Background: Oligomerization of transcription factor YY1 is not well understood.
Results: YY1 assembles homo-oligomers that bind DNAs without the consensus sequence, whose structure is studied by electron microscopy.
Conclusion: RuvBL1-RuvBL2 enhances YY1 binding to DNAs without the consensus sequence for the transcription factor.
Significance: YY1-RuvBL1-RuvBL2 complexes could contribute to functions beyond transcription, and we find this occurs during homologous recombination.
Keywords: ATPase, DNA Repair, Electron Microscopy (EM), Single Particle Analysis, Structural Biology, Transcription Factor, RuvBL1, RuvBL2, YY1
Abstract
Yin Yang 1 (YY1) is a transcription factor regulating proliferation and differentiation and is involved in cancer development. Oligomers of recombinant YY1 have been observed before, but their structure and DNA binding properties are not well understood. Here we find that YY1 assembles several homo-oligomeric species built from the association of a bell-shaped dimer, a process we characterized by electron microscopy. Moreover, we find that YY1 self-association also occurs in vivo using bimolecular fluorescence complementation. Unexpectedly, these oligomers recognize several DNA substrates without the consensus sequence for YY1 in vitro, and DNA binding is enhanced in the presence of RuvBL1-RuvBL2, two essential AAA+ ATPases. YY1 oligomers bind RuvBL1-RuvBL2 hetero-oligomeric complexes, but YY1 interacts preferentially with RuvBL1. Collectively, these findings suggest that YY1-RuvBL1-RuvBL2 complexes could contribute to functions beyond transcription, and we show that YY1 and the ATPase activity of RuvBL2 are required for RAD51 foci formation during homologous recombination.
Introduction
Yin Yang 1 (YY1)3 is a polycomb group transcription factor that regulates important cellular events through activation or repression of transcription. YY1 is ubiquitously expressed in many tissues and participates in replication, differentiation, proliferation, embryogenesis, and development (1–4). YY1 overexpression in some tumors correlates with recurrences and bad prognosis, and YY1 has been proposed as a marker for cancer progression (5). Interestingly, recent reports have broadened the spectrum of processes regulated by YY1 such as silencing of retro-elements in embryonic cells (6), regulation of long noncoding RNAs (7), inactivation of the X chromosome in the females of mammals (4), and a direct role in V(D)J recombination during early B-cell development (8).
Human YY1 is a 414-amino acid protein with a predicted molecular mass of 44 kDa and highly conserved in eukaryotes (sequence identity between human and mouse >98%) (1) (Fig. 1A). Putative structural and functional homologs in Schizosaccharomyces pombe (Iec1) (9) and Drosophila melanogaster (PHO (pleiohomeotic) and PHOL (pleiohomeotic-like)) (10) have been studied. YY1 hallmark is four C2H2-type zinc finger motifs located at its C terminus (residues 298–397), responsible for the sequence-specific recognition of a consensus DNA sequence (5′-(C/g/a)(G/t)(C/t/a)CATN(T/a)(T/g/c)-3′, where the uppercase letters indicate the preferred base for each position) being the most frequent ACAT and CCAT. This C-terminal region harbors part of the transcriptional repression activity of YY1. The co-crystal structure of residues 293–414 bound to the adeno-associated virus P5 promoter showed the four zinc fingers interacting with the DNA major groove. The structure revealed that the interaction with the consensus sequence is defined by specific contacts between YY1 side chains and bases of the DNA (11). Less is known about other segments of the protein. The N terminus has been implicated in transcriptional activation, whereas residues 170–201 participate in transcriptional repression (12, 13). Residues 201–226 form the REPO (recruitment of polycomb) domain, involved in polycomb group repression (14).
FIGURE 1.
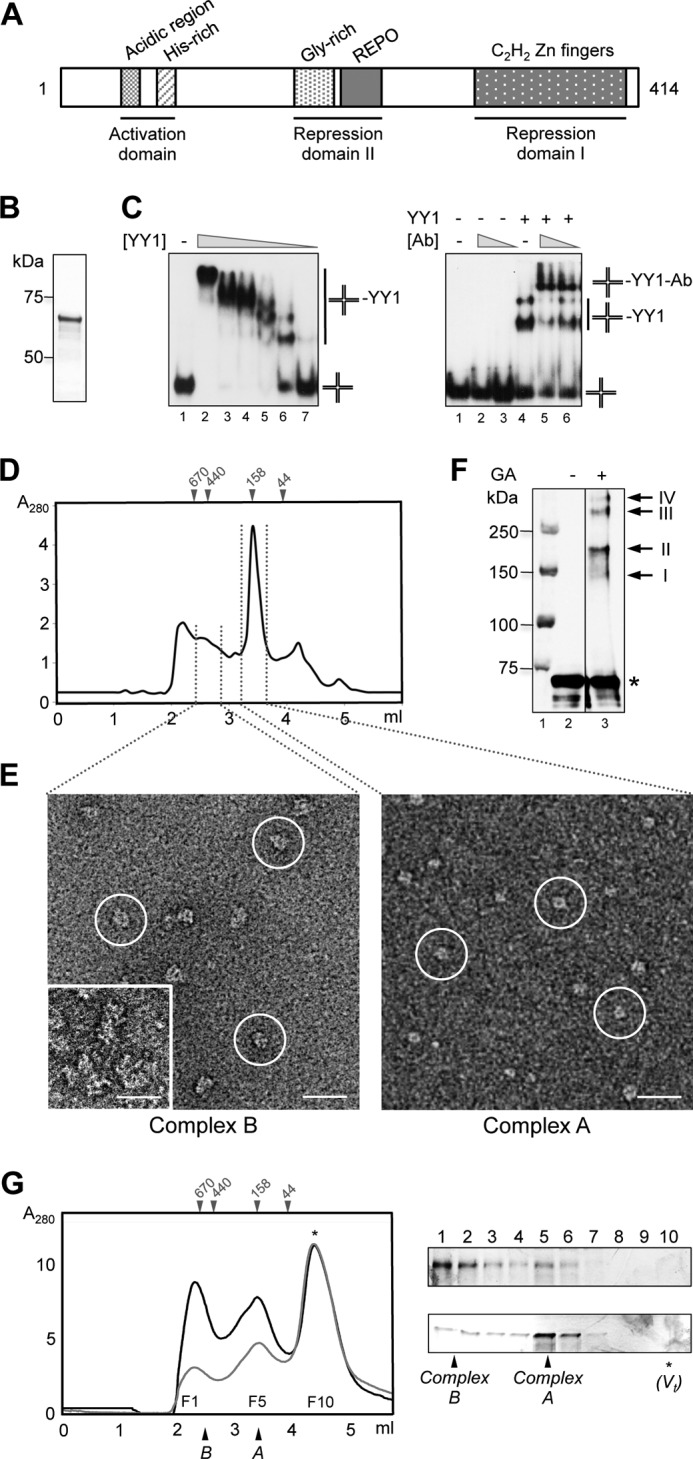
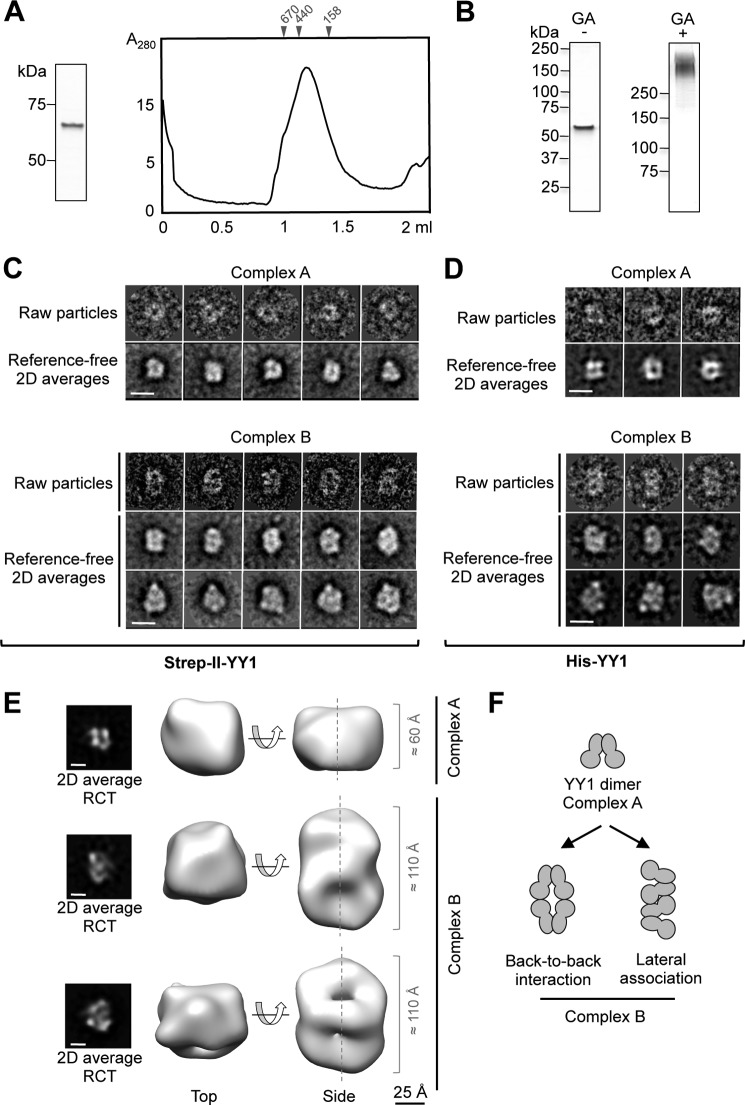
Purification and characterization of human YY1. A, scheme of human YY1 primary sequence. Relevant domains and motifs are indicated. B, SimplyBlue (Invitrogen) staining SDS-PAGE of purified His-YY1. C, left panel, His-YY1 was incubated with a synthetic HJ DNA (described as J3 under “Experimental Procedures”), and reactions were resolved by 6% non-denaturating PAGE. 0.3 nm of HJ (J3) (lane 1) was titrated with increasing amounts of YY1 (lanes 2–7, 1920, 960, 480, 240, 120, and 60 nm, respectively). Right panel, antibody super shift experiments demonstrated the specificity of the interaction. Lane 1, HJ probe alone (described as HJ_25_no_sp under “Experimental Procedures”); lanes 2 and 3, 2 and 1 μg of anti-YY1 antibody (Ab) alone, respectively; lane 4, YY1 protein alone (100 nm); lanes 5 and 6, incubation of YY1 protein (100 nm) with 2 and 1 μg of anti-YY1 antibody, respectively. Positions of unbound HJ probe, YY1-HJ, and YY1-HJ-antibody shifted-bands are indicated. D, SEC of purified His-YY1 in a BioSep-SEC-S4000 (Phenomenex) column. The column was calibrated with molecular weight standards (GE Healthcare), and the elution volume for some of the standards is shown on the top of the chromatogram. E, different fractions from the SEC in D were observed in the electron microscope, and fields from representative electron micrographs are shown. Typical molecule images are highlighted within circles. The inset in the left panel shows a view of aggregates found to elute first from the SEC column. The scale bar represents 250 Å. F, Western blot with an anti-YY1 antibody of His-YY1 after cross-linking with glutaraldehyde (GA); lane 1, molecular weight standards (Bio-Rad); lane 2, control without GA; lane 3, 30 min of incubation with GA. His-YY1 monomer migrates as a 70-kDa protein in SDS-PAGE (labeled as *). Cross-linked bands, with a relative molecular weight multiples of the YY1 monomer, are labeled with arrows. G, SEC of His-YY1 was as in D (black line). The peak fraction corresponding to complex B was re-injected (gray line) in the same column, and the fractions (F1 to F10) of both experiments were analyzed by silver-stained SDS-PAGE. The top panel corresponds to fractions from the black line and bottom panel for the gray line chromatography, respectively. Positions of the YY1 complexes A and B as well as molecular weight standards (GE Healthcare) used for column calibration are indicated. Total volume of the column (Vt) is indicated by an asterisk (*).
