Background: HIF1α is degraded under normoxic conditions by VHL. Degradation under hypoxia is poorly understood.
Results: HIF1α is degraded under hypoxia via 26 S proteasome; MDM2/E3 ligase, under the control of PI3K, mediates the hypoxic degradation of HIF1α in glioma systems.
Conclusion: MDM2 action on HIF1α under hypoxia is controlled by PI3K signaling.
Significance: Results reveal a mechanism controlling hypoxic HIF1α degradation.
Keywords: Glioblastoma, Hypoxia, Hypoxia-inducible Factor (HIF), Phosphatase and Tensin Homolog (PTEN), Phosphatidylinositide 3-Kinase (PI 3-Kinase), Proteasome
Abstract
Hypoxia-inducible factor 1 (HIF1) is a heterodimeric transcription factor containing an inducibly expressed HIF1α subunit and a constitutively expressed HIF1β subunit. Under hypoxic conditions, the HIF1α subunit accumulates because of a decrease in the rate of proteolytic degradation, and the resulting HIF1α–HIF1β heterodimers undergo post-translational modifications that promote transactivation. Previous reports suggest that amplified signaling through PI3K enhances HIF1-dependent gene expression; however, its role is controversial, and the mechanism is unclear. Using genetically engineered PTEN-deficient cell lines, we demonstrate that PTEN specifically inhibited the accumulation of HIF1α in response to hypoxia. Furthermore, we report that in glioblastoma cell lines, inhibition of PI3K pathway, using pan as well as isoform-specific PI3K inhibitors SF1126, PF4691502, BEZ-235, GDC0941, and TGX221 blocked the induction of HIF1α protein and its targets vascular endothelial growth factor, HK1, and GLUT1 mRNA in response to hypoxia. Herein, we describe the first evidence that HIF1α can be degraded under hypoxic conditions via the 26 S proteasome and that MDM2 is the E3 ligase that induces the hypoxic degradation of HIF1α. Moreover, the action of MDM2 on HIF1α under hypoxia occurs in the cytoplasm and is controlled by the PTEN-PI3K-AKT signaling axis. These data strongly suggest a new role for PTEN in the regulation of HIF1α and importantly that PI3K-AKT activation is required for the hypoxic stabilization of HIF1α and that hypoxia alone is not sufficient to render HIF1α resistant to proteasomal cleavage and degradation. Moreover, these findings suggest new therapeutic considerations for PI3K and/or AKT inhibitors for cancer therapeutics.
Introduction
The hypoxia-inducible factor-1 (HIF1)3 is the main regulator of cellular adaptation to oxygen deprivation (1, 2). Hypoxia-inducible factor-1 is a heterodimeric transcription factor composed of a constitutively expressed subunit (HIF1β or aryl hydrocarbon receptor nuclear translocator), and a regulatory subunit (HIF1α), which is continuously transcribed and translated, but protein levels are controlled by ubiquitination and proteasomal degradation (3, 4). HIF1α subunits are ubiquitinated in direct proportion to cellular O2 concentrations, providing a molecular rheostat whereby levels of hypoxia-responsive genes are finely and expeditiously regulated by O2. Under conditions of normoxia, 2-oxoglutarate-dependent prolyl hydroxylase domain-containing enzymes covalently modify the HIF1α molecule such that it will bind to the VHL protein (pVHL), an E3 ubiquitin ligase. Once hydroxylated on specific proline residues (Pro402 and Pro564), HIF1α binds tightly to pVHL, is polyubiquitinated and transported to the proteasome for rapid and complete degradation (5–7). HIF1α is also hydroxylated by FIH-1 (factor inhibiting HIF1) on asparagine residue 803 in its C-terminal transactivation domain. This prevents the HIF1α coactivator, p300/CBP, from binding and thus inhibits HIF1α-mediated transcription of target genes (8). Under hypoxic conditions, the rate of prolyl hydroxylase domain-mediated and asparaginyl hydroxylation is reduced, and pVHL will not bind, resulting in a rapid increase in HIF1α levels and function within the nucleus. Stabilized HIF1α translocates to the nucleus and dimerizes with a β subunit, and the heterodimer transactivates target genes, including vascular endothelial growth factor (VEGF) and GLUT1, and various glycolytic enzymes that help cells adapt to hypoxia (9). HIF-mediated gene expression is critical for both embryonic development (10–12) and tumor growth (13, 14).
