Background: Faithful mitochondrial DNA replication ensures functional oxidative phosphorylation.
Results: Recombination structures and replication forks are the main intermediates detected in Candida parapsilosis mtDNA.
Conclusion: Recombination driven replication initiation and not transcription primed DNA synthesis prevails in yeast mitochondria.
Significance: Our findings are essential for the understanding of yeast mitochondrial DNA metabolism.
Keywords: DNA Recombination, DNA Replication, DNA Topology, Mitochondria, Mitochondrial DNA (mtDNA), Yeast, Candida Parapsilosis
Abstract
Variation in the topology of mitochondrial DNA (mtDNA) in eukaryotes evokes the question if differently structured DNAs are replicated by a common mechanism. RNA-primed DNA synthesis has been established as a mechanism for replicating the circular animal/mammalian mtDNA. In yeasts, circular mtDNA molecules were assumed to be templates for rolling circle DNA-replication. We recently showed that in Candida albicans, which has circular mapping mtDNA, recombination driven replication is a major mechanism for replicating a complex branched mtDNA network. Careful analyses of C. albicans-mtDNA did not reveal detectable amounts of circular DNA molecules. In the present study we addressed the question of how the unit sized linear mtDNA of Candida parapsilosis terminating at both ends with arrays of tandem repeats (mitochondrial telomeres) is replicated. Originally, we expected to find replication intermediates diagnostic of canonical bi-directional replication initiation at the centrally located bi-directional promoter region. However, we found that the linear mtDNA of Candida parapsilosis also employs recombination for replication initiation. The most striking findings were that the mitochondrial telomeres appear to be hot spots for recombination driven replication, and that stable RNA:DNA hybrids, with a potential role in mtDNA replication, are also present in the mtDNA preparations.
Introduction
A number of mitochondrial diseases result from impaired DNA replication (1). This discovery revived interest in the mechanisms involved in mitochondrial DNA (mtDNA)3 replication. In several aspects, the mitochondria of simple unicellular eukaryotes have served as models for more complex organisms, because yeast and human mitochondria contain a comparable set of replication factors and they share certain common aspects of mtDNA replication (2, 3). Despite these similarities, the mitochondrial genomes exhibit substantial variability in their molecular architecture. For example, the mitochondria of various fungal species contain a wide range of topologically different mtDNA forms such as circular, multipartite, monomeric, and concatemeric molecules, linear molecules with different telomeric structures, and complex branched structures (4–8). This variability prompted us to investigate the replication strategies of such topologically different genomes.
The mitochondrial genome of bakers' yeast (Saccharomyces cerevisiae) contains sequence elements termed ori/rep with characteristic guanine and cytosine (G + C)-rich clusters that resemble the structures found at the heavy strand replication origin of mammalian mtDNA. The complete sequence of S. cerevisiae mtDNA contains up to eight ori/rep elements (9). Based on their ability to support hypersuppressivness in genetic crosses between strains with wild-type (rho+) and short truncated (rho−) mitochondrial genomes these sequences were implicated in the replication of mtDNA. Because the hypersuppressive rho− derivatives of the wild-type mtDNA contain ori/rep elements amplified as reiterated tandem repeats, these elements were thought to offer a replication and/or segregation advantage over the wild-type or neutral rho− genomes (10–12). Four of the ori/rep elements, which contain an uninterrupted canonical promoter motif, are thought to be active origins of replication. This led to the assumption that yeast mtDNA replication initiation is an RNA-primed process (13, 14). Although RNA-primed DNA strands have been detected in rho− mtDNA (12, 15, 16) they are seemingly dispensable for the hypersuppressiveness of rho− mtDNA maintenance (17) and there is no clear evidence to date showing involvement of ori/rep sequences in initiating the replication of the wild-type mtDNA. Moreover, specific rho− mutants lacking the ori/rep elements are stably maintained in yeast cells (18). In addition, ori/rep elements do not represent an evolutionary conserved feature required for mtDNA replication (19, 20) and they may originate from a mobile or selfish element that invaded and was amplified in the mitochondria of the genus Saccharomyces (21). As the G + C clusters in S. cerevisiae mtDNA represent recombinational hot spots (22, 23), the ori/rep elements may serve to promote recombination-dependent replication (RDR) and active transcription could play a stimulatory role in this process (24). Several lines of evidence indicate that rho− genomes use different mechanisms for their maintenance as the replication of the rho+ genomes requires factors like the recombination protein Mgm101, the helicase Hmi1, and the RNA polymerase Rpo41, all of which are dispensable for the replication of certain classes of rho− genomes (17, 25, 26). This implies that different, alternative replication mechanisms may operate within yeast mitochondria.
Studies that used pulsed-field gel electrophoresis and electron microscopy to investigate the structure of mtDNA suggested that yeast mtDNA is synthesized via a rolling circle replication (RCR) mechanism (4, 27, 28). This mechanism generates concatemeric linear molecules, which are heterogeneous in size, but larger than the genome unit equivalent, and may explain the amplification of oligomeric repeats in rho− strains. Similar to the mitochondria of protozoan and plant species (29–35), the phage T4-like RDR mechanism appears to at least coexist with the RCR strategy also in yeasts. The RDR mechanism has been demonstrated in the mitochondria of Candida albicans indicating that it plays a major role in mtDNA maintenance (7).
If RDR operates in C. albicans with circular mapping mtDNA, is the same mechanism involved in the replication of mtDNA with a dramatically different topology, i.e. unit sized linear mtDNA molecules with mitochondrial telomeres? In addition, the presence of linear DNA molecules terminating in specific telomeric structures in the mitochondria of phylogenetically different yeast taxa evokes intriguing questions concerning their evolutionary origin (8, 36, 37). To address these questions, we employed Candida parapsilosis that has a linear mtDNA terminating in arrays of telomeric repeats (30,923 + 2n × 738 bp, where n = 0–12) (Fig. 1A) (38, 39). The complete mitochondrial genome of C. parapsilosis has been sequenced and been shown to be organized into two major transcriptional units, rnl-nad3 and cox1-atp6, which are transcribed from a central region to the left and right telomere, respectively (38, 40). The genome lacks canonical S. cerevisiae-like ori/rep elements, but contains G + C-rich elements located in both subterminal regions.
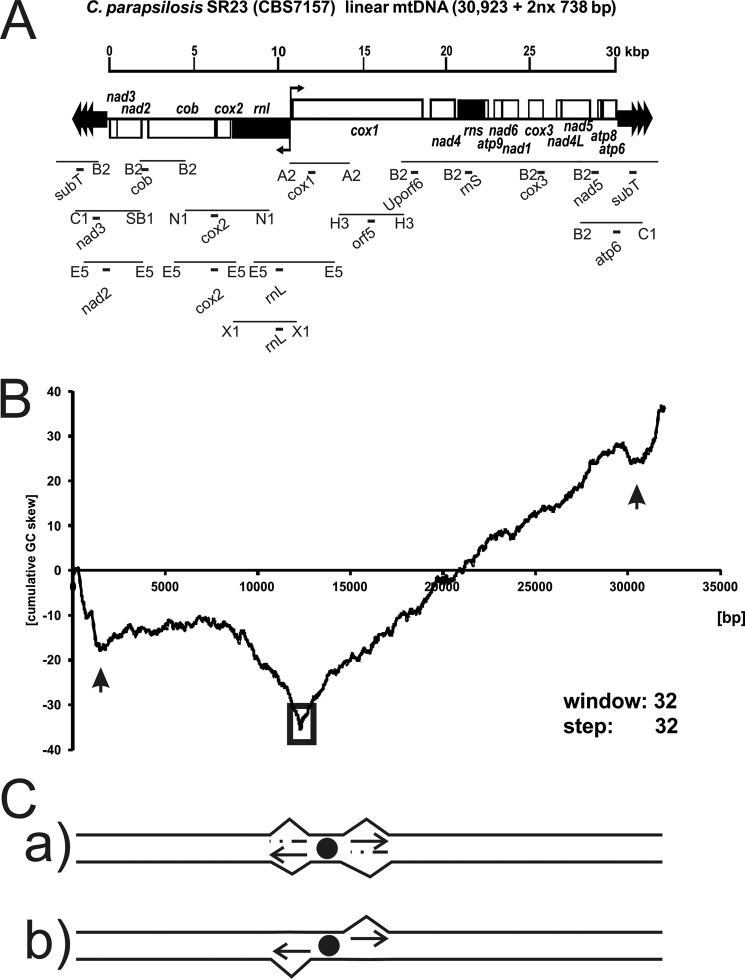
FIGURE 1.