There is evidence that YY1 is important for maintaining genome stability (15), but this could in part work through its role as a transcriptional regulator of genes involved in DNA damage responses (16, 17). Yet YY1 could also have a direct role in DNA damage responses as it has been shown that YY1-INO80 complexes may function in DNA repair by homologous recombination (HR) (15). INO80 is a multisubunit chromatin-remodeling complex that includes, in addition to YY1 and the Ino80 catalytic subunit, RuvBL1 and RuvBL2 (18, 19), two closely related (65% sequence similarity) AAA+ (ATPases associated with diverse cellular activities) proteins (20, 21). They share homology with RuvB, the prokaryotic helicase involved in Holliday junction (HJ) resolution during HR. RuvB assembles as hexameric rings, and its DNA-dependent ATPase activity provides the energy for branch migration (22). RuvBL1 and RuvBL2 are essential in multiple processes including transcription, DNA repair, and nonsense-mediated mRNA decay, and their overexpression is associated with tumorigenesis (20, 21).
X-ray crystallography (23–25) and electron microscopy (EM) (26–28) revealed that RuvBL1 and RuvBL2 are organized in three domains. Domain I (DI) and III (DIII) comprise the catalytic ATPase core, responsible for the oligomerization in homo-hexameric rings with a central channel. RuvBL1 and RuvBL2 also assemble hetero-oligomers with alternating RuvBL1 and RuvBL2 subunits (23–25). DII is a unique insertion connected to the ATPase core, resembling an OB-fold, and recombinant DII domains of RuvBL1 interact with ssDNA, dsDNA, and ssRNA in vitro (23–25). Using cryo-EM we recently solved the structure of human RuvBL1-RuvBL2 complexes assembled as hetero-dodecamers composed of two hexameric rings interacting back-to-back (27). These dodecamers co-existed in two different conformations whose functional significance is still unclear. RuvBL1 and RuvBL2 interact with YY1 in vitro (15), but these complexes have not been characterized in detail.
Although there is evidence that YY1 can oligomerize in vitro (15), little is currently known about the structure or DNA binding activity of these oligomeric complexes. Moreover, it is not known if YY1 forms oligomers in vivo. Here we have analyzed the assembly of YY1 using EM, and we determine the low resolution architecture of these YY1 oligomers, providing new structural information on full-length YY1. In addition, we also provide evidence of YY1 self-association in vivo. Interestingly, we find that YY1 oligomers bind to several DNA substrates without the consensus sequence for YY1 as a transcription factor. These oligomers also bind to RuvBL1-RuvBL2 ATPases in vitro, and we find that RuvBL1-RuvBL2 enhances DNA binding by YY1. These findings help explain some unconventional functions ascribed to YY1 recently that could hardly be understood only on the basis of its function as a transcription factor. We explore one of these, and we show that, consistent with previous results (15), YY1 and RuvBL1-RuvBL2 cooperate during DNA repair, but in addition our results indicate these proteins are required for the correct assembly of RAD51 filaments and that this function is dependent on RuvBL2 ATPase activity.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant Proteins
His-YY1 was expressed using Escherichia coli Origami (DE3) cells, and induction was performed by the addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside (final concentration) at 37 °C for 3 h. For Strep-II-YY1, BL21 (DE3) cells were used, and induction was carried out by the addition of 0.1 mm isopropyl 1-thio-β-d-galactopyranoside (final concentration) at 28 °C for 4 h. In both cases the medium was supplemented with 0.1 mm ZnCl2 (final concentration) at the time of induction, and cell lysis was performed by sonication in 100 mm Na2HPO4/NaH2PO4, 300 mm NaCl, 0.1 mm ZnCl2 containing a mixture of proteases inhibitors (Roche Applied Science). His-YY1 was purified by affinity chromatography using a HisTrap HP column (GE Healthcare) equilibrated in 100 mm Na2HPO4/NaH2PO4, 300 mm NaCl, 50 mm imidazole, 0.1 mm ZnCl2 followed by a cationic exchange Mini S PC 3.2/3 (GE Healthcare) column. Purified His-YY1 was dialyzed against 50 mm Tris-HCl, pH 7.4, 200 mm NaCl, 10% (v/v) glycerol, 0.1 mm ZnCl2 overnight and stored at −80 °C. Strep-II-YY1 was purified in one step using a StrepTrap HP column (GE Healthcare) equilibrated in 50 mm Tris-HCl, pH 7.4, 500 mm NaCl, 10% (v/v) glycerol, 0.1 mm ZnCl2, and eluted using 2.5 mm d-desthiobiotin in the same buffer. Purified Strep-II-YY1 was dialyzed in the same buffer as His-YY1 and stored at −80 °C. In all cases, size exclusion chromatography (SEC) was used as a final step of purification using a BioSep-SEC-S4000 (Phenomenex) or a Superdex 200 PC 3.2/30 column (GE Healthcare) equilibrated in 50 mm Tris-HCl, pH 7.4, 200 mm NaCl, 10% (v/v) glycerol, 0.1 mm ZnCl2.
His-RuvBL1 and His-RuvBL2 were expressed in BL21 (DE3) cells by induction with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside (final concentration) at 28 °C for 4 h. They were purified by affinity chromatography using a HisTrap HP column (GE Healthcare) equilibrated in 50 mm Tris-HCl, pH 7.4, 300 mm NaCl, 10% (v/v) glycerol, 20 mm imidazole and eluted using a gradient of 20–500 mm imidazole. Fractions containing the proteins were pooled and dialyzed overnight at 4 °C in 25 mm Tris-HCl, pH 7.4, 250 mm NaCl. In the indicated cases, SEC was performed using a Superdex 200 PC 3.2/30 (GE Healthcare) or Superdex 200 10/300 GL (GE Healthcare) column equilibrated in 50 mm Tris-HCl, pH 7.4, 250 mm NaCl, 10% (v/v) glycerol. His-RuvBL1-RuvBL2 and RuvBL1-RuvBL2 were produced as described (27).
Affinity Purification of YY1-RuvBL1-RuvBL2 Complexes
Recombinant Strep-II-YY1 was used to test interactions with His-RuvBL1, His-RuvBL2, His-RuvBL1-RuvBL2, and RuvBL1-RuvBL2 proteins in vitro. Pulldown assays were performed by incubation of purified Strep-II-YY1 with a 2-fold molar excess of the purified partner (His-RuvBL1, His-RuvBL2, His-RuvBL1-RuvBL2, or RuvBL1-RuvBL2) in 50 mm Tris-HCl, pH 7.4, 500 mm NaCl, 10% (v/v) glycerol, 0.1 mm ZnCl2 for 20 min on ice followed by the addition of StrepTactin High Performance Resin (GE Healthcare) and further incubation for 30 min at 4 °C with agitation. After 3 washes with the reaction buffer, proteins retained in the resin were eluted with 50 mm Tris-HCl, pH 7.4, 500 mm NaCl, 10% (v/v) glycerol, 0.1 mm ZnCl2, 2.5 mm d-desthiobiotin. All purification steps were monitored by SDS-PAGE, and protein detection was carried out using Oriole® Fluorescent Gel Stain (Bio-Rad) or silver staining.
Glutaraldehyde Cross-linking
Purified His-YY1 (0.1 μg/μl) was dialyzed in 100 mm Na2HPO4/NaH2PO4, 300 mm NaCl, 0.1 mm ZnCl2 for 3 h, and 13.5 μl of the protein were cross-linked in 15-μl reactions on ice using glutaraldehyde 0.005% (v/v) (final concentration) for 30 min. The reactions were stopped using 192 mm Tris-glycine (final concentration). Samples were analyzed by SDS-PAGE and Western blotting using the anti-YY1 antibody.
GraFix
Affinity-purified Strep-II-YY1 (10 μm) was stabilized in a discontinuous glycerol/glutaraldehyde gradient by the GraFix method (29). 100 μl of the sample were applied on the top of the tube. As a control, sample was also applied into a similar glycerol gradient without the cross-linking agent. Samples were centrifuged at 30,000 rpm, 4 °C for 18 h (rotor SW 55 Ti). After centrifugation, fractions were collected, and reactions stopped by the addition of 192 mm Tris-Glycine (final concentration).
Electron Microscopy and Image Processing
A few microliters of each of the samples analyzed were deposited on carbon-coated grids immediately after elution from the SEC column and stained using 2% (w/v) uranyl acetate. All observations were performed in a JEOL 1230 transmission electron microscope operated at 100 kV. Images were collected using a low-dose protocol and a 4k x 4k TVIPS CMOS detector under control of the EM-TOOLS software (TVIPS). Final magnification of the CMOS micrographs was 68222.5×. Contrast transfer function for each micrograph was estimated using CTFFIND3 (30) and corrected using BSOFT (31) before selecting particles. Particles were boxed and extracted from the micrographs using e2boxer in EMAN2 (32). 16362 particles for complex A and 52713 particles for complex B were boxed and classified using reference-free methods in EMAN (33), EMAN2 (32), and XMIPP (34). Particles were binned at 4.56 Å/pixel before further processing. Ab initio structures for each experiment were obtained by the random conical tilt (RCT) method (35) using XMIPP (34). RCT was applied on those molecule images classified as belonging to the same view of the oligomer and using the images of these same molecules after tilting the specimen holder by 45°. The resulting volumes were then used as templates for angular refinement using EMAN (33). The rotational symmetry of the YY1 complexes was determined by calculating the power rotational spectra of sections of the RCT structures generated using SPIDER (36) along the longitudinal axis of the complex, as implemented in XMIPP (34), as well as for the reference-free two-dimensional averages. Refinement was performed without assuming any symmetry, but after some rounds of refinement 2-fold rotational symmetry was imposed and refined further until convergence. The hand selected was that provided by the RCT structure. The resolution of the structure (20 Å) was estimated using Fourier shell correlation (FSC) and a 0.5 correlation coefficient. We verified that the bias of the template used for angular refinement did not affect convergence significantly. For this, we refined the same dataset from four independent templates and imposing 2-fold symmetry. Images were refined from: (i) a RCT structure; (ii) a structure obtained using the common lines method as implemented in the startAny command from EMAN (33); (iii) a model generated using the startcsym program from EMAN (33), searching for particles with the best 2-fold rotational symmetry, which are the defined as top views, and for particles with the best mirror symmetry and poor 2-fold symmetry, defined as sided views; class averages from each group and the 2-fold symmetry were used to generate a three-dimensional model; (iv) a model was generated using e2initialmodel.py in EMAN2 (32), where a random featureless blob model is used to seed a refinement using reference-free class averages. All structures were filtered according to the estimated resolution and rendered using UCSF Chimera (37).
DNA Substrates
DNAs were made by annealing combinations of oligonucleotides (Table 1). In each case one of the oligonucleotides was labeled at the 5′-end by polynucleotide kinase with [γ-32P]ATP before annealing. Holliday junction J3, which has 40-bp arms, was made from oligonucleotides J3-1 to J3-4, the duplex dsDNA_no_sp_1 (not containing the consensus sequence) from J3-2 and J3-5, and dsDNA_sp_1 (containing the consensus sequence) from 43-1 and 43-2. The 80-nt ssDNA substrate used was labeled J3-2 oligonucleotide. Holliday junction HJ_25 (with 25-bp arms) was made from oligonucleotides HJ_1 to HJ_4. Annealing was performed in 100 mm phosphate buffer, pH 7.5, with the appropriate combinations of oligonucleotides, mixing one radiolabeled oligonucleotide with cold complementary oligonucleotides in a 1:2 ratio. The annealed products were resolved on an 8% non-denaturing polyacrylamide gel. The bands containing the annealed substrates were purified as described in Zecchi et al. (38). DNA concentration was calculated by scintillation counting. DNA substrates labeled with Cy5 (Sigma) at the 5′-end were annealed as described for the γ-32P-labeled substrates. Holliday junction HJ_25_sp was made from oligonucleotides HJ_12 and HJ_34_1 and HJ_25_no_sp from HJ_12 and HJ_34_2. dsDNA containing (dsDNA_sp_2) or not (dsDNA_no_sp_2) the consensus sequence for YY1 were made from oligonucleotides DS_1 and DS_2, and DS_3 and DS_4, respectively.
