differentiation of K562 cells. C, immunoblot analysis of PDGFRA, JNK, and JUN levels; phosphorylated PDGFRA, JNK, and JUN levels; and JHEY1 and GATA-1 levels in the K562 cells transfected with miR-146b mimics or inhibitors or their controls and treated by hemin induction for 48 h. D, immunoblot analysis of PDGFRA, JNK, and JUN levels and phosphorylated PDGFRA, JNK, and JUN levels in the K562 cells transfected with miR-146b mimics or inhibitors or their controls and treated by PMA induction for 48 h. For all Western blots, GAPDH antibody was used to assess equal protein loading. E, immunoblot analysis of PDGFRA and GATA-1 levels in the K562 cells transfected with a construct carrying the PDGFRA ORF or an empty vector and with PDGFRA siRNAs or si-control and treated by hemin induction for 0, 24, and 48 h. F, immunoblot analysis of PDGFRA and GATA-1 levels in the K562 cells transfected with a construct carrying PDGFRA ORF or an empty vector and PDGFRA siRNAs or si-control and treated with PMA induction for 0, 24, and 48 h. For Western blots in E and F, GAPDH antibody was used to assess equal protein loading. The expression levels of PDGFRA and GATA-1 were calculated as a relative fold with respect to their expression in control-transfected cells before differentiation induction.
Several other factors have been reported to regulate GATA-1 expression in K562 cells. To confirm whether the miR-146b/PDGFRA/JNK/JUN/HEY1 pathway could affect GATA-1 expression, we also detected GATA-1 expression in the K562 cells transfected with pcDNA-PDGFRA or si-PDGFRA. Compared with the control cells, the overexpression of PDGFRA reduced GATA-1 levels, whereas knockdown of PDGFRA increased GATA-1 levels in the indicated time points during induced erythroid and megakaryocytic differentiation of K562 cells (Fig. 6, E and F). These findings suggest the important mediator functions of miR-146b target, PDGFRA, on GATA-1 expression.
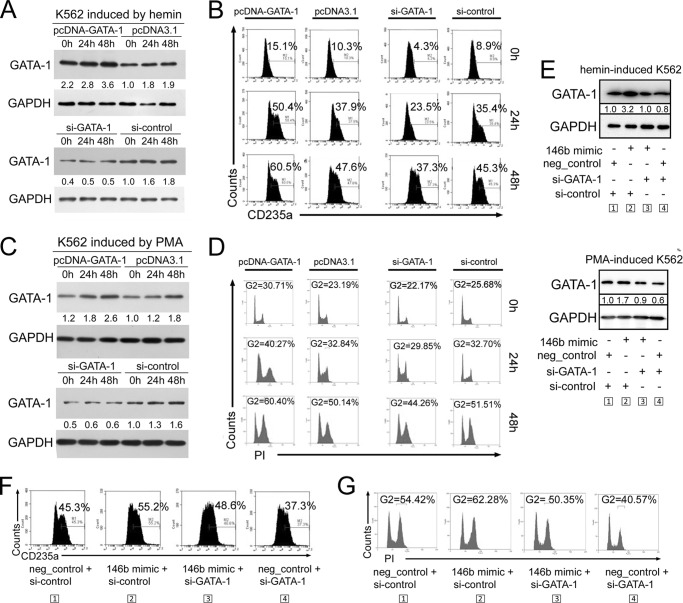
To examine whether GATA-1 levels, which were affected by miR-146b, influence erythroid and megakaryocytic differentiation, we performed gain-of-function and loss-of-function analysis of GATA-1 in K562 cells. The results demonstrate that overexpression of GATA-1 by pcDNA-GATA-1 transfection promoted hemin-induced erythroid differentiation and PMA-induced megakaryocytic differentiation, whereas knockdown of GATA-1 by si-GATA-1 transfection inhibited hemin-induced erythroid differentiation (Fig. 7, A and B) and PMA-induced megakaryocytic differentiation (Fig. 7, C and D). These findings were consistent with miR-146b overexpression and knockdown.