Organization of C. parapsilosis mtDNA. A, the mtDNA of C. parapsilosis has a unit size of 30,923 kb +2nx 738 bp, is organized into smaller and larger coding regions as defined by the opposing orientations of ORFs pointing outwards from position nt 12159. Filled dark gray arrows indicate genes. Restriction fragments are indicated by lines and letter code for the restrictase on either side of the fragment (B2, BglII; C1, ClaI; SB1, SnaBI; E5, EcoRV; N1, NdeI; X1, XbaI; A2, AvaII; H3, HindIII). Probes used in this study are indicated with black bricks and names. B, in silico analyses of mtDNA in C. parapsilosis by cumulative GC skew. The plot indicating the cumulative GC skew analysis was created using a sliding window size of 32. Two local minima are indicated by black vertical arrows (nt 1,950 and 30,800), an open box indicates a global minimum at nt 12,289–12,321. C, schematic interpretation of the originally anticipated mode of replication initiation. Primers formed at a hypothetical origin near or at nt 12,289 (GC skew) would be extended uni- or bi-directionally in a strand-coupled or uncoupled manner.
We analyzed replication intermediates of C. parapsilosis mtDNA employing two-dimensional neutral agarose gel electrophoresis (two-dimensional NAGE). We demonstrate that mtDNA intermediates in C. parapsilosis closely resemble those found in C. albicans despite the fact that the topological organization of the mtDNA is strikingly different. We only detect Y-arcs, and no initiation bubbles, in all genomic mtDNA fragments. Furthermore, we observed a characteristic presence of complex molecules that are not resolved into specific arcs or spots. These patterns are best explained by the RDR model that leaves significant fractions of molecules temporarily single-stranded. In our mtDNA preparations we also detect stable RNA:DNA hybrid molecules in the region corresponding to the starting point of the two major transcriptional units. These hybrid molecules fall under the wider definition of RITOLS (RNA incorporation throughout the lagging strand) intermediates detected in mammalian mtDNA (41, 42) and demonstrate that stable RNA:DNA hybrids can also be found in yeast mitochondria.
EXPERIMENTAL PROCEDURES
In Silico Analyses
Strand-specific asymmetry in the base composition of the C. parapsilosis mtDNA (NC_005253/X74411) was analyzed using a cumulative GC (cGC) skew diagram (Fig. 1B). The calculation was performed using the DNA base composition analysis tool with window = 32 and step = 32 settings.
Strains, Growth Conditions, and mtDNA Purification
The C. parapsilosis strain SR23 (CBS 7157, accession number AY423711) (38) was grown in YPD (1% yeast extract, 2% peptone, and 2% glucose) at 30 °C. Cells were harvested during exponential growth (A600 1–1.5) and mtDNA was purified from isolated mitochondria as described elsewhere (65).
DNA Modifications
10 μg of mtDNA were digested with 3 units/μg of restriction enzymes (Fermentas) as indicated for 3 h at 37 °C. RNase H (Fermentas) treatments were 2 units for 30 min at 37 °C, RNase 1 (Fermentas) was 10 units for 30 min at 37 °C.
Electrophoresis, Hybridizations and Quantifications
Two-dimensional NAGE was performed as previously described (7). Restriction fragments of mtDNA were separated on 0.5% agarose gels at 0.9 V/cm and 21 °C for 24 h (1st dimension) followed by 1% agarose run at 3 V/cm and 4 °C for 15 h in the presence of 300 ng/μl of EthBr (2nd dimension). Fork direction gels were prepared. First dimension gels were run in 0.4% agarose at 0.9 V/cm and 21 °C for 22 h. In gelo digests were 3 × 150 units of enzyme applied for 2 h (6 h total) at 37 °C each time. Second dimension gels were 1.5% agarose run at 3 V/cm and 4 °C for 16 h. Gels were blotted onto nylon membranes by alkaline transfer. Southern blots were hybridized to specific PCR probes as indicated (Table 1) for 3 h at 65 °C in 7% SDS, 250 mm NaHPO4, pH 7.2, 1 mm EDTA, and 0.1% BSA, washed 2 × 15 min at 65 °C with 5% SDS, 40 mm NaHPO4, pH 7.2, 1 mm EDTA. Blots were exposed to Storage Phosphor Screens for 12–72 h (GE Healthcare), detected with a TyphoonTM Trio PhorsphorImager (GE Healthcare).
TABLE 1.
Oligonucleotides used as oligo probes
Oligonucleotides are listed that were used as oligos probe or as PCR primers on purified C. parapsilosis mtDNA to generate the respective PCR products to be used as double-stranded probes.
| Sequence | |
|---|---|
| Internal | |
| TTCACTTCTTTCCAACAATCTAGATCT | NAD3 sense |
| AGATCTAGATTGTTGGAAAGAAGTGAA | NAD3 anti |
| TCATGCTGAAAAACGTTCCA | COB sense |
| AACCTTTTCCTACTTTAGCTCCA | COB anti |
| TTAGACGTGCCAACACCTTG | COX2 sense |
| TTCAAACTCAATTGATTCTCCTG | COX2 anti |
| TGGCATAAGCCGATCCTAAC | RNL sense |
| TGTGCTTTCGGGTCTATTCC | RNL anti |
| AAATCGAAAAATACCAAAGTATTGAAA | ORF5 sense |
| TTTCAATACTTTGGTATTTTTCGATTT | ORF5 anti |
| ATTCTTTGGACACCCTGAAGTATATA | upORF6 sense |
| TATATACTTCAGGGTGTCCAAAGAAT | upORF6 anti |
| GCAGCAGTGGGGAATTATTG | RNS sense |
| GGGTACCGAATCCATGTCAC | RNS anti |
| CTGATCTACTAGAATTGCTGCTATGAAA | NAD5 sense |
| TTTCATAGCAGCAATTCTAGTAGATCAG | NAD5 anti |
| CTATTTAGGTTCATCACGTTGAGGT | ATP6 sense |
| AAGAGAAATAGCACGAGAAGCATAA | ATP6 anti |
| Telomeric | |
| GTATTTATTTCTTATTCATTCTTCATTAGAAC | Oligo2 |
| GTTCTAATGAAGAATGAATAAGAAATAAATAC | Oligo2C |
| TCTTTATTTGATATCTTATCTCTTTATTATC | SUBTel |
| GATAATAAAGAGATAAGATATCAAATAAAGA | SUBTelC |
RESULTS
In Silico Analysis Reveals Three Significant Minima in a Cumulative GC Skew Diagram of C. parapsilosis mtDNA
An analysis of strand-specific asymmetry in base composition has been used to predict origins of replication in prokaryotic genomes. The leading strand of replicating DNA has a relative excess of guanine over cytosine residues and exhibits a positive value in cGC skew diagrams. The cGC skew polarity switches near the origin and the terminus of DNA replication (43). Previously, we have shown that the cGC skew calculated from the sequence of C. parapsilosis mtDNA (Fig. 1B) has a global minimum at position 12,289–12,321 localized between the rnl and cox1 genes, which contains promoters for the left and right polycistronic transcriptional units (38, 40). We initially assumed that the global minimum of the cGC skew diagram could be associated with a canonical bi-directional origin of replication. In this case, replication would proceed from this central region toward the telomeres either by coupled or uncoupled synthesis of the DNA strands (Fig. 1C). Both global and local cGC skew minima have been found at origins of replication in bacterial chromosomes. In addition to the strong global minimum, two significant local minima were observed in the C. parapsilosis mtDNA close to the 554-bp long subterminal inverted repeats. The subterminal inverted repeat regions on both sides of the linear mtDNA contain a GC-rich motif that may represent hot spots of homologous recombination involved in the recombination-driven replication of linear mtDNA and/or mitochondrial telomere maintenance. Therefore, we analyzed the replication intermediates derived from the mtDNA regions exhibiting global and local minima in the cGC skew diagrams (see below).