TABLE 1.
Sequences of oligonucleotides used for constructing several DNA substrates
| Name | Sequence 5′-3 | 5′-Label |
|---|---|---|
| J3-1 | CGCAAGCGACAGGAACCTCGAGAAGCTTCCGGTAGCAGCCTGAGCGGTGGTTGAATTCCTCGAGGTTCCTGTCGCTTGCG | |
| J3-2 | CGCAAGCGACAGGAACCTCGAGGAATTCAACCACCGCTCAACTCAACTGCAGTCTAGACTCGAGGTTCCTGTCGCTTGCG | γ-32P |
| J3-3 | CGCAAGCGACAGGAACCTCGAGTCTAGACTGCAGTTGAGTCCTTGCTAGGACGGATCCCTCGAGGTTCCTGTCGCTTGCG | |
| J3-4 | CGCAAGCGACAGGAACCTCGAGGGATCCGTCCTAGCAAGGGGCTGCTACCGGAAGCTTCTCGAGGTTCCTGTCGCTTGCG | |
| J3-5 | CGCAAGCGACAGGAACCTCGAGTCTAGACTGCAGTTGAGTTGAGCGGTGGTTGAATTCCTCGAGGTTCCTGTCGCTTGCG | |
| 43-1 | ACAGGAATTCCTCGAGGCCGCCATTTTGTGCTGTCTAGAGACT | γ-32P |
| 43-2 | AGTCTCTAGACAGCACAAAATGGCGGCCTCGAGGAATTCCTGT | |
| HJ_1 | GACGCTGCCGAATTCTGGCGTTAGGAGATACCGATAAGCTTCGGCTTAA | γ-32P |
| HJ_2 | CTTAAGCCGAAGCTTATCGGTATCTTGCTTACGACGCTAGCAAGTGATC | |
| HJ_3 | TGATCACTTGCTAGCGTCGTAAGCAGCTCGTGCTGTCTAGAGACATCGA | |
| HJ_4 | ATCGATGTCTCTAGACAGCACGAGCCCTAACGCCAGAATTCGGCAGCGT | |
| HJ_12 | GACGCTGCCGAATTCTGGCGTTAGGAGATACCGATAAGCTTCGGCTTAATTTCTTAAGCCGAAGCTTATCGGTATCTTGCTTACGACGCTAGCAAGTGATC | Cy5 |
| HJ_34_1 | TGATCACTTGCTAGCGTCGTAAGCAGCTCGTGCTGTCTAGAGACATCGATTTATCGATGTCTCTAGACAGCACGAGCCCTAACGCCAGAATTCGGCAGCGT | |
| HJ_34_2 | TGATCACTTGCTAGCGTCGTAAGCAGCTCGTGCTGTCTAGAGTATACGATTTATCGTATACTCTAGACAGCACGAGCCCTAACGCCAGAATTCGGCAGCGT | |
| DS_1 | AGGGTCTCCATTTTGAAGCG | Cy5 |
| DS_2 | CGCTTCAAAATGGAGACCCT | |
| DS_3 | CTGAAGGGGGGCTATAAAAGGGGGT | Cy5 |
| DS_4 | ACCCCCTTTTATAGCCCCCCTTCAG |
Electrophoretic Mobility Shift Assays (EMSA)
Binding of YY1 and RuvBL1-RuvBL2 to DNA was analyzed through EMSA using different radiolabeled DNA substrates (0.3 nm). Reactions were performed in 50 mm Tris-HCl, pH 7.5, 0.05 mg/ml BSA, 60 mm NaCl, 10% (v/v) glycerol, 1 mm MgCl2, 0.1 mm ZnCl2, 1 mm DTT for 30 min at 37 °C. Complexes were separated by 8 or 6% native-PAGE in Tris borate-EDTA 0.5× at 4 °C. Gels were run at 200 V for 3 h and dried before autoradiography and phosphorimaging. Apparent binding constants were determined as the protein concentration resulting in half binding to a DNA substrate at 0.3 nm by EMSA. The exact value of the half-binding point was determined from interpolation on a Hill plot. Binding constants are the average values obtained from at least three independent experiments (the results given stand within a 10% standard error).
For the supershift experiments, reactions were carried out in the same conditions as those for the binding assays, but after incubation at 37 °C for 30 min, the indicated amounts of anti-YY1 antibody or PBS 1× as a control was added to reactions and further incubated for 15 min at 37 °C. Samples were separated under the same conditions as those described before for the binding assays. Binding of YY1 fractions from the SEC to Cy5-labeled DNA (10 nm) substrates was also analyzed through EMSA. Reactions were carried out on ice for 30 min in buffer 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10% (v/v) glycerol, 10 mm MgCl2, 0.1 mm ZnCl2. Complexes were solved on 6% native-PAGE gels in Tris borate-EDTA 0.5× at 4 °C. Fluorescence of the DNA was detected with a Fujifilm FLA-3000 equipment. Native-PAGE gels were transferred to PVDF membranes, and protein was detected by Western blot with the anti-YY1 antibody.
Cell Culture and Irradiation
A549 and U2OS cells were cultured in Eagle's minimum essential medium (MEM) or DMEM (Invitrogen), respectively, supplemented with 10% FCS, l-glutamine, penicillin, and streptomycin (Invitrogen) at 37 °C in a humidified 95% air and 5% CO2 atmosphere. Cells were irradiated by exposure to a 137Cs source.
Small Interfering RNA (siRNA) Knockdown Conditions and Antibodies
siRNA-mediated knockdown was achieved using HiPerFect Transfection Reagent (Qiagen) following the manufacturer's instructions. siRNA duplexes were transfected into 4 × 105 of logarithmically growing cells per condition. Cells were harvested 24 h later, retransfected with siRNA, and then seeded and grown for 48 h. The primary antibodies used were: γH2AX (Millipore) at 1:800 for IF, replication protein A (RPA; Millipore) at 1:100 for IF, RAD51 (Santa Cruz Biotechnology) at 1:200 for IF, CENPF (Abcam) at 1:1000 for IF, YY1 (H-414, Santa Cruz Biotechnology) at 1:1000 for WB, RuvBL2 (Abcam) at 1:5000 for WB, and KAP-1 (Abcam) at 1:1000 for WB. Anti-YY1 antibody (H-10 X, Santa Cruz Biotechnologies) for supershift experiments was used. The secondary antibodies used were: FITC (Sigma) at 1:100 for IF, Cy3 (Sigma) at 1:200 for IF, AlexaFluor 488/555/350 (Invitrogen) at 1:400 for IF, goat anti-rabbit HRP at 1:10,000 for WB, rabbit anti-mouse HRP at 1:2,000 for WB. Anti-His-HRP conjugated antibody (Sigma) was used at 1:10,000 for WB.
Bimolecular Fluorescence Complementation (BiFC)
1.5 × 105 of U2OS cells were co-transfected with 0.25 μg of VN173- and VC155-derived plasmids using a NanoJuice Transfection Reagent kit (Novagen). 24 h post-transfection cells were fixed with 3% (w/v) paraformaldehyde, permeabilized with 0.5% Triton X-100, and mounted with Vectashield containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories Ltd). Slides were analyzed and imaged using an Applied Precision® Delta Vision® RT Olympus IX70 deconvolution microscope.
Immunofluorescence
Cells plated on coverslips were fixed for 10 min with fixative (2% (w/v) paraformaldehyde, 3% (w/v) sucrose, 1× PBS) and permeabilized for 3 min with 0.2% Triton X-100 in PBS. When staining for RPA/RAD51 in cells not transfected with plasmid, pre-extraction was performed by treatment with 0.2% Triton X-100 in PBS for 0.5–1 min before paraformaldehyde fixation. Cells were rinsed with PBS and incubated with primary antibody diluted in PBS + 2% (w/v) BSA for 1 h at room temperature. Cells were washed 3 times, incubated with secondary antibody (diluted in PBS + 2% (w/v) BSA) for 30 min at room temperature in the dark, incubated with DAPI for 10 min, and washed 3 times with PBS. Slides were mounted using Vectashield and visualized/analyzed using a Nikon-e400 microscope and imaged using an Applied Precision® Delta Vision® RT Olympus IX70 deconvolution microscope and softWoRx® Suite software. For γH2AX, RPA, and RAD51 foci quantification, a minimum of 30 cells were scored blindly per experiment, and error bars represent the S.D. between three experiments.
Plasmids and Constructs for in Vivo Studies
The BiFC plasmids pBiFC-VN173 (encoding the N-terminal region of Venus; Addgene no. 22010) and pBiFC-VC155 (encoding the C-terminal region of Venus; Addgene no. 22011) were obtained from Chang-Deng Hu (via Addgene (49)). The coding region of YY1 was amplified from the YY1 cDNA (Source BioScience) and cloned into the BiFC plasmids to create pYY1-VN and pYY1-VC. The RuvBL2 complete human ORF cDNA (accession number NM_006666) was purchased from Origene as GFP-tagged transfection-ready DNA. To generate siRNA-resistant RuvBL2 expression constructs, the following nucleotides were mutated: 451 G to A, 460 A to T, 466 A to T using QuikChange site-directed mutagenesis (Stratagene). The GFP-RuvBL2K83A ATPase mutant construct was generated using QuikChange site-directed mutagenesis (Stratagene) on the siRNA-resistant template.
RESULTS
YY1 Assembles Two Distinct Oligomeric Species
Human full-length His-tagged YY1 (His-YY1) was expressed in bacteria and purified to homogeneity (Fig. 1B) for structural and biochemical studies. His-YY1 migrated with the apparent mobility of a larger protein (∼65–68 kDa) due to its amino acid composition (39). We analyzed the functionality of the recombinant protein by testing the ability of His-YY1 to bind DNA using synthetic 4-way DNA substrates (HJ) by EMSA (Fig. 1C). His-YY1 bound two types of HJ with arms of 40 bp (Fig. 1C, left panel) and 25 bp (Fig. 1C, right panel). The shifted band was specifically super-shifted with an anti-YY1 antibody unable to shift DNA complexes of the RecU resolvase used as a control (not shown). There were several His-YY1 DNA complexes, suggesting the binding of multiple monomers to each DNA molecule and opening the possibility of YY1 oligomerization (15).
Confirming the work by Wu et al. (15), we found that His-YY1 behaved as an oligomer in a SEC (Fig. 1D), but in addition we observed that His-YY1 migrated as two distinct species, an ∼100–150-kDa complex (named as complex A) and a larger oligomer of about 200–300 kDa (complex B). Complex A could correspond to a 2- or 3-mer according to the calibration of the column using molecular weight standards (Fig. 1D), and this complex appeared as square-shaped molecules compatible with an oligomer by EM (Fig. 1E). Complex B appeared at the microscope as an elongated molecule with roughly double length than complex A (Fig. 1E). Some larger species were also detected eluting from the column (Fig. 1D), but they appeared as aggregates in the electron microscope (Fig. 1E, inset). Oligomerization was also detected by analyzing the electrophoretic mobility of His-YY1 after a mild cross-linking using glutaraldehyde, which showed the presence of multimers containing several subunits migrating as dimers and also larger oligomers (Fig. 1F). Several lines of evidence revealed that complex B was assembled by the oligomerization of complex A, as the ratio between complex B and A in SEC was dependent on the input sample concentration. When fractions from the SEC corresponding to complex B were re-injected in the same column, a peak containing His-YY1 that migrates as complex A and containing square-shaped molecules was detected (Fig. 1G). This was interpreted as resulting from the disassembling of the larger species, suggesting an equilibrium between the two oligomers.