HIF1α has been shown to be induced by numerous stimuli other than hypoxia, including insulin, insulin-like growth factors (IGFs), epidermal growth factor, heavy metals, and HER-2/neu activation (15–19). However, the involvement of PI3K signaling in enhancing HIF1α-mediated gene expression remains a subject of considerable debate. Several groups have implicated the role of PI3K signaling in the regulation of HIF1α (20–22). Introduction of wild type PTEN into human glioblastoma cells has been shown to inhibit the hypoxia-induced up-regulation of HIF1 target genes and expression of a luciferase reporter containing HIF1 binding sites in the promoter (22). However, the exact contribution of the PI3K pathway to HIF1α regulation remains a subject of considerable interest and controversy (23, 24).
The function of HIF1α has been shown to be modulated through direct and indirect interaction with a number of proteins that regulate passage through the cell cycle including p53, MDM2 (murine double minute 2), E2F, and pRb (25–29). p53 has been shown to decrease the transcriptional activity of HIF1α, in part through competition, in the setting of hypoxia, for binding to the transcriptional coactivators of both p53 and HIF1α, p300 and PCAF (26). Recently, the molecular interaction among HIF1α, p53, and MDM2 has been introduced as a key event in tumor promotion. HIF1α and p53 affect each other reciprocally; namely, HIF1α stabilizes p53 by blocking the MDM2-mediated p53 ubiquitination (25), but p53 destabilizes HIF1α by promoting the MDM2-mediated HIF1α ubiquitination (30). According to this hypothesis, MDM2 may function as a negative regulator of HIF1α. On the other hand, Bárdos et al. (31) demonstrated that MDM2 directly associates with HIF1α and positively regulates HIF1α expression and activation under conditions of normoxia and IGF1 stimulation. These observations raise the question of a more complex function and regulation of HIF1α in mammalian systems. Although much is known about the transcriptional and post-translational regulation of HIF1α (32, 33), there are few reports that HIF1α can be degraded under conditions of hypoxia by the 26 S proteasome (30, 34, 35). Ravi et al. (30) has suggested that p53 acts as a molecular chaperone that facilitates recruitment of HIF1α for ubiquitination by MDM2 under hypoxic conditions in a proteasome-dependent manner. Liu et al. (35) recently described oxygen-independent degradation of HIF1α by a novel HIF1α-interacting protein, RACK1 (receptor for activated protein kinase C 1). They suggested that similar to the E3 ligase, pVHL, RACK1 increased polyubiquitination of HIF1α and was unable to mediate degradation of HIF1α in the presence of proteasome inhibitor, MG132. In the same context, recent studies suggest that SHARP promotes proteasomal degradation of HIF1α independent of pVHL, hypoxia, and ubiquitination machinery (34). SHARP1 reported as a crucial regulator of metastatic phenotype in triple negative breast cancer (also known as BHLHE41 or DEC2) binds to HIFs and promotes HIF proteasomal degradation by serving as the HIF-presenting factor to the proteasome (34).
In our study, we demonstrate that PI3K inhibitors and PTEN promote degradation of HIF1α in glioma cells under hypoxic conditions. Furthermore, we have elucidated the molecular basis for how PTEN and PI3K inhibitors control the hypoxic degradation of HIF1α by the localization of MDM2 to the cytoplasm. Our data provide a new function for an old oncogene, MDM2, as an E3 ligase for HIF1α under conditions of hypoxia and limited PI3K-AKT activation.