The Major Forms of DNA Intermediates in C. parapsilosis Are Fork Structures and Recombination Intermediates
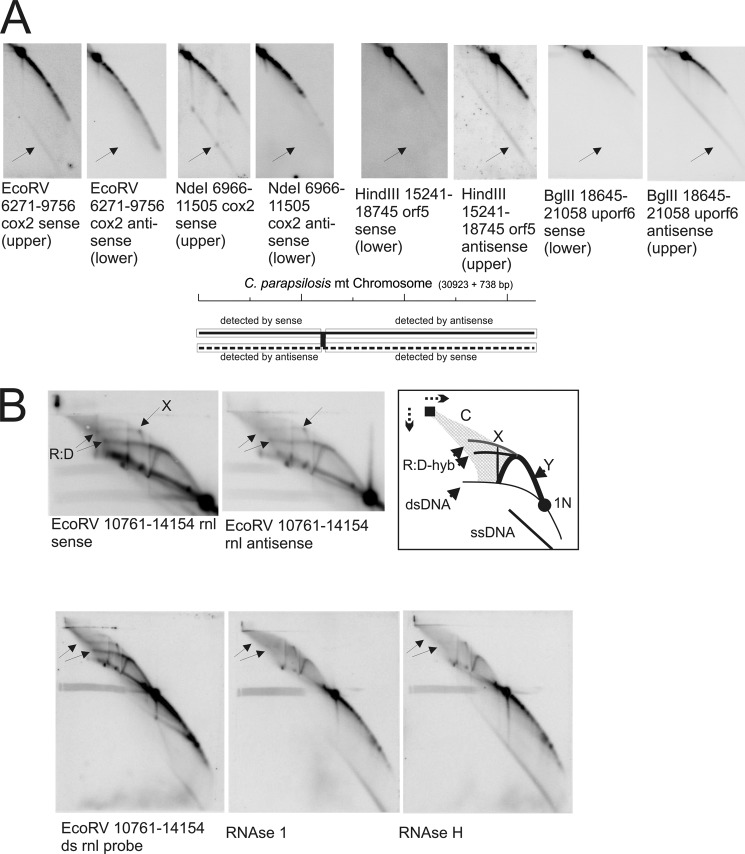
To determine and map potential replication initiation sites, the entire C. parapsilosis mtDNA was subjected to two-dimensional NAGE analysis, and restriction fragments of different sizes were chosen to overlap by several kbp (Figs. 1A and 2). Generally, restriction fragments that contain a replication origin of strand-coupled synthesis (e.g. mammalian mtDNA OH (44)) appear as “bubble arcs” (Fig. 2B, B, gray dotted line). Passing replication forks generate Y-arc patterns (Fig. 2B, Y) that extend from the 1N spot of non-replicating molecules to the 2N spot of almost fully replicated molecules. X-arcs emerging vertically from the 2N (Fig. 2B, X) comprise four-stranded DNA structures (Holliday junctions) (45, 46). C-arcs, named after a cloud-like halo spreading from the well of the first dimension gel to the Y-arc, comprise of complex branched molecules that contain single-stranded stretches (7). D- or R-loop-based initiation bubble intermediates are expected to form an arc that runs between the Y-arc and double-stranded linear molecules (47).
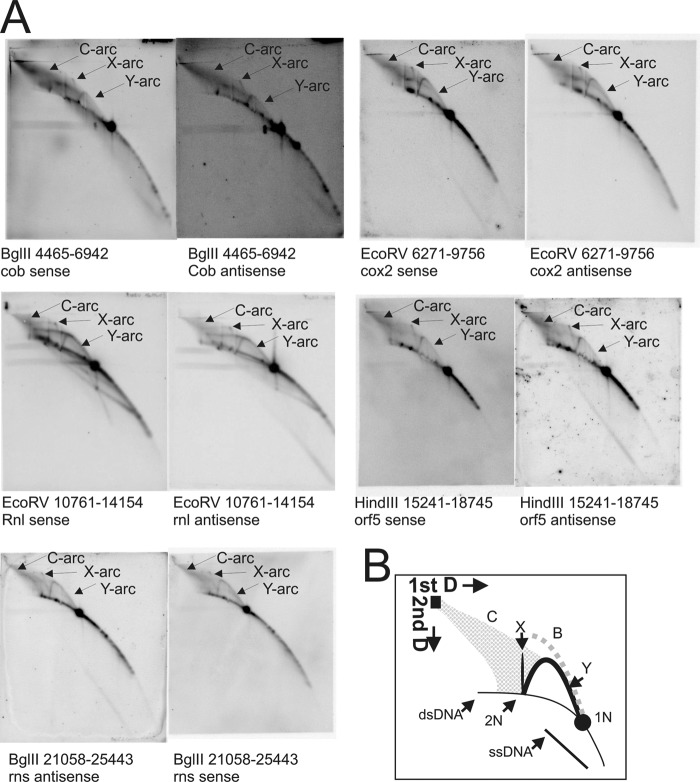
FIGURE 2.
Topological analyses of the C. parapsilosis mtDNA by two-dimensional NAGE shows significant formation of recombination intermediates. A, two-dimensional radiographs as examples from both coding regions to show Y-, X-, and C-arcs present in all of the coding regions. B, interpretation of two-dimensional AGE radiographs where: C, cloud-arc; X, X-arc; Y, Y-arc; 1N, non-replicated restriction fragment; 2N, almost fully replicated intermediates. The dotted gray line named B indicates the expected pattern that would be observed by bubble-shaped intermediates. For positions of restriction enzyme cleavage and probe positions refer to Fig. 1A.
The comprehensive analysis of C. parapsilosis mtDNA did not reveal bubble arcs, but showed Y-arcs on all restriction fragments, as for example, on fragments BglII (nt 4,465–6,942) probed for cob, EcoRV (nt 6,271–9,756) probed for cox2, EcoRV (nt 10,761–14,154) probed for rnl, HindIII (nt 15,241–18,745) probed for cox1-i1-orf306, and BglII (nt 21,058–25,433) probed for rns (Fig. 2A). Also X-arcs and C-arcs (7) that consist of ssDNA-rich complex molecules were detected, both arc-types indicating homologous recombination throughout the mtDNA. The presence of Y-arcs in the absence of bubble-shaped intermediates was previously proposed to indicate RCR of yeast mtDNA (48, 49), in other words a form of strand displacement synthesis. The absence of circular unit size mtDNA molecules in C. parapsilosis does not allow for RCR to explain the obtained results. However, the prominent C-arcs and X-arcs are indicative of recombination initiated DNA replication. The C-arc is diagnostic of multiple invasion structures. These are turned into Holliday junctions (X-arcs) and further into Y-arcs to proceed with DNA synthesis. Taking into consideration the similarities with replication intermediates found in C. albicans mitochondria (7), we propose recombination-based coupled and -uncoupled mechanisms for mtDNA replication in C. parapsilosis. RC replication of C. parapsilosis mtDNA is seemingly not possible in the absence of circular template molecules of the main genome, but appears to be employed for telomere maintenance, i.e. t-circle replication (50).