YY1 Forms Multimers in Vivo
Whether the oligomerization of YY1 found for the recombinant protein also occurs in vivo is unknown. Thus we used BiFC to detect YY1 self-association in vivo. We created two YY1 fusion constructs with complementary fragments (VN or VC) of the YFP fluorescent reporter protein Venus (Fig. 2A). Fluorescence occurs when the complementary fragments of Venus are in close proximity, providing readout of protein-protein interactions in vivo. We co-transfected cells with the YY1-VN and YY1-VC fusion constructs. As controls, cells were transfected with YY1-VN in combination with the VC empty vector (YY1-VN and VC), YY1-VC with the VN empty vector (VN and YY1-VC), or the two empty vectors (VN and VC). In all of the control cells, we saw pan-nuclear fluorescence. However, when the YY1 constructs were co-transfected, we saw distinct foci present in the cells (Fig. 2B), suggesting that YY1 forms multimers in vivo.
FIGURE 2.
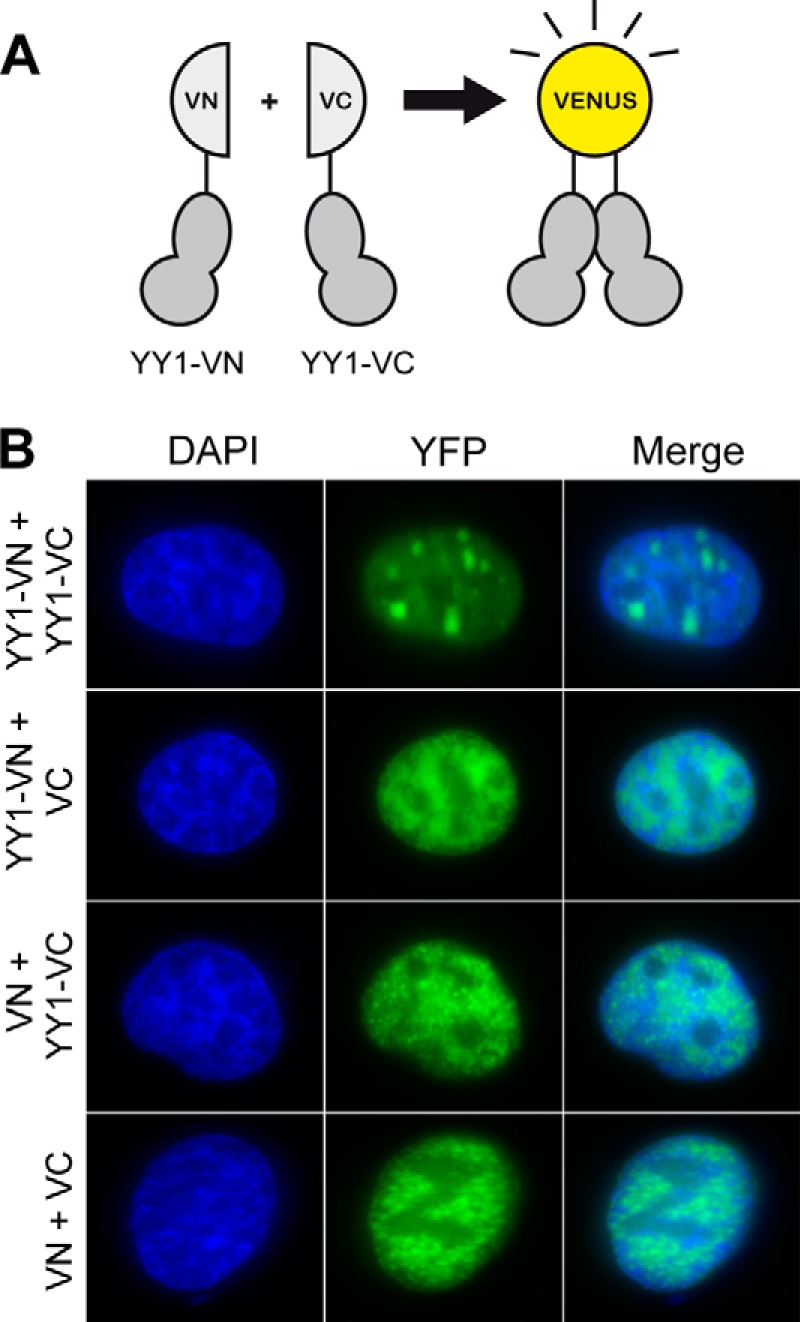
YY1 multimerizes in vivo. A, scheme of the expression constructs used in BiFC experiments. The coding sequence of YY1 was fused to either the N-terminal (YY1-VN) or C-terminal (YY1-VC) domain of Venus. B, U2OS cells transiently co-transfected with plasmids expressing the fusions in A were analyzed by microscopy. Co-transfection of YY1-VN with VC or YY1-VC with VN displayed no difference in pan-nuclear fluorescence compared with cells co-transfected with the empty vectors (VC and VN). Only cells co-transfected with both YY1 fusion constructs (YY1-VN and YY1-VC) had distinct foci present in the nuclei.
YY1 Assembles a Bell-shaped Core Dimer
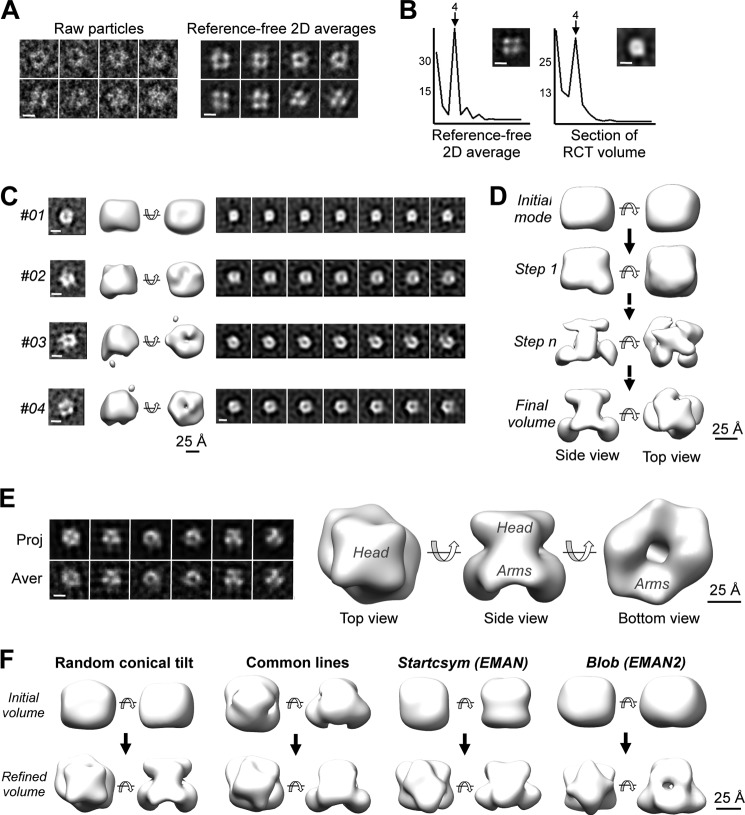
To further understand the structure of the YY1 multimers, an homogenous preparation of complex A from the peak fraction of a SEC experiment was observed by EM, revealing a square-shaped molecule with approximate dimensions of 60 × 50 Å, a low density region at its center (Fig. 3A), and a clear maximum in 4-fold rotational power spectra compatible with the oligomerization of 2 or 4 identical YY1 subunits (Fig. 3B, left panel). Several low resolution ab initio structures of YY1 complex A were obtained for homogenous sub-classes using the RCT method (Fig. 3C, four representative RCT structures are shown). Sections along the longitudinal axis of these ab initio structures revealed a square shape (Fig. 3C), with a 4-fold maximum in the rotational power spectra (Fig. 3B, right panel). Given that the SEC experiment suggested that YY1 oligomers are either 2- or 3-mers but never 4-mers, the images in the electron microscope suggested the assembly of an YY1 dimer.
FIGURE 3.
Structure of the human YY1 dimer. A, single molecule images and two-dimensional reference-free averages of complex A of His-YY1 show a typical square shape. The scale bar represents 5 nm. B, left panel, analysis of the rotational power spectra of a representative reference-free two-dimensional (2D) average of YY1. Right panel, rotational power spectra of sections along the longitudinal axis of the YY1 RCT structure. One representative section is displayed that revealed a strong component with 4-fold symmetry compatible with a dimeric or tetrameric complex. Scale bars represent 5 nm. C, four representative ab initio low resolution structures of YY1 complex A obtained by the RCT method. Two-dimensional reference-free averages used for each three-dimensional reconstruction are shown in the left panels and sections of each of the models (right panels). The scale bar represents 25 Å. D, angular refinement of YY1 dimer (complex A). Initial refinement was performed without imposing any symmetry (steps 1 and n) using the RCT model #01 shown in C as the initial template, and after convergence, c2 rotational symmetry was imposed and further refined (Final volume). Volumes are shown rendered at a threshold representing around 75% of the protein mass for visualization of structural features. The scale bar represents 25 Å. E, three-dimensional structure of the YY1 dimer (complex A) obtained after angular refinement. The scale bar represents 25 Å. Characteristic regions of the complex, defined as head and arm, are labeled. The left panel shows the comparison of the computational projections (Proj.) of the refined volume with the averages (Aver.) of the single molecule images assigned to each orientation. The scale bar represents 5 nm. F, angular refinement of YY1 complex A using several initial volumes as templates to discard a significant model bias. Refinement was performed applying 2-fold rotational symmetry and using as templates: a random conical tilt structure, a reconstruction obtained by common lines, or using the Startcsym command in EMAN (33) and a featureless blob using methods defined in EMAN2 (32). In all cases refinements converged to very similar structures (Refined volume). The scale bar represents 25 Å.
Dimerization of YY1 was further supported by solving the structure of these oligomers using 16,362 images obtained in the electron microscope (Fig. 3E). First, a three-dimensional reconstruction of YY1 was obtained by image processing methods and using an ab initio RCT structure as the initial template (model #01 in Fig. 3C) for refinement and applying no symmetry constrains (Fig. 3E). The estimated molecular mass of this non-symmetrized reconstruction, defined by the number of voxels occupied by the reconstruction and assuming an average density for proteins, was ∼100 kDa, which only matched a dimeric species. Then, 2-fold symmetry was applied to obtain the structure of YY1 complex A at 20 Å resolution (Fig. 3E). The structure of the YY1 revealed two regions (Fig. 3E). The top region (“head”) was compact and displayed a square shape and was therefore responsible for the strong component of 4-fold symmetry detected in the averages (Fig. 3B). The bottom region (the “arms”) was slightly elongated and fitted better to 2-fold symmetry. To rule out a significant bias of the initial template during image processing, we confirmed that the data converged to very similar reconstructions when using several initial models obtained by distinct methods: RCT, common lines, use of rotational symmetry, and a featureless blob (see “Experimental Procedures” for details) (Fig. 3F).