FIGURE 7.
miR-146b mediates erythropoiesis and megakaryocytopoiesis via modulating GATA-1 levels. A, immunoblot analysis of GATA-1 levels in the K562 cells transfected with GATA-1 siRNAs or si-control and a construct carrying GATA-1 ORF (pcDNA-GATA-1) or an empty vector (pcDNA3.1) and induced with hemin for 0, 24, and 48 h. B, flow cytometry analysis of erythroid differentiation of K562 cells treated as described in A. C, immunoblot analysis of GATA-1 levels in the K562 cells transfected with GATA-1 siRNAs or si-control and pcDNA-GATA-1 or pcDNA3.1 and induced with PMA for 0, 24, and 48 h. D, PI staining analysis of megakaryocytic differentiation of K562 cells treated as described in C. E–G, rescue assays demonstrated that miR-146b promoted erythroid and megakaryocytic differentiation by modulating GATA-1 levels. K562 cells were transfected with miR-146b mimic or scrambled mimic controls. After 24 h of transfection, the cells were subsequently re-transfected with control siRNAs or siRNAs specific to GATA-1 and treated with hemin or PMA for 48 h. E, immunoblot analysis of GATA-1 in the K562 cells treated as described above. F, flow cytometry analysis of erythroid differentiation of K562 cells treated as described above. G, PI staining analysis of megakaryocytic differentiation of K562 cells treated as described above.
Furthermore, we performed rescue analyses. First, miR-146b mimic transfection resulted in increased GATA-1 levels (Fig. 7E, miR-146b mimic + si-control versus neg_control + si-control), which was accompanied by the increase in percentages of differentiated erythroid (CD235a+ cells) (Fig. 7F) and differentiated megakaryocytic cells (G2 cells) (Fig. 7G). However, re-introduction of si-GATA-1 reduced the increase in GATA-1 from miR-146b mimic transfection (Fig. 7E, miR-146b mimic + si-GATA-1 versus miR-146b mimic + si-control). This reduction decreased the promotive effects on erythroid (Fig. 7F) and megakaryocytic (Fig. 7G) differentiation resulting from miR-146b mimic transfection. These results confirmed that miR-146b mediated both erythropoiesis and megakaryocytopoiesis by modulating GATA-1 levels.
Verification of the Function and Mechanism of miR-146b in Erythroid and Megakaryocytic Differentiation of Human HSPCs
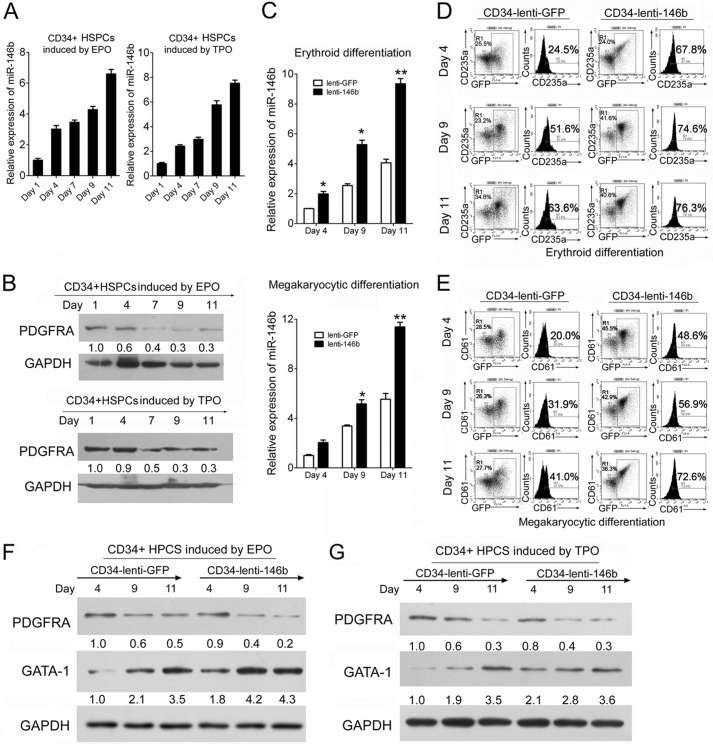
The aforementioned study was performed using hemin- and PMA-induced K562 cells as the differentiation models. To examine the function and mechanism of miR-146b in erythropoiesis and megakaryocytopoiesis in the normal hematopoietic program, we first analyzed the expression of miR-146b and its target in the erythroid and megakaryocytic induction cultures of CD34+ HSPCs derived from umbilical cord blood. A gradual increase in miR-146b expression (Fig. 8A) and a decrease in PDGFRA expression (Fig. 8B) were detected during the induction cultures. The HSPCs were infected with a recombinant lentivirus that expresses miR-146b (Lenti-146b) and were induced to either erythroid or megakaryocytic cultures. Cells were collected at days 4, 9, and 11 in the induction cultures to analyze miR-146b, PDGFRA, and GATA-1 expression and cell differentiation. Real time qPCR revealed a 2–3-fold increase in miR-146b expression in the induction cultures of HSPCs infected with Lenti-146b