The polarity of the replication forks of coupled synthesis was assessed by a modified two-dimensional NAGE technique (see e.g. Ref. 7). DNA intermediates of restriction enzyme-digested DNA samples are separated on the first dimension gel and then cleaved with an additional restriction enzyme in gelo (Fig. 3A). As depicted in Fig. 3B, in gelo cleavage of replication forks can produce 2 distinguishable patterns: forks are either entering the restriction fragment of interest from left to right, in which case the double-stranded linear portion of the Y-structure closer to the 1N spot is cleaved off the fork. In this case, a shorter horizontal arc is produced, from which the remaining Y-structures extend (Fig. 3B, Yb). Alternatively, forks are entering from right to left, resulting in remaining Y-structures closer to 1N and double-stranded linear fragments forming a horizontal arc further away from the 1N and Y-structure fragments (Fig. 3B, Ya). There is, however, the possibility of observing both types in analyzed fragments, indicating forks of both possible polarities, i.e. bi-directional replication. Our analysis showed that replication forks pass the analyzed restriction fragments from both possible directions (Fig. 3C, apexes of both Yb and Ya are indicated with arrows). If a single, bi-directional origin was initiating replication from the center of the C. parapsilosis mtDNA, analyses of fork polarity on either side of it would produce uni-polar forks. This is to say a left-to-right fork in fragments situated right of the origin and right-to-left forks to the left of it. Observing forks of both possible polarities on any of the analyzed fragments thus provides two alternative, but not mutually exclusive possibilities. The initiation of DNA synthesis can be a recombination driven process that uses ssDNA annealing or strand invasions anywhere in the genome (Fig. 6). Alternatively, the genome can be replicated from both ends of the linear mtDNA molecule by taking advantage of mechanisms used for mitochondrial telomere maintenance, which depends on recombination with telomeric circles (t-circles) (50). Similarly to C. albicans, C. parapsilosis also has mtDNA molecules migrating on the ds linear arc of sizes smaller than 1N (Figs. 2A and 4A). As proposed for C. albicans mtDNA, these may provide precursors for generating invasive 3′-ssDNA termini that would be suitable for priming RDR (7, 51, 52).
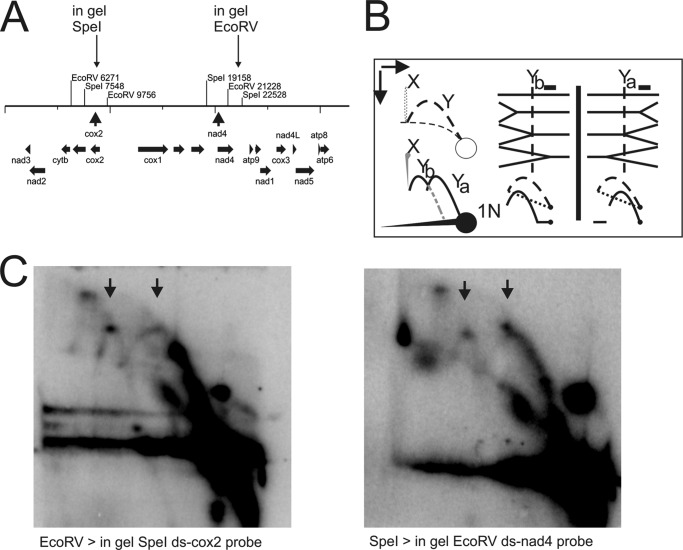
FIGURE 3.
Fork direction two-dimensional AGE of C. parapsilosis mtDNA. A, restriction map indicating cleavage sites for the initial mtDNA digest and showing the position of the site used for in gel cleavage of replication intermediates. B, interpretation of potentially forming Y-arcs depending on the polarity of the analyzed replication fork. Dashed lines indicate the original two-dimensional NAGE pattern, heavy lines depict the detectable patterns after in gelo treatment. The position of the probe is indicated by a black filled box above the Y-structures. The vertical dotted line crossing the Y-structures mimics the position of in gelo cleavage. C, mtDNA digested with EcoRV or SpeI was separated on a first-dimension gel, digested with SpeI or EcoRV in gelo and then separated on a second-dimension gel. Y-arc patterns of both possible fork polarities are seen on both radiographs, thus showing that replication forks pass the fragments bi-directionally.
FIGURE 6.
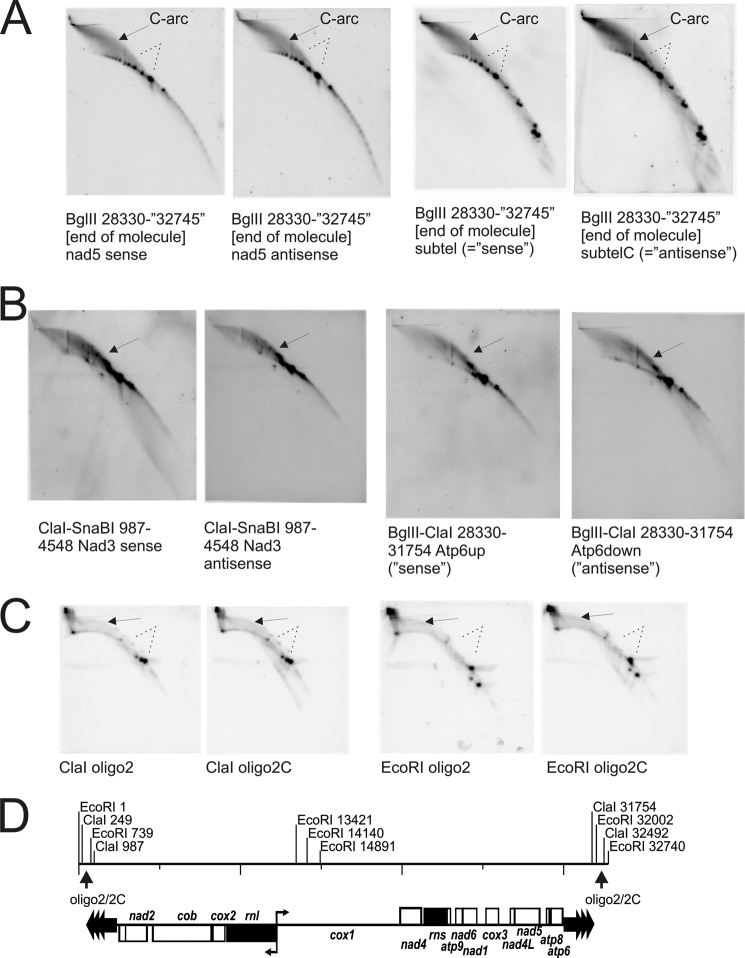
Analyses of mtDNA intermediates within and close to the telomeric repeats. No Y-arcs are detected but the C-arc is very dominant and even stronger than in fragments from the coding region. A, BglII-digested C. parapsilosis mtDNA probed for nad5 in the vicinity of subtelomeric and TR, and probed for subtel (non-coding sequences between coding regions and TR) do not reveal Y- or X-arcs (dotted lines) but show strong C-arcs (arrows). Dots on dsDNA on BglII pictures may reflect n-times repeated TR sequence (with n = ≥1). B, more detailed analyses of fragments containing only TR sequence (ClaI-SnaBI probed for nad3 and BglII-ClaI probed for atp6) confirm that no complete Y-arcs are observed. However, it can be seen that Y-like arcs are extending from 1N (arrows). These distorted Y-arc-like signals suggest strand invasions transforming into complex C-arc molecules (reasoning see Ref. 7), formation of Yel and Yes therein). C, probing exclusively for TR sequences on ClaI and EcoRI fragments (restriction map in D) that strictly show intermediates of TR-mtDNA by detection with oligo2 and oligo2C probes (refer also to Fig. 1 A) again shows C-arcs (arrows) but neither complete nor distorted Y-arcs or X-arcs (dotted lines). In conclusion, we have formation of complex molecules in TR that are being resolved into replication forks when replication proceeds toward the coding region of chloroplast mtDNA.
FIGURE 4.