YY1 Multimerizes by the Association of Dimers
The structural characterization of YY1 complex B was first addressed by concentrating His-YY1 to enrich the complex, but this resulted in its aggregation. We then produced an N-terminal Strep-II-tagged version (Strep-II-YY1) that yielded a major peak composed of abundant complex B in SEC (Fig. 4A). Strep-II-YY1 behaved as a discrete high molecular weight band in SDS-PAGE after stabilization with a mild concentration of glutaraldehyde using the GraFix method (29) (Fig. 4B), clearly indicative of a defined oligomeric species. Images of Strep-II-YY1 complex B revealed an elongated molecule (Fig. 4C), and similar images were obtained for the complex after GraFix (not shown) and for His-YY1 complex B (Fig. 4D). A minor percentage of Strep-II-YY1 complex A co-existed in the same micrographs (20% of the data set) (Fig. 4C), and ab initio RCT structures for each complex were obtained by the RCT method to deal with this heterogeneity. Complex B (80% of the data set) appeared as elongated structures of roughly 100 Å in length, approximately double in size than the dimers (Fig. 4E). We were unable to refine these structures to higher resolutions despite rigorous attempts. Major difficulties were found to assign each molecule image to a certain conformation as well as the ambiguities to define the axis of rotational symmetry. Nonetheless, the two-dimensional averages of the complexes and their RCT structures suggested that these were distinct oligomeric species assembled by the association of dimers, and several models could be possible (Fig. 4F).
FIGURE 4.
YY1 multimerizes in larger complexes by the association of YY1 dimers. A, purification of Strep-II-YY1. Left panel, SDS-PAGE and SimplyBlue (Invitrogen) staining of purified Strep-II-YY1. Right panel, SEC of Strep-II-YY1 using a Superdex 200 PC 3.2/30 (GE Healthcare) column. Molecular weight standards (GE Healthcare) used for column calibration are indicated on the top of the chromatogram. B, silver staining SDS-PAGE of the Strep-II-YY1 protein purified and stabilized by the GraFix method in a glycerol/glutaraldehyde (GA) gradient (right panel) and in a glycerol gradient (left panel). C, selected raw molecule images and reference-free averages of Strep-II-YY1 (complexes A and B). The scale bar represents 10 nm. 2D averages, two dimensional averages. D, raw molecule images and reference-free averages of His-YY1 (complexes A and B). Images for both samples (His- and Strep-II-tagged YY1) were similar. The scale bar represents 10 nm. E, representative RCT structures obtained from images and averages of Strep-II-YY1. The peak from the SEC contained a mixture of complexes A and B. The scale bar represents 25 Å. The two-dimensional averages of the images used for each RCT reconstructions are shown in the left panels. The scale bar represents 5 nm. F, hypothetical models for the association of YY1. Two potential ways of association of YY1 dimers into larger oligomers, based on the two-dimensional averages and the RCT structures of complexes A and B, are proposed.
YY1 Oligomers Can Bind DNA with and without the Consensus Sequence
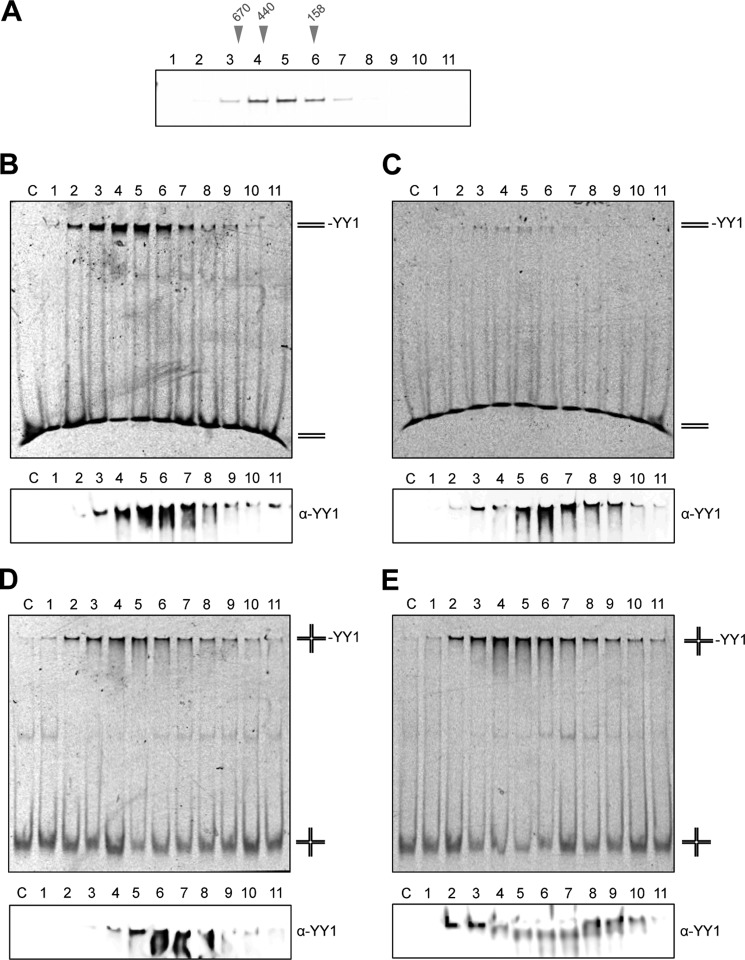
Zinc finger motifs of YY1 define its function as a transcription factor by recognizing a consensus DNA sequence. We verified that Strep-II-YY1 oligomeric fractions obtained by SEC (Fig. 5A) recognize a short dsDNA (20-bp) and 4-way HJ, containing a consensus sequence (CCAT) by EMSA (Fig. 5, B and C, Table 1). Unexpectedly, we found that YY1 oligomers could also bind a 25-bp dsDNA without a consensus sequence (although with less affinity than the dsDNA with the consensus sequence) and a HJ without a consensus sequence (Fig. 5, B–E), suggesting these oligomers could have acquired new DNA binding properties. The DNA mobility shifts observed correlated with YY1 present in the fractions as revealed when the native gels were analyzed by Western blot with an anti-YY1 antibody (Fig. 5, B–E, bottom panels). Maximum binding was detected for the peak fraction enriched in YY1 oligomers, mainly complex B. These results revealed that YY1 can bind DNA after oligomerization, and remarkably these oligomers seem to have the potential to recognize some DNA substrates even when lacking the consensus sequence.
FIGURE 5.
YY1 oligomers recognize DNA with and without the consensus sequence. A, purified Strep-II-YY1 was fractionated as in Fig. 4A and analyzed by SDS-PAGE and silver-staining. Molecular weight markers used for SEC calibration are indicated above the gel. B, EMSA using the fractions from the SEC experiment shown in A and a dsDNA containing the consensus sequence for YY1 (CCAT) (described as dsDNA_sp_2 under “Experimental Procedures”). C, EMSA as in B but using a dsDNA that does not contain the consensus sequence (described as dsDNA_no_sp_2 under “Experimental Procedures”). D, EMSA as in B but using a synthetic 4-way HJ containing the consensus sequence for YY1 (CCAT) (described as HJ_25_sp under “Experimental Procedures”). E, EMSA as in D but using a synthetic 4-way HJ that does not contain the consensus sequence (described as HJ_25_no_sp under “Experimental Procedures”). Lanes labeled as C (B–E) show the nucleic acid in each case in the absence of protein. The bottom panels (B–E) show the same gels, run for longer, and where the presence of YY1 in the shifted DNA was detected by Western blot with an anti-YY1 antibody (α-YY1).
RuvBL1-RuvBL2 Interacts with YY1 and Enhances DNA Binding
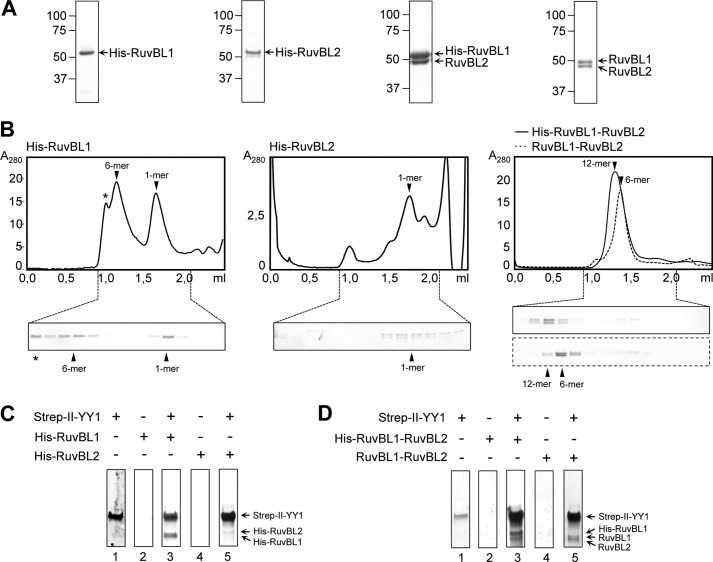
YY1 has been shown to bind RuvBL1 and RuvBL2 (18, 19), and thus we decided to analyze if the oligomeric forms of YY1 also interacted with these ATPases and how this could affect DNA binding. RuvBL1 and RuvBL2 assemble homo-hexameric rings on their own, whereas its co-expression results in a mixture of hetero-hexameric and dodecameric complexes that are believed to be functional forms of these ATPases in the cell (23–25). We purified His-RuvBL1, His-RuvBL2, and His-RuvBL1-RuvBL2 complexes and also RuvBL1-RuvBL2 where the tag was removed (27) (Fig. 6A), and the oligomeric state of each sample was characterized by SEC (Fig. 6B). Purified His-RuvBL1 was resolved as a mixture of oligomers and free subunits, whereas His-RuvBL2 did not oligomerize under our experimental conditions. On the other hand, His-RuvBL1-RuvBL2 and RuvBL1-RuvBL2 formed hetero-oligomeric complexes with an approximate 1:1 ratio that we interpreted as hexamers and dodecamers based on previous information (27). We found that Strep-II-YY1 interacted with His-RuvBL1 but not His-RuvBL2 under our experimental conditions after pulling down the Strep-II tag and eluting the retained His-RuvBL1 or His-RuvBL2 with d-desthiobiotin (Fig. 6C, lanes 3 and 5). Similar experiments performed using purified His-RuvBL1 hexamers or monomers retrieved identical results (not shown), indicating that Strep-II-YY1 interacts with these two forms of His-RuvBL1. Also, we observed that RuvBL1-RuvBL2 (either containing or not the His tag) was also eluted in complex with Strep-II-YY1 (Fig. 6D; note that His-RuvBL1 and RuvBL1 run differently on SDS-PAGE).
FIGURE 6.
Analysis of the interaction of purified YY1 with RuvBL1 and RuvBL2. A, SDS-PAGE and SimplyBlue (Invitrogen) staining of purified His-RuvBL1, His-RuvBL2, His-RuvBL1-RuvBL2, and RuvBL1-RuvBL2. B, chromatograms and SDS-PAGE analysis of the fractions from a SEC of His-RuvBL1, His-RuvBL2, and His-RuvBL1-RuvBL2 (solid line) and RuvBL1-RuvBL2 (dash line) using a Superdex 200 PC 3.2/30 (GE Healthcare) column. Proteins were stained using SimplyBlue (Invitrogen) (His-RuvBL1, His-RuvBL1-RuvBL2, and RuvBL1-RuvBL2) or by silver staining (His-RuvBL2). The positions for the different oligomeric species (12-mer, dodecamer; 6-mer, hexamer; 1-mer, monomer) are indicated in each case and were determined by comparison with molecular weight standards. The asterisk (*) in the case of His-RuvBL1 SEC indicates a peak of aggregated material as observed by EM. C, pulldown experiments of Strep-II-YY1 and His-RuvBL1 or His-RuvBL2. Strep-II-YY1 was incubated without (lane 1) or with His-RuvBL1 (lane 3) or His-RuvBL2 (lane 5) and affinity purified using the Strep-II-tag present in YY1. As a control, His-RuvBL1 (lane 2) or His-RuvBL2 (lane 4) was purified in the same conditions but in the absence of YY1. D, pulldown experiments of Strep-II-YY1 and His-RuvBL1-RuvBL2 or RuvBL1-RuvBL2. Strep-II-YY1 was incubated without (lane 1) or with His-RuvBL1-RuvBL2 (lane 3) or RuvBL1-RuvBL2 (lane 5) and affinity-purified as in A. As a control, His-RuvBL1-RuvBL2 (lane 2) or RuvBL1-RuvBL2 (lane 4) was pull downed in the same conditions as those before but in the absence of YY1.