compared with that in cells transfected with Lenti-GFP-control at each of the aforementioned time points (Fig. 8C). The generation of more mature erythroid cells (CD235a-positive cells) in GFP-positive subsets was consistently faster for Lenti-146b-transduced HSPCs compared with that for Lenti-GFP-transduced control (43% at day 4, 23% at day 9, and 13% at day 11; Fig. 8D). Similarly, the levels of the megakaryocytic marker CD61 were significantly higher in the cells with excess miR-146b (Fig. 8E). A decrease in PDGFRA and an increase in GATA-1 protein levels were also detected in the induction cultures of HSPCs infected with Lenti-146b compared with those in cells transfected with Lenti-GFP-control at each of the aforementioned time points (Fig. 8, F and G). These results demonstrated that miR-146b negatively regulated PDGFRA expression and increased GATA-1 expression, which is involved in the regulation of erythropoiesis and megakaryocytopoiesis of HSPCs.
FIGURE 8.
miR-146b promotes erythroid and megakaryocytic differentiation in CD34+ HSPCs via regulation of its target PDGFRA. A and B, qPCR analysis of miR-146b level in CD34+ HSPCs undergoing erythroid (A) and megakaryocytic (B) differentiation at days 1, 4, 7, 9, and 11. C, qPCR analysis of miR-146b levels in CD34+ HSPCs infected with lentivirus expressing mature miR-146b (Lenti-146b) or a control virus (Lenti-GFP), and cultured in erythroid (upper panels) or megakaryocytic (lower panels) induction medium for the indicated times. Error bars represent the standard deviation obtained from three independent experiments. *, p < 0.05; **, p < 0.01. D, FACS analysis of the erythroid culture of CD34+ HSPCs transduced with Lenti-146b or Lenti-GFP at the indicated induction culture days. E, flow cytometry analysis of the megakaryocytic induction culture of CD34+ HSPCs transduced with Lenti-146b or Lenti-GFP at the indicated induction culture days. F and G, immunoblot analysis of PDGFRA expression in the erythroid (F) and megakaryocytic (G) induction cultures of CD34+ HSPCs transduced with Lenti-GFP control and Lenti-146b. GAPDH antibody was used to assess equal protein loading. The expression levels of PDGFRA and GATA-1 were calculated as a relative fold with respect to their expression at day 4 in the induction cultures of Lenti-GFP-infected CD34+ HSPCs.
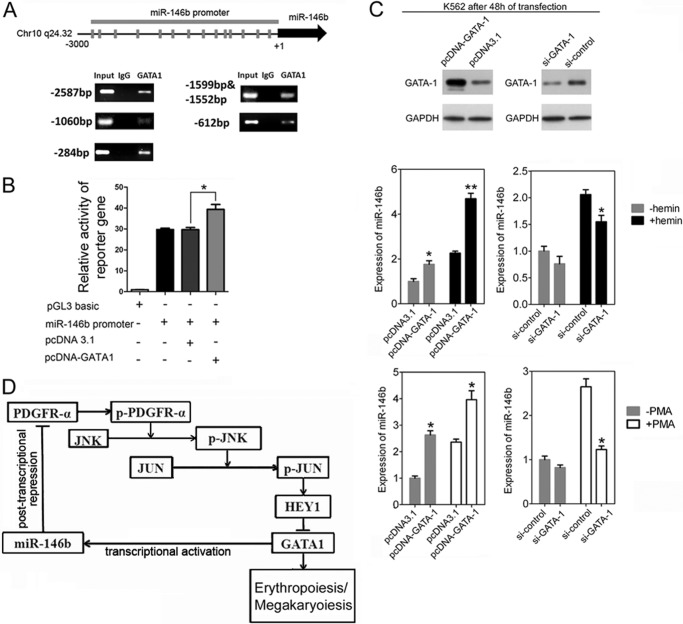
Transcription of miR-146 Gene Is Positively Regulated by GATA-1
In our attempt to investigate the potential factors responsible for miR-146b gene activation during erythroid differentiation and megakaryocytic differentiation, we performed transcription element search system-mediated sequence analysis. The results revealed 14 putative GATA-1 sites scattered within the promoter region of the human miR-146b locus (Fig. 9A). ChIP-PCR results support the binding of GATA-1 upstream of the miR-146b gene (Fig. 9A). The effect of GATA-1 on miR-146b promoter activity was examined by luciferase reporter assay following co-transfection with pcDNA-GATA-1, which expresses GATA-1, and the wild-type pGL-3 promoter construct in 293T cells. Increased GATA-1 levels successfully increased reporter activity by 30%, suggesting the occurrence of GATA-1-directed positive regulation of the miR-146b locus (Fig. 9B).