EXPERIMENTAL PROCEDURES
Tissue Culture Conditions and Reagents
The LN229-HRE-AP cell line was provided by Dr. Erwin Van Meir. The LN229-HRE-AP reporter cell line for HIF transcriptional activity was created by stably transfecting LN229 cells with the pACN188 plasmid, which contains an alkaline phosphatase gene driven by six hypoxia response elements (HREs) derived from the VEGF promoter (36). PRK5-HA-MDM2 plasmid is a kind gift from Dr. David Schlaepfer. S166A and S166E FLAG-MDM2 plasmids were a gift from Dr. David Donner. p53−/− and p53−/− MDM2−/− MEFs were a kind gift from Dr. Inder Verma. Glioblastoma cell lines derived from U87MG containing inducible mutant PTEN, 23.24GE (lipid phosphatase inactive because of a G129E mutation) and 23.44GR (both lipid and protein phosphatase inactive because of a G129R mutation) were cultured as previously described (37, 38). In these cell lines, PTEN was induced using 0.5 μmol/liter muristerone. GFAP V12 Ras Tg mice develop high grade malignant gliomas in vivo (39). In this animal model described by Ding et al. (39), 100% of mice develop glial tumors. The tumors are evident from abnormal neurological function of the mice necessitating euthanasia or by routine histopathological analysis of neurologically normal mice. Glial tumors ranging from low grade to high grade gliomas were described with significant mouse strain effects noted. We bred these mice with PTENfl/fl animals obtained from Dr. Ramon Parsons (Columbia University, NY). We isolated normal astrocytes and tumor-derived glioma cells from PTEN-deficient mice. The GFAP V12 Ras Tg mice have been successfully bred with PTENfl/fl mice to generate a PTENfl/fl × GFAP V12 Ras glioma cell line. The GFAP V12 Ras PTENfl/fl glioma cells are then transduced with a retroviral vector, pMIEG-cre, which contains cre recombinase and internal ribosome entry site-directed enhanced GFP. This allowed us to delete one or both alleles of PTEN and develop isogenic PTEN−/−, PTEN+/−, or PTEN+/+ GFAP V12 Tg positive glioma cell lines. U87MG, LN229-HRE-AP, MEF PTEN+/+, MEF PTEN−/−, GFAP V12 Ras PTENfl/fl, and GFAP V12 Ras PTENfl/fl cre+ cell lines were propagated in Dulbecco's modified Eagle's medium (Cellgro), 10% FBS, 20 mm HEPES, 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin (all from Invitrogen). Cells were routinely cultured in 95% O2 and 5% CO2 at 37 °C and made hypoxic by placing them in hypoxic chamber (Billups chamber) at 1% O2, 5% CO2, and 94.9% nitrogen. Cells were in all cases serum-starved for 4 h and then pretreated with indicated drug and concentration for 30 min prior to IGF and/or hypoxia treatment. BEZ-235, BKM 120, PF4691502, GDC-0941, and TGX-221 were purchased from Selleck Chemicals. SF1126 was obtained from SignalRx Pharmaceuticals (40). Drugs were diluted into medium to the appropriate concentration before adding to cells to ensure that all treated dishes received the same concentration of DMSO that was added to control or untreated dishes at a final dilution of 1:1000.
Western Blots
For all Western blots, 2 × 106 cells were plated in 10-cm tissue culture dishes such that the density of the cells at the time of lysis was 70–80% confluent. Cells were allowed to adhere overnight and were then starved for 4 h in 0% FBS and pretreated for 30 min with the indicated drug or DMSO control. The cells were then stimulated with IGF and under hypoxia or normoxia for 4 h. Lysis was performed immediately at the bench in ambient air. For HIF1α Western blots, nuclear extracts were prepared using a modified Dignam protocol (41). Whole cell lysates were prepared using WCE buffer: 150 mm NaCl, 50 mm Tris, pH 7.4, 5 mm EDTA, 0.1% SDS, 20 mm β-glycerophosphate, 10 mm NaF, 250 μm NaVO4, 1 mm phenylmethylsulfonyl fluoride, and complete protease inhibitor (Roche Molecular Biochemicals). Extracts were electrophoresed, transferred, and immunoblotted according to standard protocols or the manufacturer's instructions using 5% nonfat dry milk in Tris-buffered saline/Tween 20. Αntibodies against human HIF1α were purchased from Transduction Laboratories (catalog number 610959). Antibodies against total and phosphorylated AKT (Ser473) (catalog number 9271) and PTEN (catalog number 9552) were purchased from Cell Signaling Technologies.
HIF1α-Alkaline Phosphatase Reporter Assay
LN229-HRE-AP cells (4 × 105/well) were plated onto 6-well plates, serum-starved, and incubated with or without inhibitors for 24 h under hypoxic or normoxic condition. Cells were washed with TBS, and p-nitrophenyl phosphate (Sigma) was added into each well. The plates were incubated at 37 °C for 30 min to detect and quantify AP enzymatic activity by measuring optical density values at 405 nm in a spectrophotometer.