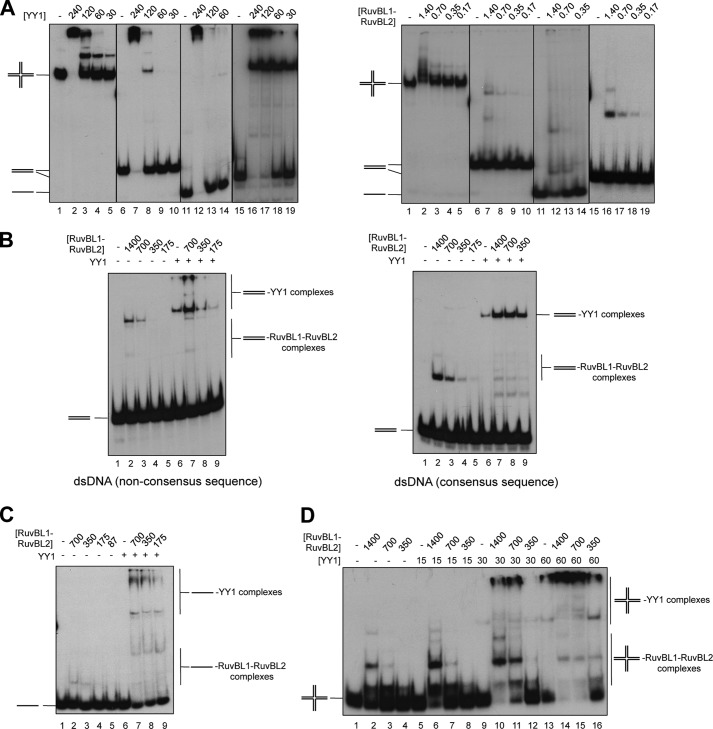
We analyzed how the interaction between YY1 and RuvBL1-RuvBL2 affected DNA binding by EMSA, and we focused on two dsDNAs, one containing and another one missing the consensus sequence for YY1 (Fig. 7), and compared its binding affinity to the binding to a HJ DNA without a consensus sequence. We first measured the affinity of each protein for these forms of DNA and included also ssDNA in our analysis (Fig. 7A, Table 2). The interaction of YY1 with both dsDNAs mainly revealed two types of complexes, a single band of shifted DNA and a complex migrating at the gel well at high protein concentrations (Fig. 7A, left panel). YY1 bound 4–5 times better to the dsDNA containing the consensus sequence, but the difference in binding affinity between the specific DNA and HJ DNA was only 2–3 times (Table 2). Binding to ssDNA was also observed. The analysis of the binding of RuvBL1-RuvBL2 to DNA showed that the preferred substrate was HJ (J3) followed by the 80-nt ssDNA (Fig. 7A, right panel; Table 2). RuvBL1-RuvBL2 bound DNA forming several complexes. Interestingly, when RuvBL1-RuvBL2 and YY1 were incubated together in the presence of a dsDNA with or without the consensus sequence, we observed that the top bands corresponding to DNA shifted by YY1 were enhanced significantly (Fig. 7B, compare lane 6 for YY1 alone with lanes 7–9 for YY1 incubated with RuvBL1-RuvBL2). Experiments were done at a YY1 concentration that resulted in <10% of DNA shifted in the absence of RuvBL1-RuvBL2, and at the highest RuvBL1-RuvBL2 concentration used (700 nm), a 5-fold increase in the amount of YY1 complexes with the consensus sequence and a 7-fold for YY1 complexes with dsDNA without the consensus sequence was observed. Similar effects were found for ssDNA (Fig. 7C) and a HJ (Fig. 7D). Control experiments were performed where the different DNA substrates were incubated with YY1 and an excess of BSA to demonstrate that the enhancement in binding is not due to a “protein concentration” stabilizing effect (data not shown).
FIGURE 7.
YY1 and RuvBL1-RuvBL2 cooperate in DNA binding. A, left panel, EMSA assays showing binding of Strep-II-YY1 to several DNA substrates (described under “Experimental Procedures”) and represented as a schematic in each gel. Binding reactions contained 0, 240, 120, 60, or 30 nm protein and 0.3 nm DNA species: HJ (J3) (lanes 1–5), dsDNA non-consensus sequence (dsDNA_no_sp_1) (lanes 6–10), 80-nt ssDNA (lanes 11–14), and dsDNA consensus sequence (dsDNA_sp_1) (lanes 15–19). Right panel, EMSA assays showing binding of His-RuvBL1-RuvBL2 to the same DNA substrates. Binding reactions contained 0, 1.4, 0.7, 0.35, or 0.17 μm protein and 0.3 nm DNA species: HJ (J3) (lanes 1–5), dsDNA non-consensus sequence (dsDNA_no_sp_1) (lanes 6–10), 80-nt ssDNA (lanes 11–14), and dsDNA consensus sequence dsDNA_sp_1) (lanes 15–19). Reactions were performed as described under “Experimental Procedures,” and protein-DNA complexes were visualized by 6% PAGE and autoradiography. B, RuvBL1-RuvBL2 enhances the binding of YY1 to dsDNA either containing (right panel, dsDNA_sp_1) or not the consensus sequence (left panel, dsDNA_no_sp_1). Reactions assembled on ice contained combinations of a decreasing concentration of His-RuvBL1-RuvBL2 (1.4, 0.7, 0.35 or 0.17 μm) and a fixed concentration of Strep-II-YY1 (120 nm in the left panel and 15 nm in the right panel). After DNA addition (0.3 nm), samples were incubated for 30 min at 37 °C before electrophoresis. C, YY1 and RuvBL1-RuvBL2 also cooperate in ssDNA binding. Shown is a similar experiment as B but using a 80-nt ssDNA (0.3 nm) (lanes 1–9). His-RuvBL1-RuvBL2 concentrations were varied as indicated from 700 to 87 nm, and the fixed concentration of Strep-II-YY1 used was 120 nm. D, EMSA of the enhancement in binding of Strep-II-YY1 to HJ (J3) (0.3 nm) in the presence of the indicated concentrations His-RuvBL1-RuvBL2 (concentrations are expressed in nm).
TABLE 2.
DNA binding affinities of YY1 and RuvBL1-RuvBL2
| DNA | Kd appa YY1 | Kd app RuvBL1-RuvBL2 |
|---|---|---|
| nm | μm | |
| dsDNA_sp_1 | 40 | >2b |
| dsDNA_no_sp_1 | 170 | >2b |
| 80-nt ssDNA | 180 | 1.8 |
| Holliday junction J3 | 90 | 1 |
a The apparent binding constant (Kd app) for each substrate was determined from a compilation of at least three separate gel shift experiments, each with a dilution series of protein and a DNA substrate concentration fixed at 0.3 nm.
b The Kd app for this substrate is estimated, because at the maximal protein concentration tested (2 μm) <25% of the DNA was retained.
Taken together, these results are a strong indication that YY1 and RuvBL1-RuvBL2 can form a complex with increased affinity for DNA. Interestingly, these effects are found to occur also when the DNA does not contain the consensus sequence for YY1.
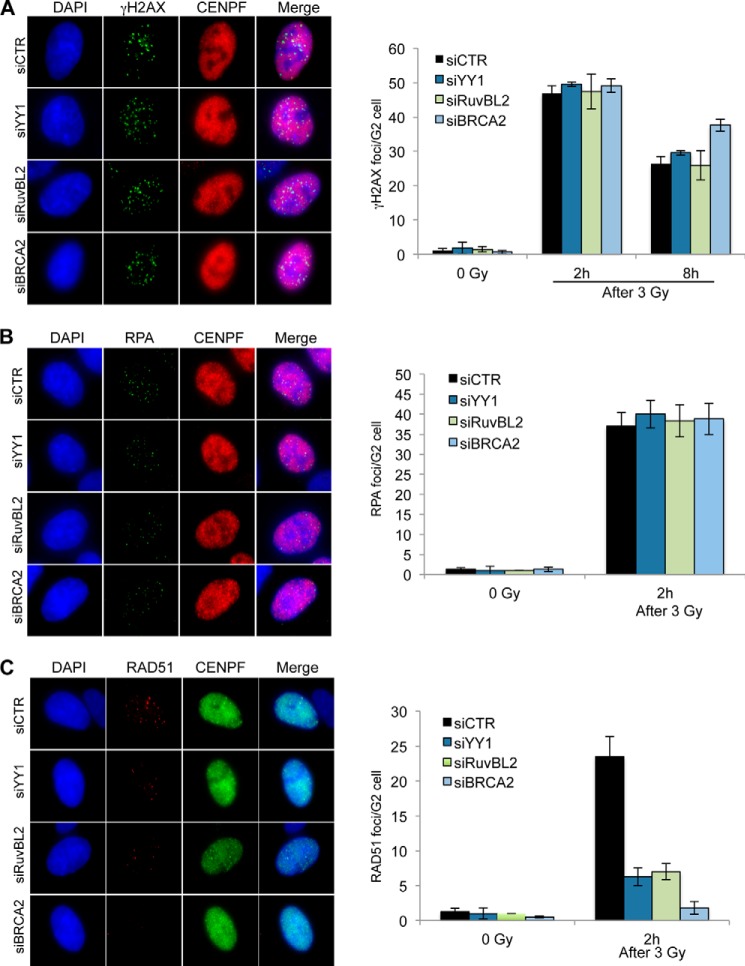
YY1 and RuvBL1-RuvBL2 Cooperate in Vivo to Promote RAD51 Foci Formation during Homologous Recombination
YY1 functions in HR as part of the INO80 complex (containing RuvBL1 and RuvBL2 subunits), and although the mechanism for this is unknown, it has been suggested that it could occur through the recognition of Holliday junction-like structures (15). Our findings showing that the interaction of YY1 and RuvBL1-RuvBL2 enhances binding to ssDNA and dsDNA prompted us to further define their role during HR. We explored if these proteins cooperate at several stages of the HR process where HJ substrates are not available and also the requirement of catalytic activity of the RuvBL1-RuvBL2 ATPases. To investigate this, we transfected cells with siRNA targeting YY1 and examined them after treatment with ionizing radiation (IR) (Fig. 8). HR is restricted to the S and G2 phases of the cell cycle in mammalian cells, so we immunostained for CENPF to identify G2 phase cells and then measured the accumulation of DNA damage response factors into IR induced foci. We found that the number of phosphorylated H2AX (γH2AX)-containing foci at the 2-h time point was unaffected by depletion of YY1 (Fig. 8A), suggesting that the initial detection and signaling of the DNA breaks are unaltered. At 8 h after treatment, the BRCA2 (breast cancer type 2 susceptibility protein)-depleted cells, which are HR-defective, showed a modest increase in foci at this time point (Fig. 8A).
FIGURE 8.
YY1 and RuvBL2 function during G2 to promote RAD51 foci formation, but not γH2AX or RPA foci, after exposure to IR. A, A549 cells were transfected with scrambled siRNA (siCTR) or siRNA directed against YY1, RuvBL2, or BRCA2, irradiated (3 Gy) and harvested 2 or 8 h later and immunostained for γH2AX and CENPF (to identify G2 phase cells). Average γH2AX foci per G2 cell were quantified (right panel). B, cells transfected as in A were irradiated (3 Gy), harvested after 2 h, and immunostained for RPA and CENPF. Average RPA foci per G2 cell were quantified (right panel). C, cells transfected as in A were immunostained for RAD51 and CENPF, and average RAD51 foci per G2 cell were quantified (right panel). All data (A–C) are the means of >3 experiments; error bars, S.D.