FIGURE 9.
Transcription of miR-146b gene is positively regulated by GATA-1. A, ChIP-PCR analysis of the GATA-1 hit upstream of miR-146b locus in K562 cells. B, functional activity of GATA-1 on the miR-146b promoter in luciferase reporter analysis. Error bars represent the standard deviation obtained from three independent experiments. *, p <0.05. C, qPCR analysis of miR-146b expression in the transfected K562 cells. The cells were transfected with pcDNA3.1, pcDNA3.1-GATA1, si-GATA-1, and si-control for 24 h, respectively, and then were either induced by hemin or PMA for another 24 h. Upper panel, immunoblot analysis of GATA-1 expression in the transfected K562 cells. Lower panel, qPCR analysis of miR-146b expression in the transfected K562 cells. Error bars represent the standard deviation obtained from three independent experiments. *, p < 0.05. D, schematic representation of the regulatory circuit comprising GATA-1, miR-146b, and PDGFRA in erythroid and megakaryocytic differentiation.
To determine whether GATA-1 influences the expression of miR-146b, miR-146b levels were evaluated in K562 cells transfected with siRNAs specific to GATA-1 or pcDNA-GATA-1 (Fig. 9C). Overexpression of GATA-1 enhanced miR-146b expression before and after hemin or PMA treatment (Fig. 9C), whereas GATA-1 knockdown reduced miR-146b levels (Fig. 9C). Overall, our results suggest that a feed-forward circuit containing miR-146b, PDGFRA, and GATA-1 may exist, positively regulating erythroid and megakaryocytic differentiation (Fig. 9D).
DISCUSSION
Hematopoiesis is highly orchestrated by the interaction between transcription factors and noncoding regulators; this process drives pluripotent precursors to differentiate toward mature blood cells (26, 27). Increasing evidence suggested that this differentiation, along with various hematopoietic lineages, including erythropoiesis and megakaryocytopoiesis, is largely regulated by miRNAs. For example, miR-451 is required for erythrocyte maturation in both zebrafish and murine development (28–31); miR-221 and miR-222 inhibit normal erythropoiesis and erythroleukemic cell growth by down-regulating the Kit protein (32); and miR-210 increases the expression of the γ-globin gene in differentiating erythroid cells (33). Our previous work also demonstrated that miR-223, miR-103, and miR-376a inhibit erythroid differentiation by targeting different protein-coding genes (34–36). In this study, we demonstrated that miR-146b promoted erythroid and megakaryocytic development of K562 cells and CD34+ HSPCs via gain- and loss-of-function analyses. We also identified PDGFRA mRNA, which encodes a receptor tyrosine kinase that functions in cell survival, proliferation, and differentiation in many tissues, as a direct target of miR-146b.