Plasmid Constructs, Transient Transfections, and shRNA
Transient transfections were done using Lipofectamine Plus (Invitrogen) for MEFs and FuGENE (Roche) for LN229-HRE-AP cells according to the manufacturer's instructions. Briefly, cells were split into 10-cm dishes so that 24 h later they were 70% confluent. At this time, each dish was transfected 10 μg of plasmid using FuGENE or Lipofectamine Plus. After 48 h of transfection, lysates were prepared. The short hairpin RNA was purchased from Dharmacon Research (Lafayette, CO). shRNA-transfected LN229-HRE-AP cells with different concentrations of shRNA according to instruction manual were allowed to select on puromycin for multiple passages, until we detect a stable knockdown of PTEN in each cell line.
Site-directed Mutagenesis
The QuikChange site-directed mutagenesis kit (Stratagene) was used to mutate ring finger domain site cysteine 464 to alanine in pcDNA S166A MDM2 plasmid. Oligonucleotides used for mutagenesis were CAGGACATCTTATGGCCTGCTTTACAGCCGCAAAGAAGCTAAAGAAAAG and CTTTTCTTTAGCTTCTTTGCGGCTGTAAAGCAGGCCATAAGATGTCCTG (C464A). Dideoxynucleotide sequencing ensured successful mutagenesis of C464A in FLAG tag pcDNA S166A MDM2 plasmid.
Confocal Microscopy
p53−/− MDM2−/− MEFs were transfected with 1 μg of HA-MDM2 or S166A or S166E FLAG-MDM2. Forty-eight hours after transfection, cells transfected with HA-MDM2 or FLAG-MDM2 plasmid were serum-starved for 4 h, followed by treatment with 500 nm PF4691502 for half an hour. Cells were fixed, permeated with 0.1% Triton X-100, and blocked with 2% BSA. A FLAG or HA primary antibody was added, followed by Alexa 568-conjugated goat anti-mouse secondary antibody. The nucleus was detected by staining with DAPI. Excitation of the stains was performed on a Nikon STORM confocal super resolution system. Double-blind analyses of cells in numerous fields were done to study cytoplasmic or nuclear localization of MDM2 in the presence of constructs or treatment conditions.
RT-PCR Analyses
Total RNA was isolated from cell lines using the Qiagen RNAeasy kit (Qiagen) according to manufacturer's instructions. cDNA was prepared from 1 μg of RNA sample using an iScript cDNA synthesis kit (Bio-Rad). cDNA (2 μl) was amplified by RT-PCRs with 1× SYBR green supermix (Bio-Rad) in 96-well plates on an CFX96 real time system (Bio-Rad), using the following program: 5 min at 95 °C, then 40 cycles of 20 s at 95 °C, 1 min at 58 °C, and 30 s at 72 °C. The specificity of the produced amplification product was confirmed by examination of dissociation reaction plots. Relative expression levels were normalized to Gapdh expression according to the formula < 2̂(Ct gene of interest-Ct Gapdh) (42).