After recognition, DNA breaks are resected to leave 3′ ssDNA overhangs that are bound by RPA. Subsequent to resection, BRCA2 mediates the replacement of RPA with RAD51 filaments on the ssDNA overhang, and the RAD51 filament performs the homology search and strand invasion steps (40). When we examined RPA foci formation in G2 cells, we found no difference in the number of foci in the cells depleted for YY1 when compared with control cells (Fig. 8B). Whereas it is possible that the extent of resection is affected, this result suggests that there is no defect in the number of resected DNA breaks in these cells. Cells depleted for BRCA2 show normal γH2AX and RPA foci but defective RAD51 foci after IR (Fig. 8, B and C). Similar to loss of BRCA2, we found that there were significantly fewer RAD51 foci in the YY1 and RuvBL2 depleted cells, although the severity of the defect was not as pronounced as in the BRCA2-depleted cells (Fig. 8C). Notably, we found that depletion of RuvBL2 recapitulated HR defects after YY1 depletion. Specifically, the numbers of γH2AX and RPA foci were largely unaffected by depletion of RuvBL2, but the number of RAD51 foci was reduced to similar levels as those observed in YY1-depleted cells (Fig. 8).
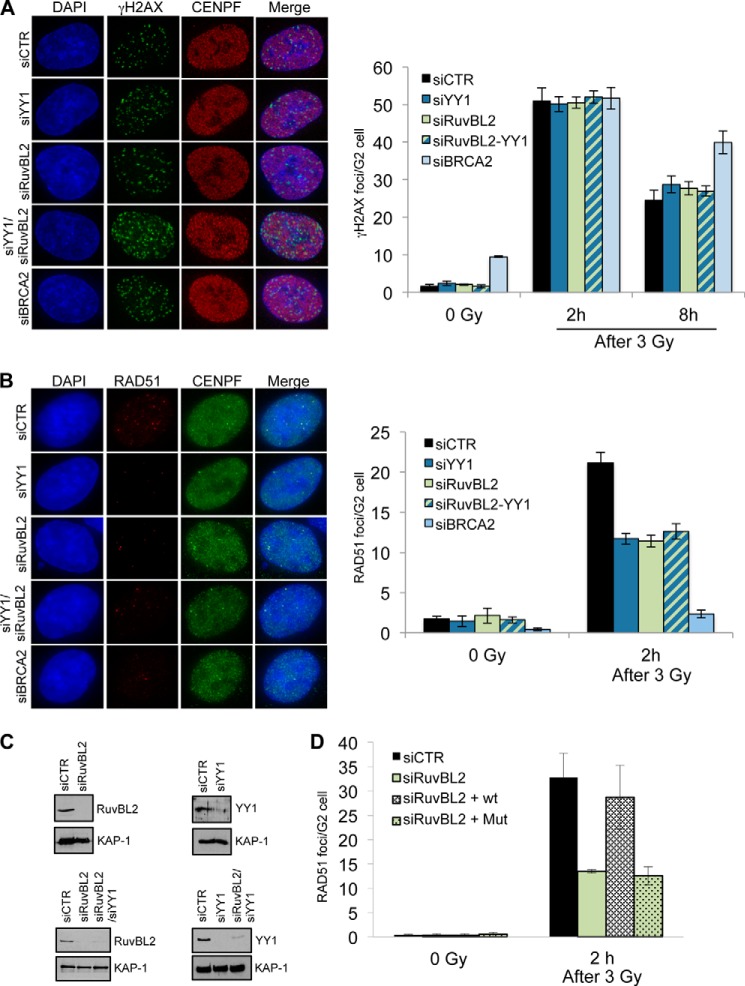
These results showed that depletion of YY1 or RuvBL2 resulted in comparable HR defects, suggesting that YY1 and RuvBL2 could cooperate to promote or stabilize RAD51 filament formation. To investigate whether this was the case, we analyzed the formation of γH2AX, RPA, and RAD51 foci in G2 cells following IR exposure after depletion of YY1, RuvBL2, or both (Fig. 9A). If these subunits were functioning at separate steps during HR, we would predict an increase in unrepaired breaks when they are co-depleted, leading to an increase in γH2AX foci. Furthermore, if YY1 and RuvBL2 were promoting or stabilizing RAD51 foci formation via distinct mechanisms, depletion of both proteins would result in a greater defect in RAD51 foci relative to cells with depletion of a single subunit. However, we found that both γH2AX and RAD51 foci formations were similar in cells depleted of both proteins to cells depleted of either individual subunits (Fig. 9B), indicating that these genes function on the same pathway to promote HR.
FIGURE 9.
RuvBL2 cooperates with YY1 to promote RAD51 foci formation, and the ATPase activity of RuvBL2 is required for this activity. A, A549 cells were transfected with scrambled siRNA (siCTR) or siRNAs directed against YY1, RuvBL2, or BRCA2, as indicated, irradiated (3 Gy), harvested 2 or 8 h later, and immunostained for γH2AX and CENPF (to identify G2 phase cells). Average γH2AX foci per G2 cell were quantified (right panel). B, cells treated as in A were immunostained for RAD51 and CENPF, and average RAD51 foci per G2 cell were quantified (right panel). All data (A and B) are the mean of >3 experiments; error bars, S.D. C, top, Western blot analysis of whole cell extracts prepared from A549 cells transfected with the indicated siRNA. Bottom, Western blot analysis of whole cell extracts prepared from A549 cells transfected with single or double siRNA constructs as indicated. Anti-KAP1 is shown as a loading control. D, U2OS cells transfected with siRNA directed against RuvBL2 and either GFP, GFP-RuvBL2, or GFP-RuvBL2-K83A were irradiated (3 Gy), harvested, and immunostained for RAD51 and CENPF. Average RAD51 foci per GFP positive G2 cell were quantified and are presented as the means of three experiments; error bars, S.D.
We made use of a point mutation within the Walker A motif of RuvBL2 that impairs ATP binding but does not disrupt its folding (41) to determine whether the ATPase activity of RuvBL2 is required for the observed effect on RAD51 foci formation after IR exposure. To do this we generated a siRNA-resistant GFP-tagged RuvBL2 expression construct and then introduced the K83A point mutation into this construct (Fig. 9C). We then transfected the wild type and mutant constructs (along with a GFP control) alongside siRNA directed against RuvBL2 and analyzed γH2AX and RAD51 foci formation in GFP-positive G2 cells. We found that the wild type construct was able to complement the RAD51 foci formation defect of the RuvBL2-depleted cells, whereas the mutant construct was not able to complement the defect (Fig. 9D), indicating that the ATPase activity of RuvBL2 is required during this step of HR. All these data suggest that the epistatic function of YY1 and RuvBL1-RuvBL2 during HR requires the catalytic activity of these ATPases, at least in the case of RuvBL2.
DISCUSSION
Recombinant YY1 was previously shown to behave as an oligomer in SEC and glutaraldehyde cross-linking experiments (15). Our structural characterization of the transcription factor YY1 not only confirms those results using His-YY1 and Strep-II-YY1, but in addition it offers a deeper understanding of the architecture of these oligomers. We determine that YY1 assembles as dimers that can associate into larger oligomeric complexes, and we characterize the structural organization of these oligomers using EM. Moreover, we show that the self-association of YY1 also occurs in vivo. The most characteristic structural feature for the transcription factor YY1 is the C-terminal C2H2-type zinc finger motifs (11). Although the main function of these motifs is to interact with nucleic acids, they have been also implicated in protein-protein interactions, including in some cases homo-dimerization (42–44). One example is Ikaros, a protein containing six C2H2-type zinc fingers, four of which are involved in DNA binding and the two C-terminal fingers are responsible for the assembly of a homodimer (45). Thus, it is conceivable that YY1 could use, at least in part, these multifunctional zinc fingers to homo-dimerize. Interestingly, dimerization of Ikaros dramatically increased its affinity for DNA, and a similar effect could be taking place in the case of YY1 (see below).
YY1 zinc finger motifs are responsible for specifically binding to a consensus DNA sequence (5′-(C/g/a)(G/t)(C/t/a)CATN(T/a)(T/g/c)-3′) found at the promoters of target genes (11). DNA binding by the oligomeric forms of YY1 is not well characterized, and the work by Wu et al. (15) used nucleic acid substrates with a consensus sequence for YY1, where it could be difficult to identify interactions with DNA distinct to those as a transcription factor. In this context, one significant new finding of our study is that these oligomers can interact with several DNAs that do not contain the consensus sequence, at least in vitro. This is consistent with the finding that YY1 binds to RNA during inactivation of one X chromosome in mammalian females independent of a consensus motif (4). What could be the molecular/structural basis for these unconventional interactions between YY1 and nucleic acids? C2H2-type zinc finger domains can bind, in addition to DNA, to several nucleic acids such as ssRNA, dsRNA, and DNA-RNA hetero-duplexes (44), although the structural basis for these properties are poorly understood. One hypothesis might be that the oligomerization of YY1 could expand its properties to bind DNA, RNA, and/or other proteins by the combination of zinc fingers motifs from several monomers. This opens the possibility that, when in the form of larger oligomers and/or as part of larger complexes, YY1 could contribute to functions unrelated to those of a transcription factor. Interestingly, recent reports describe roles of YY1 in distinct functions such as proviral silencing in embryonic cells (6), the regulation of the expression of long noncoding RNAs during myogenesis (7), and in V(D)J somatic rearrangement of Ig loci during B-cell development by a mechanism that involves long distance DNA interactions (46).
One of these unconventional functions is the link between YY1 and DNA repair (15, 47). We predict that the functions of YY1 in DNA repair should be independent of the recognition of specific sequences, and our results indicate that YY1 has the potential to do so. We corroborate that YY1 functions in HR, but in addition, our data suggest that defects in HR observed in the absence of YY1 may be due to, at least in part, an inability to efficiently promote formation or stabilization of the RAD51 filament.
A direct interaction of YY1 with RuvBL1 and RuvBL2 has been previously described (15). These AAA+ ATPases are implicated in several cellular processes as part of large macromolecular assemblies, such as INO80, SWR1, TIP60, and R2TP complexes, among others (20, 21). Despite both proteins being essential, not much is known about their mechanism of action, and the functional significance of the several oligomeric forms of RuvBL1 and RuvBL2 is still unclear (20, 21). We tested the interaction of YY1 with the RuvBL1 and RuvBL2 ATPases, showing that this interaction takes place also with purified YY1 oligomers. In addition, we observed that YY1 binds RuvBL1-RuvBL2 complexes, but YY1 interacts preferentially with RuvBL1, and it associates to RuvBL2 mostly when this is in complex with RuvBL1. We cannot rule out that RuvBL2 could bind YY1 at higher concentrations.
Unexpectedly, we found that when YY1 and RuvBL1-RuvBL2 are incubated together with several DNA substrates, they interact with the nucleic acids with enhanced affinity compared with each protein on their own, and this effect is especially relevant when considering those DNAs without a consensus sequence. YY1 forms a complex with DNA that is detected as a lower migrating band in EMSA experiments, and the abundance of this complex is enhanced 7-fold in the presence of RuvBL1-RuvBL2. These results suggest that YY1 and RuvBL1-RuvBL2 cooperate to enhance binding to DNA. Similar findings have been recently described for another transcription factor, GATA-binding protein 3 (Gata3), which controls differentiation of T cells (48). Gata3 forms a complex with RuvBL2, and it was proposed that the interaction promoted the DNA binding activity of Gata3.