PDGFR is one of many growth factor receptors, and it has a domain organization consisting of five extracellular immunoglobulin-like domains, a single-spanning transmembrane domain, and intracellular split kinase domain (37, 38). After ligand binding, the fully activated phosphorylated kinase domains from PDGFRA then phosphorylate multiple tyrosine residues of the receptor cytoplasmic part, which act as docking sites for Src homology 2 domains of various signal transduction proteins, including signal transducers and activators of transcription (STATs), phospholipase Cγ, phosphatidylinositol 3-kinase, SRC family kinases, and SHP2 phosphatase (39–42). These pathways lead to the regulation of numerous transcription factors that regulate cell growth and survival, such as c-MYC, AP1, FOXO, or SREBP (43–46). The PDGFR-mediated pathways are crucial for embryonic development, cell proliferation, cell migration, and angiogenesis (44). These pathways have also been linked to several diseases, such as atherosclerosis, fibrosis, and malignant diseases (44). However, the function of PDGFRA in hematopoiesis remains unclear. PDGFRA is expressed in bone marrow but not in blood leukocytes (47). Knock-out mice for PDGF-B or PDGFRβ exhibit anemia and thrombocytopenia (48). Moreover, various rare chromosomal rearrangements of the PDGFRA gene have been associated with myeloproliferative neoplasms, chronic myelomonocytic leukemia, atypical chronic myelogenous leukemia, and chronic eosinophilic leukemia (49). For example, the fusion of FIP1L1 with PDGFRA results from a cryptic internal deletion of 800 kb on the chromosome, which is found in about 10% of patients with idiopathic hypereosinophilia (50). A few cases of systemic mastocytosis and acute myeloid leukemia with eosinophilia have also been described (50). The FIP1L1-PDGFRA fusions provide a proliferative advantage to granulocyte/monocyte progenitors, as well as erythroid progenitors, indicating the requirement of PDGFR in the regulation of proliferation of hematopoietic progenitor cells (50). In this study, we found that PDGFRA continuously decreased during erythroid and megakaryocytic differentiation of both K562 cells and CD34+ HSPCs. Knockdown of PDGFRA in K562 cells efficiently increased the proportion of differentiated cells, because reduced PDGFRA is required for cells to withdraw from the hyperproliferative progenitor state during the terminal stage of differentiation. Our finding that miR-146b regulated PDGFRA suggests that miR-146b may be a potentially therapeutic target for some types of leukemia.
The megakaryocytic and erythroid lineages are tightly associated during differentiation, and both lineages are generated from a bipotent megakaryocyte-erythroid progenitor. Although several key differences exist in their regulatory networks in megakaryocytic development and erythroid development from the megakaryocyte-erythroid progenitor, these two closely related hematopoietic lineages share many regulators, including transcription factors, such as GATA-1, GFI-1B, c-Kit, and ZNF-16 (51, 52), and signaling molecules, such as JAK2 and STAT5 (51). Although the function of miR-146b or the proposed regulatory circuit may not be exactly parallel in erythroid and megakaryocytic differentiation, we observed a consistent tendency that increased miR-146b expression down-regulated the expression of the target PDGFRA and then silenced the JNK-JUN-HEY1 pathway, resulting in increased transcription of GATA-1, which could be negatively regulated by HEY1. Thus, erythroid and megakaryocytic differentiation was promoted.
GATA-1 has been demonstrated to be essential for erythroid and megakaryocytic development (53). In this study, we also found that GATA-1 combined to the chromatin sites of the miR-146b promoter and promoted its transcriptional activation during erythroid and megakaryocytic differentiation. These results suggest the involvement of a feed-forward circuit containing miR-146b, PDGFRA, and GATA-1 in erythroid and megakaryocytic differentiation regulation.
This work was supported by National Natural Science Foundation of China Grants 30871249 and 31171311.
- miRNA
- microRNA
- miR-146b
- microRNA-146b-5p
- PDGFR
- platelet-derived growth factor receptor
- PDGFRA
- PDGFR α
- GATA-1
- GATA-binding factor 1
- HSPC
- hematopoietic stem/progenitor cells
- qPCR
- quantitative real time PCR
- PMA
- phorbol 12-myristate 13-acetate
- PI
- propidium iodide.