Specific formation of ssDNA and RNA:DNA hybrids are observed in preparations of C. parapsilosis mtDNA. A, examples of ssDNA formation in both coding regions show that overall (almost always) the upper strand is detected as ssDNA. B, RNase-sensitive arcs are observed near the putative transcription start site around approximately nt 12,200. RNase 1 significantly reduces the structures, but RNase H removes them indicating that they are mainly RNA:DNA hybrids with RNA overhangs. Therefore we call them pseudo-RITOLS because they are most likely directly related to transcription, whereas an obvious connection to replication cannot be made. For positions of restriction enzyme cleavage and probe positions refer to Fig. 1A.
ssDNA Molecules of Different Sizes Almost Exclusively Consisting of One Specific Strand Are Omnipresent in the mtDNA in C. parapsilosis
Initial experiments had revealed prominent single-stranded DNA arcs (ssDNA) in mtDNA preparations from C. parapsilosis (data not shown). Together with the observed cGC skew and the resulting working hypothesis predicting the initiation of replication between the two transcriptional units, we considered the possibility of uncoupled bi-directional replication. In this case, the specific detection of an exposed lagging strand in the ssDNA form would be expected.
We employed strand-specific oligonucleotide probes in the two-dimensional NAGE experiments to analyze the polarity of ssDNA species. This strand-specific detection revealed that the observed ssDNA mostly originates from the upper strand of the C. parapsilosis mtDNA (Fig. 4A). As indicated by the schematic in Fig. 4A, oligonucleotide probes called “sense” detect the upper strand if they are directed against sequences within the first third of the C. parapsilosis mtDNA (nt 1–12,000). However, beyond this point (nt 12,000–30,923) the upper strand is detected by oligonucleotide probes called “antisense.” An exception is the EcoRV rnl fragment, where the ssDNA originates from the lower strand.
Based on RNase treatments (Fig. 4B) and previous studies (7) we could exclude that the ssDNA-arc contains or comprises RNA. According to the initial working hypothesis of bi-directional replication from the region around nt 12,000, we expected to find little ssDNA in the case of strand-coupled replication or the respective lagging strand in the case of uncoupled synthesis (upper strand up to about nt 12,000, lower strand on from about nt 12,000). The formation of the ssDNA is therefore independent of what is expected to be formed as a result of uncoupled replication initiation from the hypothetical replication origin around nt 12,000.
The detection of lower strand ssDNA by uncoupled or strand displacement replication as it was observed in the present study can be explained by replication initiation close to or at the telomeric repeats (nt 1–1,100) with lagging strand synthesis initiation at several sites along the mtDNA. Remarkably, the length of ssDNA molecules varies in different regions of the mtDNA (Figs. 2A and 4A). The size difference could reflect the time a particular region is single-stranded and this can be dependent on the distance of the particular probe from a lagging strand initiation site.
D-loop-like Structures Emerge from Various Fragments Supporting the Formation of ssDNA during Uncoupled Replication
Several analyzed fragments revealed D-loop-like arcs (47) emerging from the Y-arc of passing replication forks and reaching into the C-arc of complex and ss stretch-rich molecules (examples shown in Fig. 4B, EcoRV nt 10,761–14,154, and Fig. 2A, BglII nt 4,465–6,942 or EcoRV nt 6,271–9,756). Remarkably, these arcs were more clearly seen on radiographs that revealed shorter ssDNA arcs. Accordingly, radiographs showing longer ssDNA arcs had more pronounced signals within the C-arc. In addition, D-loop-like arcs were best detected approaching the telomeric repeats and close to nt 12,000, the putative start site of the two major transcriptional units. However the D-loops were not detected within the telomeric repeat (TR) region. D-loops close to nt 12,000 were more closely investigated and EcoRV fragments (nt 10,761–14,154) were subjected to RNase 1 and RNase H treatments. RNase 1 preferentially cleaves ssRNA over dsRNA and RNase H uses RNA:DNA hybrids as a substrate. These treatments showed clearly that the arc of the single-stranded nucleic acids is not degraded, meaning that it consists of DNA. On the other hand, the D-loop-like structure emerging from the apex of the Y-arc (R:D-hyb in Fig. 4B) was significantly reduced in size.
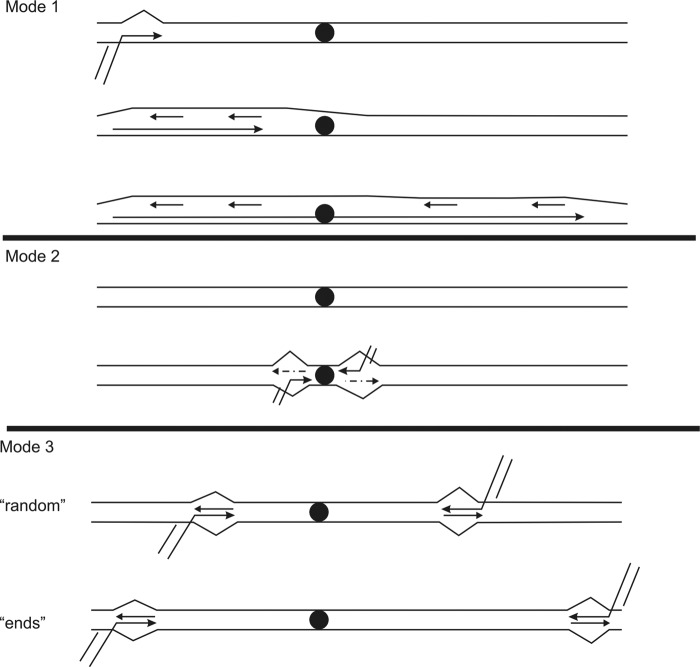
The D-loop-like structures suggest a mode of uncoupled synthesis on certain molecules within the population analyzed. We interpret their occurrence as intermediates of uncoupled replication initiation by recombination or strand invasion, which could be turned into coupled synthesis via a defined D-loop-like intermediate in the course of replication progression (Fig. 5, mode 1). Accordingly, D-loop-like structures would form from invasion of a homologous DNA strand. As DNA synthesis continues, the lagging strand template would be displaced, thus also explaining the observed formation of ssDNA species (Figs. 2 and 4). Lagging strand synthesis might initiate at certain points of the exposed template thus explaining the differing length of detected ssDNA species as described above. As previously seen with C. albicans mtDNA, passing forks of strand-coupled synthesis are at least as prominent as the D-loop or C-arc features, thus indicating the parallel occurrence of coupled and uncoupled replication. Importantly, the partial RNase sensitivity of D-loop-like structures identifies them as rather stable RNA:DNA hybrids (41, 42). These structures could be RITOLS-like intermediates. Because RNA:DNA hybrids are mainly formed at the major transcription initiation site, we consider it to be very likely that they consist of stabilized transcription intermediates that serve here to facilitate recombination and thus to provide primers for subsequent RDR (Fig. 5, mode 2). In this way, an alternative replication mode, differing from the above described asynchronous replication from telomeric repeats could be employed by C. parapsilosis. Whether this is locally restricted to the observed region or might be true in other parts of the mtDNA in C. parapsilosis remains to be investigated.
FIGURE 5.
Proposed modes of replication that might co-exist in C. parapsilosis mitochondria. The filled black circle indicates the switching point of the two major transcriptional units. In mode 1, unidirectional replication initiates from the left TR exposing ssDNA of the upper strand. Proceeding through the molecule, lagging-strand synthesis initiates at different locations thus explaining the occurrence of ssDNA of different lengths along the mtDNA of C. parapsilosis. Mode 2 accounts for the possibility of RDR facilitated by transcription (RNA is depicted as broken line). Transcription can support strand invasions to initiate strand-coupled and/or bidirectional replication. Mode 3 is proposed based on observations as presented in Figs. 1 and 3; strand invasions (due to homologous recombination by t-circles or self-invasion of main-molecules) may provide primers for replication initiation at random locations across the C. parapsilosis mtDNA or from the TR toward the respective opposite ends. As soon as the replisome reaches the coding regions, replication proceeds in a strand-coupled mode (Fig. 1 formation of Y-arcs).