Thus, the interaction of YY1 with RuvBL1 and RuvBL2 either alone or as part of a larger complex could represent a mechanism to modify the functionality of this transcription factor. The molecular/structural bases for this remain to be explored, but RuvBL1-RuvBL2 contains two regions with the potential to bind nucleic acids. Domain II shows a structure reminiscent of RPA, and recombinant DII domains from RuvBL1 in isolation bind ssRNA, ssDNA and dsDNA (24). Similar to other AAA+ family members, RuvBL1 and RuvBL2 assemble hexameric complexes with a central channel that could potentially bind ssDNA (24), although this has not been demonstrated experimentally. YY1 could cooperate with any of these two potential DNA binding sites in RuvBL1-RuvBL2 to enhance the affinity for nucleic acids, somehow mimicking the behavior of the bacterial homolog RuvB, which has poor DNA binding itself and needs to interact with RuvA to bind DNA structures (22). The mechanism of how exactly YY1, RuvBL1, and RuvBL2 proteins cooperate to promote steps during HR and/or other cellular events needs to be further explored. Here we provide evidence suggesting an important role of the RuvBL1-RuvBL2 ATPases in enhancing the DNA binding properties of YY1, which could be directly linked to the regulation of different cellular processes, including transcription and DNA repair.
Acknowledgments
We thank Andreas Kakarougkas (Sussex) and Martín Alcorlo (Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas) for technical assistance.
This work was supported by the Spanish Government (SAF2011-22988 (to O. L.), BES-2009-014133 (to A. L.-P.), BFU2012-39879-C02-02 (to S. A.)) and by Cancer Research UK (CEA-C7905 (to J. A. D.)).
- YY1
- Yin Yang 1
- HR
- homologous recombination
- HJ
- Holliday junction
- SEC
- size exclusion chromatography
- RCT
- random conical tilt
- RPA
- replication protein A
- WB
- Western blot
- IF
- immunofluorescence
- BiFC
- bimolecular fluorescence complementation
- IR
- ionizing radiation
- Gata3
- GATA-binding protein 3
- Gy
- gray
- nt
- nucleotide
- H2AX
- histone 2AX
- CENPF
- centromere protein F.
REFERENCES
- 1. Gordon S., Akopyan G., Garban H., Bonavida B. (2006) Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25, 1125–1142 [DOI] [PubMed] [Google Scholar]
- 2. Calame K., Atchison M. (2007) YY1 helps to bring loose ends together. Genes Dev. 21, 1145–1152 [DOI] [PubMed] [Google Scholar]
- 3. He Y., Casaccia-Bonnefil P. (2008) The Yin and Yang of YY1 in the nervous system. J. Neurochem. 106, 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeon Y., Lee J. T. (2011) YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nicholson S., Whitehouse H., Naidoo K., Byers R. J. (2011) Yin Yang 1 in human cancer. Crit. Rev. Oncog. 16, 245–260 [DOI] [PubMed] [Google Scholar]
- 6. Schlesinger S., Lee A. H., Wang G. Z., Green L., Goff S. P. (2013) Proviral silencing in embryonic cells is regulated by Yin Yang 1. Cell Rep. 4, 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu L., Sun K., Chen X., Zhao Y., Wang L., Zhou L., Sun H., Wang H. (2013) Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. 32, 2575–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu H., Schmidt-Supprian M., Shi Y., Hobeika E., Barteneva N., Jumaa H., Pelanda R., Reth M., Skok J., Rajewsky K. (2007) Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 21, 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hogan C. J., Aligianni S., Durand-Dubief M., Persson J., Will W. R., Webster J., Wheeler L., Mathews C. K., Elderkin S., Oxley D., Ekwall K., Varga-Weisz P. D. (2010) Fission yeast Iec1-ino80-mediated nucleosome eviction regulates nucleotide and phosphate metabolism. Mol. Cell. Biol. 30, 657–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown J. L., Fritsch C., Mueller J., Kassis J. A. (2003) The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130, 285–294 [DOI] [PubMed] [Google Scholar]
- 11. Houbaviy H. B., Usheva A., Shenk T., Burley S. K. (1996) Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl. Acad. Sci. U.S.A. 93, 13577–13582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lewis B. A., Tullis G., Seto E., Horikoshi N., Weinmann R., Shenk T. (1995) Adenovirus E1A proteins interact with the cellular YY1 transcription factor. J. Virol. 69, 1628–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang W. M., Inouye C., Zeng Y., Bearss D., Seto E. (1996) Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. U.S.A. 93, 12845–12850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilkinson F. H., Park K., Atchison M. L. (2006) Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl. Acad. Sci. U.S.A. 103, 19296–19301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu S., Shi Y., Mulligan P., Gay F., Landry J., Liu H., Lu J., Qi H. H., Wang W., Nickoloff J. A., Wu C. (2007) A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat. Struct. Mol. Biol. 14, 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doetsch M., Gluch A., Poznanovi G., Bode J., Vidakovi M. (2012) YY1-binding sites provide central switch functions in the PARP-1 gene expression network. PLoS ONE 7, e44125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ishii H., Hulett M. D., Li J. M., Santiago F. S., Parish C. R., Khachigian L. M. (2012) Yin Yang-1 inhibits tumor cell growth and inhibits p21WAF1/Cip1 complex formation with cdk4 and cyclin D1. Int. J. Oncol. 40, 1575–1580 [DOI] [PubMed] [Google Scholar]
- 18. Bao Y., Shen X. (2011) SnapShot: chromatin remodeling: INO80 and SWR1. Cell 144, 158–158.e152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen L., Cai Y., Jin J., Florens L., Swanson S. K., Washburn M. P., Conaway J. W., Conaway R. C. (2011) Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J. Biol. Chem. 286, 11283–11289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nano N., Houry W. A. (2013) Chaperone-like activity of the AAA+ proteins Rvb1 and Rvb2 in the assembly of various complexes. Philos. Trans R Soc. Lond. B. Biol. Sci. 368, 20110399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rosenbaum J., Baek S. H., Dutta A., Houry W. A., Huber O., Hupp T. R., Matias P. M. (2013) The emergence of the conserved AAA+ATPases Pontin and Reptin on the signaling landscape. Sci. Signal. 6, mr1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yamada K., Miyata T., Tsuchiya D., Oyama T., Fujiwara Y., Ohnishi T., Iwasaki H., Shinagawa H., Ariyoshi M., Mayanagi K., Morikawa K. (2002) Crystal structure of the RuvA-RuvB complex: a structural basis for the Holliday junction migrating motor machinery. Mol. Cell 10, 671–681 [DOI] [PubMed] [Google Scholar]
- 23. Gorynia S., Bandeiras T. M., Pinho F. G., McVey C. E., Vonrhein C., Round A., Svergun D. I., Donner P., Matias P. M., Carrondo M. A. (2011) Structural and functional insights into a dodecameric molecular machine: the RuvBL1/RuvBL2 complex. J. Struct. Biol. 176, 279–291 [DOI] [PubMed] [Google Scholar]
- 24. Matias P. M., Gorynia S., Donner P., Carrondo M. A. (2006) Crystal structure of the human AAA+ protein RuvBL1. J. Biol. Chem. 281, 38918–38929 [DOI] [PubMed] [Google Scholar]
- 25. Petukhov M., Dagkessamanskaja A., Bommer M., Barrett T., Tsaneva I., Yakimov A., Quéval R., Shvetsov A., Khodorkovskiy M., Käs E., Grigoriev M. (2012) Large-scale conformational flexibility determines the properties of AAA+ TIP49 ATPases. Structure 20, 1321–1331 [DOI] [PubMed] [Google Scholar]
- 26. Cheung K. L., Huen J., Houry W. A., Ortega J. (2010) Comparison of the multiple oligomeric structures observed for the Rvb1 and Rvb2 proteins. Biochem. Cell Biol. 88, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. López-Perrote A., Muñoz-Hernández H., Gil D., Llorca O. (2012) Conformational transitions regulate the exposure of a DNA-binding domain in the RuvBL1-RuvBL2 complex. Nucleic Acids Res. 40, 11086–11099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Torreira E., Jha S., López-Blanco J. R., Arias-Palomo E., Chacón P., Cañas C., Ayora S., Dutta A., Llorca O. (2008) Architecture of the pontin/reptin complex, essential in the assembly of several macromolecular complexes. Structure 16, 1511–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kastner B., Fischer N., Golas M. M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E., Uchtenhagen H., Urlaub H., Herzog F., Peters J. M., Poerschke D., Lührmann R., Stark H. (2008) GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 [DOI] [PubMed] [Google Scholar]
- 30. Mindell J. A., Grigorieff N. (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 [DOI] [PubMed] [Google Scholar]
- 31. Heymann J. B., Belnap D. M. (2007) Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 [DOI] [PubMed] [Google Scholar]
- 32. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 33. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 34. Scheres S. H., Núñez-Ramírez R., Sorzano C. O., Carazo J. M., Marabini R. (2008) Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 3, 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Radermacher M., Wagenknecht T., Verschoor A., Frank J. (1987) Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc. 146, 113–136 [DOI] [PubMed] [Google Scholar]
- 36. Shaikh T. R., Gao H., Baxter W. T., Asturias F. J., Boisset N., Leith A., Frank J. (2008) SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3, 1941–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goddard T. D., Huang C. C., Ferrin T. E. (2007) Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 [DOI] [PubMed] [Google Scholar]
- 38. Zecchi L., Lo Piano A., Suzuki Y., Cañas C., Takeyasu K., Ayora S. (2012) Characterization of the Holliday junction resolving enzyme encoded by the Bacillus subtilis bacteriophage SPP1. PLoS ONE 7, e48440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shi Y., Lee J. S., Galvin K. M. (1997) Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys. Acta 1332, F49–F66 [DOI] [PubMed] [Google Scholar]
- 40. Thompson L. H. (2012) Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res. 751, 158–246 [DOI] [PubMed] [Google Scholar]
- 41. Maslon M. M., Hrstka R., Vojtesek B., Hupp T. R. (2010) A divergent substrate-binding loop within the pro-oncogenic protein anterior gradient-2 forms a docking site for Reptin. J. Mol. Biol. 404, 418–438 [DOI] [PubMed] [Google Scholar]
- 42. Brayer K. J., Kulshreshtha S., Segal D. J. (2008) The protein-binding potential of C2H2 zinc finger domains. Cell Biochem. Biophys. 51, 9–19 [DOI] [PubMed] [Google Scholar]
- 43. Brayer K. J., Segal D. J. (2008) Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem. Biophys. 50, 111–131 [DOI] [PubMed] [Google Scholar]
- 44. Iuchi S. (2001) Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci. 58, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun L., Liu A., Georgopoulos K. (1996) Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 15, 5358–5369 [PMC free article] [PubMed] [Google Scholar]
- 46. Atchison M. L. (2014) Function of YY1 in long-distance DNA interactions. Front Immunol. 5, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jiang Y., Wang X., Bao S., Guo R., Johnson D. G., Shen X., Li L. (2010) INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc. Natl. Acad. Sci. U.S.A. 107, 17274–17279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hosokawa H., Tanaka T., Kato M., Shinoda K., Tohyama H., Hanazawa A., Tamaki Y., Hirahara K., Yagi R., Sakikawa I., Morita A., Nagira M., Poyurovsky M. V., Suzuki Y., Motohashi S., Nakayama T. (2013) Gata3/Ruvbl2 complex regulates T helper 2 cell proliferation via repression of Cdkn2c expression. Proc. Natl. Acad. Sci. U.S.A. 110, 18626–18631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shyu Y. J., Suarez C. D., Hu C. D. (2008) Visualization of ternary complexes in living cells by using a BiFC-based FRET assay. Nat. Protoc. 3, 1693–1702 [DOI] [PubMed] [Google Scholar]