REFERENCES
- 1. Gregory R. I., Chendrimada T. P., Cooch N., Shiekhattar R. (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640 [DOI] [PubMed] [Google Scholar]
- 2. Bushati N., Cohen S. M. (2007) microRNA functions. Annu. Rev. Cell Dev. Biol. 23, 175–205 [DOI] [PubMed] [Google Scholar]
- 3. Ameres S. L., Zamore P. D. (2013) Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14, 475–488 [DOI] [PubMed] [Google Scholar]
- 4. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 5. Bernstein E., Kim S. Y., Carmell M. A., Murchison E. P., Alcorn H., Li M. Z., Mills A. A., Elledge S. J., Anderson K. V., Hannon G. J. (2003) Dicer is essential for mouse development. Nat. Genet. 35, 215–217 [DOI] [PubMed] [Google Scholar]
- 6. Yang W. J., Yang D. D., Na S., Sandusky G. E., Zhang Q., Zhao G. (2005) Dicer is required for embryonic angiogenesis during mouse development. J. Biol. Chem. 280, 9330–9335 [DOI] [PubMed] [Google Scholar]
- 7. O'Connell R. M., Zhao J. L., Rao D. S. (2011) MicroRNA function in myeloid biology. Blood 118, 2960–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Labbaye C., Testa U. (2012) The emerging role of MIR-146A in the control of hematopoiesis, immune function and cancer. J. Hematol. Oncol. 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sonkoly E., Ståhle M., Pivarcsi A. (2008) MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin. Cancer Biol. 18, 131–140 [DOI] [PubMed] [Google Scholar]
- 10. Perry M. M., Moschos S. A., Williams A. E., Shepherd N. J., Larner-Svensson H. M., Lindsay M. A. (2008) Rapid changes in microRNA-146a expression negatively regulate the IL-1β-induced inflammatory response in human lung alveolar epithelial cells. J. Immunol. 180, 5689–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jazdzewski K., Murray E. L., Franssila K., Jarzab B., Schoenberg D. R., de la Chapelle A. (2008) Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl. Acad. Sci. U.S.A. 105, 7269–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Y., Vandenboom T. G., 2nd, Wang Z., Kong D., Ali S., Philip P. A., Sarkar F. H. (2010) miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 70, 1486–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao J. L., Rao D. S., Boldin M. P., Taganov K. D., O'Connell R. M., Baltimore D. (2011) NF-κB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. U.S.A. 108, 9184–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Curtale G., Citarella F., Carissimi C., Goldoni M., Carucci N., Fulci V., Franceschini D., Meloni F., Barnaba V., Macino G. (2010) An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood 115, 265–273 [DOI] [PubMed] [Google Scholar]
- 15. Dai R., Phillips R. A., Zhang Y., Khan D., Crasta O., Ahmed S. A. (2008) Suppression of LPS-induced interferon-γ and nitric oxide in splenic lymphocytes by select estrogen-regulated microRNAs: a novel mechanism of immune modulation. Blood 112, 4591–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kafasla P., Karakasiliotis I., Kontoyiannis D. L. (2012) Decoding the functions of post-transcriptional regulators in the determination of inflammatory states: focus on macrophage activation. Wiley Interdiscip. Rev. Syst. Biol. Med. 4, 509–523 [DOI] [PubMed] [Google Scholar]
- 17. Crosby J. R., Seifert R. A., Soriano P., Bowen-Pope D. F. (1998) Chimaeric analysis reveals role of Pdgf receptors in all muscle lineages. Nat. Genet. 18, 385–388 [DOI] [PubMed] [Google Scholar]
- 18. Ostman A. (2004) PDGF receptors-mediators of autocrine tumor growth and regulators of tumor vasculature and stroma. Cytokine Growth Factor Rev. 15, 275–286 [DOI] [PubMed] [Google Scholar]
- 19. Boström H., Willetts K., Pekny M., Levéen P., Lindahl P., Hedstrand H., Pekna M., Hellström M., Gebre-Medhin S., Schalling M., Nilsson M., Kurland S., Törnell J., Heath J. K., Betsholtz C. (1996) PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell 85, 863–873 [DOI] [PubMed] [Google Scholar]
- 20. Boström H., Gritli-Linde A., Betsholtz C. (2002) PDGF-A/PDGF α-receptor signaling is required for lung growth and the formation of alveoli but not for early lung branching morphogenesis. Dev. Dyn. 223, 155–162 [DOI] [PubMed] [Google Scholar]