The Mitochondrial Telomeres of C. parapsilosis Do Not Exhibit Typical Intermediates of Strand-coupled Synthesis
Analyses of restriction fragments, which cover the telomeric repeats (nt 1–1,348 and 31,398–32,745), either exclusively or as parts of larger DNA fragments containing unique coding sequences alongside them revealed a significantly different picture in comparison with mtDNA intermediates from the coding region. These fragments did not contain Y-arcs of passing strand-coupled replication forks (Fig. 6). The examples shown here are BglII (28,330–32,745) probed for nad5 or subtelC (Fig. 6A) probed by oligo2/2C. The predominant feature on these radiographs is made up of strong C-arcs of complex ssDNA-rich molecules (7). The numerous dots along the arc dsDNA (linear) represent the telomeric repeats, which can occur at (n = 0, 1, 2, 3 …). These are not observed for EcoRI (nt 1–739/32,002–32,740, respectively) and ClaI (nt 249–987/31,754–32,492, respectively) probed for oligo2/2c (Fig. 6B) because these enzymes cleave the repeat sequence itself, thus exclusively generating 1N fragments of the repeat sequence(s). These small regions, exclusively containing TR sequences, still show C-arcs, and even the expected pattern of ssDNA formation as described above, but no Y-arcs. Interestingly, fragments containing the subtelomeric sequence and unique coding parts, like the ClaI-SnaBI (nt 987–4,548) fragment probed for nad3 and BglII-ClaI (nt 28,330–31,754) (Fig. 6C), show again strong C-arcs and imperfect Y-arcs. The imperfect Y-arcs resemble extra small and extra large Y-arcs (YES and YEL), as observed in special non-coding parts of the C. albicans mtDNA (7), and therefore indicate strand invasion events similar to those described for C. albicans mitochondria. We have previously shown that t-circles are involved in mitochondrial telomere maintenance in C. parapsilosis (50), and proposed the recombination of t-circles into the termini of the main chromosome as the underlying mechanism. As described for C. albicans mtDNA, C-arcs are indicative of complex structures, which result from recombination and imperfect Y-structures, which are readily initiated by strand invasion during homologous recombination. These observations made in C. albicans combined with the model for recombination-driven telomere maintenance in C. parapsilosis imply that RDR mechanisms are combined with the initiation of replication and the maintenance of chromosomal termini within the telomeric repeats as discussed hereafter.
DISCUSSION
With the data presented here, we produced further evidence for the general importance of homologous recombination in mtDNA replication in yeast. We suggest models for replication initiation and progression, which are likely to co-occur in the mitochondria of C. parapsilosis (Fig. 5). We and others have previously discussed the universal impact of RDR in organelle DNA maintenance with examples from yeasts, plants, and human heart (7, 51–56). A link between topology and maintenance mechanisms has been drawn for chloroplast DNA (54), and this concept applies for C. albicans mtDNA as well. However, these organelle DNAs have mainly a complex, branched topology with random or undefined ends.
In contrast, C. parapsilosis contains a linear unit size mtDNA carrying specific telomeric structures at both ends. A putative replication origin located between the two postulated major transcription units (Fig. 1, A and B) was predicted by in silico analyses, and together this seemed to provide a working hypothesis for mapping a typical bi-directional origin to the given region (Fig. 1C). However, the analyses of C. parapsilosis mtDNA replication intermediates did not reveal bubble arcs indicating replication origins that use RNA priming for strand-coupled DNA synthesis. The overall observation was rather in line with our previous findings for C. albicans, namely mtDNA intermediates that are predicted to result from coupled and uncoupled DNA synthesis accompanied by different recombination intermediates. Despite the significant differences in genome organization and mtDNA topology between these two yeasts, they seem to share similar replication strategies. This emphasizes the hypothesis that RDR is indeed a universally applicable mechanism for mtDNA replication in yeast and, as demonstrated for plant organelles and human heart, obviously has an important role in organelle DNA maintenance across the eukaryotes.
RC replication has long been proposed for mtDNA replication in yeast. As we discussed previously (7) and as was commented on elsewhere (Ref. 57 and references therein), this replication model does not seem to hold for yeast mitochondria. In yeasts like S. cerevisiae or Schizosaccharomyces pombe circular mtDNA molecules are extremely rare. In C. albicans such molecules are not detectable (if present at all) and C. parapsilosis clearly consists of unit size linear molecules. Although a comprehensive study of S. cerevisiae wt mtDNA is yet to be conducted, the data presented here as well as for S. pombe (48) and C. albicans (7) share striking similarities. Despite a different interpretation of the topological data from S. pombe, our conclusion is that yeast mtDNA replicates via RDR, which can generate a strand-coupled and an uncoupled mode of replication progression (Fig. 5). As outlined before, the D-loop-like structures, the C-arcs, and the formation of ssDNA arcs with different lengths can be explained by an uncoupled replication mechanism that involves strand displacement and initiation of lagging strand DNA synthesis at different locations along the mtDNA (Fig. 5, mode 1). The presence of Y-arcs clearly shows that strand-coupled replication occurs in parallel. As depicted in Fig. 5, mode 3, we propose that strand invasions either close to TR sequences (Fig. 5, mode 3, ends) but also possibly at random positions along the mtDNA (Fig. 5, mode 3, random) initiate this mode of replication. It remains to be elucidated, if both modes operate simultaneously on the same molecules (which we regard as unlikely though not impossible), on sister molecules within the same cell/mitochondrial network, or cell specifically, and also whether the mode of replication depends on the cell cycle or the age of individual cells.
Strikingly, we found evidence of RITOLS-like intermediates of C. parapsilosis mtDNA (Fig. 4B, interpreted in Fig. 5, mode 2). Because these intermediates are observed at the predicted transcription start site, we propose that RNA transcription facilitates strand invasion of homologous DNA. DNA synthesis primed this way may progress either by coupled or uncoupled mechanisms that remain to be elucidated. Based on data acquired from cultured mammalian cells that were depleted of their mtDNA and allowed to re-amplify (58), we are tempted to speculate that there is a possibility of observing strand-coupled replication in yeast mitochondrial networks that are predominantly amplifying mtDNA, but not in networks that are mainly transcribing the genome and replicating DNA as a side product of DNA strand opening in a RITOLS-like manner. We do not rule out, however, the possibility that both modes operate in parallel within the same network. Postulating both a high demand of newly synthesized mtDNA and a high requirement of mitochondrial gene expression in quickly doubling cells with a high energy demand, a subset of mtDNA units in C. parapsilosis could ensure a high DNA yield via strand-coupled replication, whereas uncoupled or even RITOLS-like coupled replication would suit to maintain the integrity of DNA strand exposure during transcription. The latter could readily explain the observed cGC skew pattern because, nonetheless, exposure of DNA, especially during elevated energy production, could impose extensive exposure to reactive oxygen species (ROS) and thus damage repair. The resulting cGC skew might be reflected in our in silico data, whereas it is further possible that the uncoupled synthesis observed (or the RITOLS mode) could account for DNA repair synthesis.
A particular feature of C. parapsilosis mtDNA, among the various kinds of yeast mtDNA genomes, is the presence of telomeric repeats (38, 50). Our studies to date have shown that telomeric loop (t-loop) structures formed at the ends of a subset of linear molecules and t-circles are involved in mitochondrial telomere maintenance (50, 59, 60). In the proposed model, t-circles recombine with terminal sequences to prevent shortening of the main chromosome due to the “end replication problem” (61, 62). Herein, double hairpin elements in mtDNA can be involved in rearrangements and may serve as sites for recombinational events (63, 64). Because the C. parapsilosis mtDNA contains DHE-like elements in both sub-terminal regions these sites may be involved in recombination-dependent telomere lengthening as proposed by Ref. 50 and serve as initiation sites for RDR of the whole mitochondrial DNA (Fig. 5, mode 3, ends). Indeed, the topological data acquired here is in strong support of this model (Fig. 6).