RESULTS
PTEN Regulates Expression of HIF1α Protein and Downstream HIF1α Transcriptional mRNA Targets
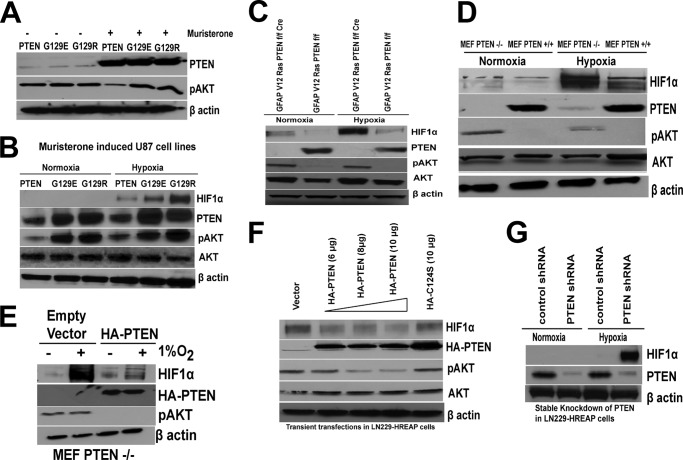
The tumor suppressor PTEN is an antagonist of PI3K signaling and functions by removing the phosphate at the D3 position of phosphatidylinositol trisphosphate and phosphatidylinositol bisphosphate. To determine the role of PTEN in the regulation of HIF1α, we used four different genetically engineered PTEN-deficient cell lines. Glioblastoma cell lines derived from U87MG containing inducible WT or mutant PTEN (G129E and G129R) were cultured as previously described (37). In these U87MG cell lines, expression of PTEN (or mutants of PTEN) is under the control of a muristerone responsive promoter (38). First, we verified the biochemical induction of PTEN and suppression of p-AKT in these cell lines when induced with muristerone (Fig. 1A). The U87 cell line containing muristerone-inducible WT PTEN, or mutant PTEN (G129E, G129R), MEF PTEN+/+, MEF PTEN−/−, GFAP V12 Ras PTENfl/fl, and GFAP V12 Ras PTENfl/fl cre+ cell lines were serum-starved for 4 h and then exposed to 4 h of hypoxia. The mutation or loss of PTEN induced a high level of HIF1α stabilization in U87 PTEN G129E, U87 PTEN G129R, GFAP V12 Ras PTENfl/fl cre+, and MEF PTEN−/− cell lines (Fig. 1, B–D) compared with isogenic PTEN-positive cells. We observed that U87 glioblastoma cell lines containing muristerone-inducible WT PTEN block HIF1α accumulation, whereas cell lines expressing muristerone-inducible PTEN mutants, G129E and G129R, were unable to block HIF1α accumulation (Fig. 1B). These results were also verified in MEFs where the hypoxic accumulation of HIF1α was higher in PTEN−/− MEFs (Fig. 1D), and this effect was abrogated when PTEN−/− MEFs were transfected with HA-PTEN plasmid (Fig. 1E). To further support the role of PTEN in HIF1α stabilization, we transfected WT PTEN or a lipid phosphatase dead mutant C124S PTEN into LN229-HRE-AP cells. The transient transfection of PTEN blocked HIF1α stabilization in LN229-HRE-AP cells, whereas there is no effect on HIF1α stability when these cells were transfected with the catalytic dead PTEN C124S mutant (Fig. 1F). These results were further confirmed by increased HIF1α stabilization when PTEN was knocked down by stable transfection of PTEN shRNA in LN229-HRE-AP cells (Fig. 1G).
FIGURE 1.
PTEN regulates expression of HIF1α protein. A, muristerone-inducible PTEN or G129E and G129R mutants of PTEN engineered in the U87MG cell lines were used to detect the expression of PTEN and pAKT before and after induction of 0.5 μmol/liter muristerone. After 36 h of muristerone induction, cell lysates were prepared and used for Western blot analysis of PTEN and pAKT. B–D, muristerone induced expression of PTEN in the U87 cell line including wild type PTEN orG129E and G129R PTEN mutant expression. B–D, GFAP V12 Ras PTENfl/fl (B) or GFAP V12 Ras PTENfl/fl cre cell lines (C) and MEF PTEN+/+ and MEF PTEN−/− cell lines (D) were serum-starved for 4 h and were kept in normoxic (21% O2) or hypoxic (1% O2) for 4 h followed by preparation of nuclear extracts (for HIF1α) as well as WCE and Western blot analysis. E, MEF PTEN−/− cells were transfected with 10 μg of empty vector or HA-PTEN plasmid using Lipofectamine Plus. 48 h after transfection, cells were serum-starved for 4 h followed by hypoxia (1% O2) for 4 h. Cells were used for preparation of nuclear extracts (for HIF1α) and whole cell extracts for HA and pAKT for Western blot. Cell lysates were run on SDS-PAGE, and proteins were transferred on nitrocellulose membrane and probed with HIF1α, HA or pAKT (Ser473) antibodies. F, LN229-HRE-AP cell lines were transfected with different concentrations of HA-PTEN (6, 8, and 10 μg), C124S (10 μg) mutant, and control plasmid (10 μg) using FuGENE transfection reagent. After 48 h of transfection, cells were serum-starved for 4 h and exposed to hypoxia (1% O2) for 4 h followed by preparation of nuclear extracts and whole cell extracts for Western blot analysis. G, LN229-HRE-AP cell lines stably transfected with PTEN shRNA or a control construct as described under “Experimental Procedures” were serum-starved for 4 h followed by hypoxia (1% O2) for 4 h and preparation of nuclear extracts and Western blot analysis. The experiments were repeated three times with similar results.