- 21. Yu J., Wang F., Yang G. H., Wang F. L., Ma Y. N., Du Z. W., Zhang J. W. (2006) Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem. Biophys. Res. Commun. 349, 59–68 [DOI] [PubMed] [Google Scholar]
- 22. Yu J., Ryan D. G., Getsios S., Oliveira-Fernandes M., Fatima A., Lavker R. M. (2008) MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc. Natl. Acad. Sci. U.S.A. 105, 19300–19305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marshall C. J. (1995) Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80, 179–185 [DOI] [PubMed] [Google Scholar]
- 24. Jin N., Hatton N., Swartz D. R., Xia Xl, Harrington M. A., Larsen S. H., Rhoades R. A. (2000) Hypoxia activates jun-N-terminal kinase, extracellular signal-regulated protein kinase, and p38 kinase in pulmonary arteries. Am. J. Respir. Cell Mol. Biol. 23, 593–601 [DOI] [PubMed] [Google Scholar]
- 25. Elagib K. E., Xiao M., Hussaini I. M., Delehanty L. L., Palmer L. A., Racke F. K., Birrer M. J., Shanmugasundaram G., McDevitt M. A., Goldfarb A. N. (2004) Jun blockade of erythropoiesis: role for repression of GATA-1 by HERP2. Mol. Cell. Biol. 24, 7779–7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith C. (2003) Hematopoietic stem cells and hematopoiesis. Cancer Control 10, 9–16 [DOI] [PubMed] [Google Scholar]
- 27. Martin C. S., Moriyama A., Zon L. I. (2011) Hematopoietic stem cells, hematopoiesis and disease: lessons from the zebrafish model. Genome Med. 3, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhan M., Miller C. P., Papayannopoulou T., Stamatoyannopoulos G., Song C. Z. (2007) MicroRNA expression dynamics during murine and human erythroid differentiation. Exp. Hematol. 35, 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dore L. C., Amigo J. D., Dos Santos C. O., Zhang Z., Gai X., Tobias J. W., Yu D., Klein A. M., Dorman C., Wu W., Hardison R. C., Paw B. H., Weiss M. J. (2008) A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl. Acad. Sci. U.S.A. 105, 3333–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pase L., Layton J. E., Kloosterman W. P., Carradice D., Waterhouse P. M., Lieschke G. J. (2009) miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood 113, 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bruchova-Votavova H., Yoon D., Prchal J. T. (2010) miR-451 enhances erythroid differentiation in K562 cells. Leukemia Lymphoma 51, 686–693 [DOI] [PubMed] [Google Scholar]
- 32. Felli N., Fontana L., Pelosi E., Botta R., Bonci D., Facchiano F., Liuzzi F., Lulli V., Morsilli O., Santoro S., Valtieri M., Calin G. A., Liu C. G., Sorrentino A., Croce C. M., Peschle C. (2005) MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl. Acad. Sci. U.S.A. 102, 18081–18086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bianchi N., Zuccato C., Lampronti I., Borgatti M., Gambari R. (2009) Expression of miR-210 during erythroid differentiation and induction of γ-globin gene expression. BMB Rep. 42, 493–499 [DOI] [PubMed] [Google Scholar]
- 34. Yuan J. Y., Wang F., Yu J., Yang G. H., Liu X. L., Zhang J. W. (2009) MicroRNA-223 reversibly regulates erythroid and megakaryocytic differentiation of K562 cells. J. Cell. Mol. Med. 13, 4551–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang G. H., Wang F., Yu J., Wang X. S., Yuan J. Y., Zhang J. W. (2009) MicroRNAs are involved in erythroid differentiation control. J. Cell. Biochem. 107, 548–556 [DOI] [PubMed] [Google Scholar]
- 36. Wang F., Yu J., Yang G. H., Wang X. S., Zhang J. W. (2011) Regulation of erythroid differentiation by miR-376a and its targets. Cell Res. 21, 1196–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schneider R., Schweiger M. (1991) A novel modular mosaic of cell adhesion motifs in the extracellular domains of the neurogenic trk and trkB tyrosine kinase receptors. Oncogene 6, 1807–1811 [PubMed] [Google Scholar]