As such, the telomeric ends of C. parapsilosis mtDNA would simultaneously mark replication origins, thus fulfilling a dual function. It has been discussed for maize chloroplast DNA that the molecule ends lie near putative replication origins and such ends are suitable for RDR initiation (52, 54). Our data are in full accordance with these observations. In addition, our cGC skew analysis did not only reveal a strong global minimum between the first third and the second two-thirds of the C. parapsilosis mtDNA, but also two significant local minima near nt 1,950 and 30,800 positions (Fig. 1B). Because these are likely to indicate recombination hot spots similarly to what we observed in C. albicans (7), they provide another piece of evidence in favor of RDR initiated from the telomeric repeat in C. parapsilosis. This would at the same time be a means of rescuing end shortening from previous rounds of replication. The polarity of replication forks of coupled synthesis is then in favor of unidirectional strand-coupled replication from either telomeric repeat or both (Fig. 5, mode 3).
In conclusion, our work presented here underlines and reinforces the significance of RDR for mtDNA maintenance in yeast and probably organelle DNA maintenance in general. A novel adjunct is the dual function of RDR in the replication initiation of mtDNA and mitochondrial telomere maintenance.
Acknowledgments
We are grateful to Prof. Howy Jacobs and the members of our laboratories for fruitful and idea generating discussions on our data. We are very grateful to Dr. Helen Cooper for revision of the manuscript.
This work was supported by Estonian Science Foundation Grants ETF 8449 and 8845, the Estonian Institutional Funding Grant IUT14021, Slovak Research and Development Agency Grants APVV 0123-10 and 0035-11, and Slovak Grant Agency Grants VEGA 1/0405/11 and 1/0311/12.
- mtDNA
- mitochondrial DNA
- RDR
- recombination driven replication
- RCR
- rolling circle DNA replication
- NAGE
- neutral agarose gel electrophoresis
- RITOLS
- RNA incorporation throughout the lagging strand
- nt
- nucleotide
- ssDNA
- single-stranded DNA
- cGC
- cumulative GC
- dsDNA
- double-stranded DNA
- TR
- telomeric repeat.
REFERENCES
- 1. Copeland W. C. (2008) Inherited mitochondrial diseases of DNA replication. Annu. Rev. Med. 59, 131–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lecrenier N., Foury F. (2000) New features of mitochondrial DNA replication system in yeast and man. Gene 246, 37–48 [DOI] [PubMed] [Google Scholar]
- 3. Spelbrink J. N. (2010) Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life 62, 19–32 [DOI] [PubMed] [Google Scholar]
- 4. Maleszka R., Skelly P. J., Clark-Walker G. D. (1991) Rolling circle replication of DNA in yeast mitochondria. EMBO J. 10, 3923–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fukuhara H., Sor F., Drissi R., Dinouël N., Miyakawa I., Rousset S., Viola A. M. (1993) Linear mitochondrial DNAs of yeasts: frequency of occurrence and general features. Mol. Cell. Biol. 13, 2309–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fricova D., Valach M., Farkas Z., Pfeiffer I., Kucsera J., Tomaska L., Nosek J. (2010) The mitochondrial genome of the pathogenic yeast Candida subhashii: GC-rich linear DNA with a protein covalently attached to the 5′ termini. Microbiology 156, 2153–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerhold J. M., Aun A., Sedman T., Jõers P., Sedman J. (2010) Strand invasion structures in the inverted repeat of Candida albicans mitochondrial DNA reveal a role for homologous recombination in replication. Mol. Cell 39, 851–861 [DOI] [PubMed] [Google Scholar]
- 8. Valach M., Farkas Z., Fricova D., Kovac J., Brejova B., Vinar T., Pfeiffer I., Kucsera J., Tomaska L., Lang B. F., Nosek J. (2011) Evolution of linear chromosomes and multipartite genomes in yeast mitochondria. Nucleic Acids Res. 39, 4202–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Foury F., Roganti T., Lecrenier N., Purnelle B. (1998) The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440, 325–331 [DOI] [PubMed] [Google Scholar]
- 10. de Zamaroczy M., Baldacci G., Bernardi G. (1979) Putative origins of replication in the mitochondrial genome of yeast. FEBS Lett. 108, 429–432 [DOI] [PubMed] [Google Scholar]
- 11. de Zamaroczy M., Marotta R., Faugeron-Fonty G., Goursot R., Mangin M., Baldacci G., Bernardi G. (1981) The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature 292, 75–78 [DOI] [PubMed] [Google Scholar]
- 12. Baldacci G., Chérif-Zahar B., Bernardi G. (1984) The initiation of DNA replication in the mitochondrial genome of yeast. EMBO J. 3, 2115–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baldacci G., Bernardi G. (1982) Replication origins are associated with transcription initiation sequences in the mitochondrial genome of yeast. EMBO J. 1, 987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Zamaroczy M., Faugeron-Fonty G., Baldacci G., Goursot R., Bernardi G. (1984) The ori sequences of the mitochondrial genome of a wild-type yeast strain: number, location, orientation and structure. Gene 32, 439–457 [DOI] [PubMed] [Google Scholar]
- 15. Xu B., Clayton D. A. (1992) Assignment of a yeast protein necessary for mitochondrial transcription initiation. Nucleic Acids Res. 20, 1053–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Graves T., Dante M., Eisenhour L., Christianson T. W. (1998) Precise mapping and characterization of the RNA primers of DNA replication for a yeast hypersuppressive petite by in vitro capping with guanylyltransferase. Nucleic Acids Res. 26, 1309–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fangman W. L., Henly J. W., Brewer B. J. (1990) RPO41-independent maintenance of [rho−] mitochondrial DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fangman W. L., Henly J. W., Churchill G., Brewer B. J. (1989) Stable maintenance of a 35-base pair yeast mitochondrial genome. Mol. Cell. Biol. 9, 1917–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jõers P., Gerhold J. M., Sedman T., Kuusk S., Sedman J. (2007) The helicase CaHmi1p is required for wild-type mitochondrial DNA organization in Candida albicans. FEMS Yeast Res. 7, 118–130 [DOI] [PubMed] [Google Scholar]
- 20. Petersen R. F., Langkjaer R. B., Hvidtfeldt J., Gartner J., Palmen W., Ussery D. W., Piskur J. (2002) Inheritance and organisation of the mitochondrial genome differ between two Saccharomyces yeasts. J. Mol. Biol. 318, 627–636 [DOI] [PubMed] [Google Scholar]
- 21. Piskur J. (1989) Transmission of the yeast mitochondrial genome to progeny: the impact of intergenic sequences. Mol. Gen. Genet. 218, 161–168 [DOI] [PubMed] [Google Scholar]
- 22. Zinn A. R., Pohlman J. K., Perlman P. S., Butow R. A. (1988) In vivo double-strand breaks occur at recombinogenic G + C-rich sequences in the yeast mitochondrial genome. Proc. Natl. Acad. Sci. U.S.A. 85, 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dieckmann C. L., Gandy B. (1987) Preferential recombination between GC clusters in yeast mitochondrial DNA. EMBO J. 6, 4197–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williamson D. (2002) The curious history of yeast mitochondrial DNA. Nat. Rev. Genet. 3, 475–481 [DOI] [PubMed] [Google Scholar]
- 25. Sedman T., Kuusk S., Kivi S., Sedman J. (2000) A DNA helicase required for maintenance of the functional mitochondrial genome in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 1816–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zuo X. M., Clark-Walker G. D., Chen X. J. (2002) The mitochondrial nucleoid protein, Mgm101p, of Saccharomyces cerevisiae is involved in the maintenance of ρ+ and ori/rep-devoid petite genomes but is not required for hypersuppressive ρ− mtDNA. Genetics 160, 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maleszka R. (1992) Electrophoretic profiles of mitochondrial plasmids in Neurospora suggest they replicate by a rolling circle mechanism. Biochem. Biophys. Res. Commun. 186, 1669–1673 [DOI] [PubMed] [Google Scholar]
- 28. Maleszka R., Clark-Walker G. D. (1992) In vivo conformation of mitochondrial DNA in fungi and zoosporic moulds. Curr. Genet. 22, 341–344 [DOI] [PubMed] [Google Scholar]
- 29. Preiser P. R., Wilson R. J., Moore P. W., McCready S., Hajibagheri M. A., Blight K. J., Strath M., Williamson D. H. (1996) Recombination associated with replication of malarial mitochondrial DNA. EMBO J. 15, 684–693 [PMC free article] [PubMed] [Google Scholar]
- 30. Backert S. (2002) R-loop-dependent rolling-circle replication and a new model for DNA concatemer resolution by mitochondrial plasmid mp1. EMBO J. 21, 3128–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oldenburg D. J., Bendich A. J. (2001) Mitochondrial DNA from the liverwort Marchantia polymorpha: circularly permuted linear molecules, head-to-tail concatemers, and a 5′ protein. J. Mol. Biol. 310, 549–562 [DOI] [PubMed] [Google Scholar]
- 32. Backert S., Dörfel P., Lurz R., Börner T. (1996) Rolling-circle replication of mitochondrial DNA in the higher plant Chenopodium album (L.). Mol. Cell Biol. 16, 6285–6294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Backert S. (2000) Strand switching during rolling circle replication of plasmid-like DNA circles in the mitochondria of the higher plant Chenopodium album (L.). Plasmid 43, 166–170 [DOI] [PubMed] [Google Scholar]
- 34. Backert S., Lurz R., Börner T. (1996) Electron microscopic investigation of mitochondrial DNA from Chenopodium album (L.). Curr. Genet. 29, 427–436 [DOI] [PubMed] [Google Scholar]
- 35. Backert S., Börner T. (2000) Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 37, 304–314 [DOI] [PubMed] [Google Scholar]
- 36. Nosek J., Tomáska L. (2003) Mitochondrial genome diversity: evolution of the molecular architecture and replication strategy. Curr. Genet. 44, 73–84 [DOI] [PubMed] [Google Scholar]
- 37. Nosek J., Tomáska L., Fukuhara H., Suyama Y., Kovác L. (1998) Linear mitochondrial genomes: 30 years down the line. Trends Genet. 14, 184–188 [DOI] [PubMed] [Google Scholar]
- 38. Nosek J., Novotna M., Hlavatovicova Z., Ussery D. W., Fajkus J., Tomaska L. (2004) Complete DNA sequence of the linear mitochondrial genome of the pathogenic yeast Candida parapsilosis. Mol. Genet. Genomics 272, 173–180 [DOI] [PubMed] [Google Scholar]
- 39. Nosek J., Dinouël N., Kovac L., Fukuhara H. (1995) Linear mitochondrial DNAs from yeasts: telomeres with large tandem repetitions. Mol. Gen. Genet. 247, 61–72 [DOI] [PubMed] [Google Scholar]
- 40. Kosa P., Valach M., Tomaska L., Wolfe K. H., Nosek J. (2006) Complete DNA sequences of the mitochondrial genomes of the pathogenic yeasts Candida orthopsilosis and Candida metapsilosis: insight into the evolution of linear DNA genomes from mitochondrial telomere mutants. Nucleic Acids Res. 34, 2472–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang M. Y., Bowmaker M., Reyes A., Vergani L., Angeli P., Gringeri E., Jacobs H. T., Holt I. J. (2002) Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell 111, 495–505 [DOI] [PubMed] [Google Scholar]
- 42. Yasukawa T., Reyes A., Cluett T. J., Yang M. Y., Bowmaker M., Jacobs H. T., Holt I. J. (2006) Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 25, 5358–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grigoriev A. (1998) Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26, 2286–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Holt I. J., Lorimer H. E., Jacobs H. T. (2000) Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 100, 515–524 [DOI] [PubMed] [Google Scholar]
- 45. Lockshon D., Zweifel S. G., Freeman-Cook L. L., Lorimer H. E., Brewer B. J., Fangman W. L. (1995) A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell 81, 947–955 [DOI] [PubMed] [Google Scholar]
- 46. MacAlpine D. M., Perlman P. S., Butow R. A. (1998) The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc. Natl. Acad. Sci. U.S.A. 95, 6739–6743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pohjoismäki J. L., Holmes J. B., Wood S. R., Yang M. Y., Yasukawa T., Reyes A., Bailey L. J., Cluett T. J., Goffart S., Willcox S., Rigby R. E., Jackson A. P., Spelbrink J. N., Griffith J. D., Crouch R. J., Jacobs H. T., Holt I. J. (2010) Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 397, 1144–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Han Z., Stachow C. (1994) Analysis of Schizosaccharomyces pombe mitochondrial DNA replication by two-dimensional gel electrophoresis. Chromosoma 103, 162–170 [DOI] [PubMed] [Google Scholar]
- 49. Backert S., Meissner K., Börner T. (1997) Unique features of the mitochondrial rolling circle-plasmid mp1 from the higher plant Chenopodium album (L.). Nucleic Acids Res. 25, 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nosek J., Rycovska A., Makhov A. M., Griffith J. D., Tomaska L. (2005) Amplification of telomeric arrays via rolling-circle mechanism. J. Biol. Chem. 280, 10840–10845 [DOI] [PubMed] [Google Scholar]
- 51. Oldenburg D. J., Bendich A. J. (2004) Changes in the structure of DNA molecules and the amount of DNA per plastid during chloroplast development in maize. J. Mol. Biol. 344, 1311–1330 [DOI] [PubMed] [Google Scholar]
- 52. Oldenburg D. J., Bendich A. J. (2004) Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J. Mol. Biol. 335, 953–970 [DOI] [PubMed] [Google Scholar]
- 53. Pohjoismäki J. L., Goffart S., Tyynismaa H., Willcox S., Ide T., Kang D., Suomalainen A., Karhunen P. J., Griffith J. D., Holt I. J., Jacobs H. T. (2009) Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J. Biol. Chem. 284, 21446–21457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bendich A. J. (2004) Circular chloroplast chromosomes: the grand illusion. Plant Cell 16, 1661–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ling F., Hori A., Shibata T. (2007) DNA Recombination-initiation plays a role in the extremely biased inheritance of yeast [rho−] mitochondrial DNA that contains the replication origin ori5. Mol. Cell. Biol. 27, 1133–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shibata T., Ling F. (2007) DNA recombination protein-dependent mechanism of homoplasmy and its proposed functions. Mitochondrion 7, 17–23 [DOI] [PubMed] [Google Scholar]
- 57. Bendich A. J. (2010) The end of the circle for yeast mitochondrial DNA. Mol. Cell 39, 831–832 [DOI] [PubMed] [Google Scholar]
- 58. Yasukawa T., Yang M. Y., Jacobs H. T., Holt I. J. (2005) A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell 18, 651–662 [DOI] [PubMed] [Google Scholar]
- 59. Tomaska L., Makhov A. M., Griffith J. D., Nosek J. (2002) t-Loops in yeast mitochondria. Mitochondrion 1, 455–459 [DOI] [PubMed] [Google Scholar]
- 60. Tomaska L., Nosek J., Makhov A. M., Pastorakova A., Griffith J. D. (2000) Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res. 28, 4479–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Olovnikov A. M. (1973) A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41, 181–190 [DOI] [PubMed] [Google Scholar]
- 62. Watson J. D. (1972) Origin of concatemeric T7 DNA. Nat. New Biol. 239, 197–201 [DOI] [PubMed] [Google Scholar]
- 63. Paquin B., Laforest M. J., Lang B. F. (2000) Double-hairpin elements in the mitochondrial DNA of allomyces: evidence for mobility. Mol. Biol. Evol. 17, 1760–1768 [DOI] [PubMed] [Google Scholar]
- 64. Bullerwell C. E., Leigh J., Forget L., Lang B. F. (2003) A comparison of three fission yeast mitochondrial genomes. Nucleic Acids Res. 31, 759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Visacka K., Gerhold J. M., Petrovicova J., Kinsky S., Jõers P., Nosek J., Sedman J., Tomaska L. (2009) Novel subfamily of mitochondrial HMG box-containing proteins: functional analysis of Gcf1p from Candida albicans. Microbiology 155, 1226–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]