- 38. Lokker N. A., O'Hare J. P., Barsoumian A., Tomlinson J. E., Ramakrishnan V., Fretto L. J., Giese N. A. (1997) Functional importance of platelet-derived growth factor (PDGF) receptor extracellular immunoglobulin-like domains. Identification of PDGF binding site and neutralizing monoclonal antibodies. J. Biol. Chem. 272, 33037–33044 [DOI] [PubMed] [Google Scholar]
- 39. Vignais M. L., Gilman M. (1999) Distinct mechanisms of activation of Stat1 and Stat3 by platelet-derived growth factor receptor in a cell-free system. Mol. Cell. Biol. 19, 3727–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lakner A. M., Moore C. C., Gulledge A. A., Schrum L. W. (2010) Daily genetic profiling indicates JAK/STAT signaling promotes early hepatic stellate cell transdifferentiation. World J. Gastroenterol. 16, 5047–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xiong W., Cheng B. H., Jia S. B., Tang L. S. (2010) Involvement of the PI3K/Akt signaling pathway in platelet-derived growth factor-induced migration of human lens epithelial cells. Curr. Eye Res. 35, 389–401 [DOI] [PubMed] [Google Scholar]
- 42. Lin C. C., Lee I. T., Chi P. L., Hsieh H. L., Cheng S. E., Hsiao L. D., Liu C. J., Yang C. M. (2014) c-Src/Jak2/PDGFR/PKCdelta-dependent MMP-9 induction is required for thrombin-stimulated rat brain astrocytes migration. Mol. Neurobiol. 49, 658–672 [DOI] [PubMed] [Google Scholar]
- 43. Besançon F., Atfi A., Gespach C., Cayre Y. E., Bourgeade M. F. (1998) Evidence for a role of NF-κB in the survival of hematopoietic cells mediated by interleukin 3 and the oncogenic TEL/platelet-derived growth factor receptor β fusion protein. Proc. Natl. Acad. Sci. U.S.A. 95, 8081–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tsatsanis C., Spandidos D. A. (2000) The role of oncogenic kinases in human cancer (review). Int. J. Mol. Med. 5, 583–590 [DOI] [PubMed] [Google Scholar]
- 45. Erovic B. M., Harris L., Jamali M., Goldstein D. P., Irish J. C., Asa S. L., Mete O. (2012) Biomarkers of parathyroid carcinoma. Endocr. Pathol. 23, 221–231 [DOI] [PubMed] [Google Scholar]
- 46. Guida T., Anaganti S., Provitera L., Gedrich R., Sullivan E., Wilhelm S. M., Santoro M., Carlomagno F. (2007) Sorafenib inhibits imatinib-resistant KIT and platelet-derived growth factor receptor β gatekeeper mutants. Clin. Cancer Res. 13, 3363–3369 [DOI] [PubMed] [Google Scholar]
- 47. Wernig G., Mercher T., Okabe R., Levine R. L., Lee B. H., Gilliland D. G. (2006) Expression of Jak2V617F causes a polycythemia vera-like disease with associated myelofibrosis in a murine bone marrow transplant model. Blood 107, 4274–4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cao Y. (2005) Direct role of PDGF-BB in lymphangiogenesis and lymphatic metastasis. Cell Cycle 4, 228–230 [DOI] [PubMed] [Google Scholar]
- 49. Ozawa T., Brennan C. W., Wang L., Squatrito M., Sasayama T., Nakada M., Huse J. T., Pedraza A., Utsuki S., Yasui Y., Tandon A., Fomchenko E. I., Oka H., Levine R. L., Fujii K., Ladanyi M., Holland E. C. (2010) PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 24, 2205–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pardanani A., Ketterling R. P., Brockman S. R., Flynn H. C., Paternoster S. F., Shearer B. M., Reeder T. L., Li C. Y., Cross N. C., Cools J., Gilliland D. G., Dewald G. W., Tefferi A. (2003) CHIC2 deletion, a surrogate for FIP1L1-PDGFRA fusion, occurs in systemic mastocytosis associated with eosinophilia and predicts response to imatinib mesylate therapy. Blood 102, 3093–3096 [DOI] [PubMed] [Google Scholar]
- 51. Doré L. C., Crispino J. D. (2011) Transcription factor networks in erythroid cell and megakaryocyte development. Blood 118, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen J., Li X. B., Su R., Song L., Wang F., Zhang J. W. (2014) ZNF16 (HZF1) promotes erythropoiesis and megakaryocytopoiesis via its regulation on c-KIT gene. Biochem. J. 458, 171–183 [DOI] [PubMed] [Google Scholar]
- 53. Suzuki M., Moriguchi T., Ohneda K., Yamamoto M. (2009) Differential contribution of the Gata1 gene hematopoietic enhancer to erythroid differentiation. Mol. Cell. Biol. 29, 1163–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]