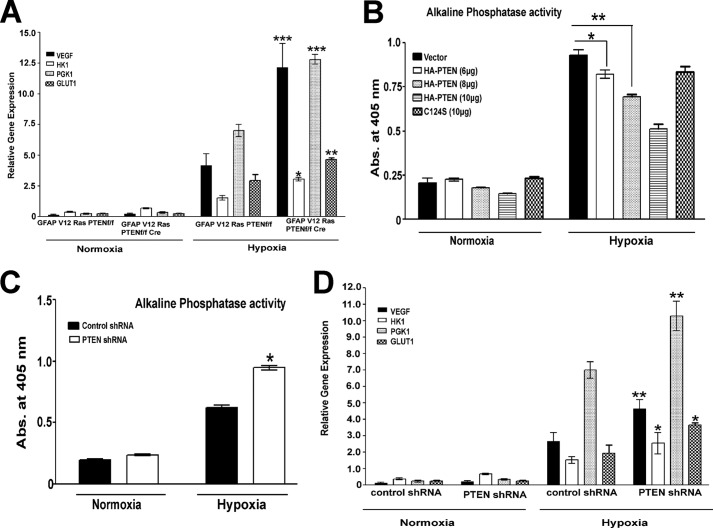
Our next point of focus was to investigate the role of PTEN in the transcriptional activation of HIF1α target genes. GFAP V12 PTENfl/flcre+ cell lines display a significant increase in the transcription of VEGF, GLUT1, and HK1 (Fig. 2A) as compared with the isogenic GFAP V12 PTENfl/fl (PTEN-positive) cell line. Moreover, HIF1α transcriptional activity was reduced in the LN229-HRE-AP cell lines transiently transfected with PTEN (Fig. 2B). This effect was dose-dependent because it was correlated with levels of PTEN, p-AKT suppression, and HIF1α protein expression (Fig. 1F). Moreover, the transcription of HIF1α target genes were increased by knockdown of PTEN in LN229-HRE-AP cells as evidenced by increased HRE-dependent alkaline phosphatase activity and real time PCR detection of VEGF, HK1, PGK1, and GLUT1 mRNA (Fig. 2, C and D).
FIGURE 2.
PTEN regulates HIF1α-dependent target gene expression. A, mRNA was isolated from GFAP V12 RasPTEN f/fl or GFAP V12 Ras PTENfl/fl cre cell lines and the expression of VEGF, HK1, PGK-1, and GLUT1 probes were analyzed by real time PCR. B, different concentrations of HA-PTEN plasmid vector (6, 8, and 10 μg) DNA or its mutant C124S (10 μg) were transfected into serum-starved LN229-HRE-AP cells. After 48 h of transfection, cells were exposed to normoxia (21% O2) or hypoxia (1% O2) for 24 h, followed by preparation of lysates, and HRE-mediated AP activity was read at 405 nm using p-nitrophenyl phosphate as a substrate. Cells transfected with pcDNA were used as a control. C, LN229-HRE-AP cell lines stably transfected with PTEN shRNA or a control construct were subjected to normoxia (21% O2) or hypoxia (1% O2) for 24 h, and HRE-mediated AP activity was read at 405 nm. D, RNA was isolated form LN229-HRE-AP cell lines stably transfected with PTEN shRNA and its control constructs in normoxic (21% O2) and hypoxic (1% O2) conditions and used to quantitate expression of VEGF, HK1, PGK-1, and GLUT1 genes by real time PCR. The data are representative of two or three independent experiments. Graphs present means ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus GFAP V12 Ras PTENfl/fl (A), vector (B), and control shRNA (C and D) in hypoxic conditions as determined using pair wise two-sided Student's t test. Please note that the expression values of all genes (A and D) and alkaline phosphatase activity (B and C) in hypoxic conditions are significant compared with normoxic conditions.
Inhibition of PI3K Pathway Blocks Hypoxic Accumulation of HIF1α and Its Targets but Not HIFα mRNA
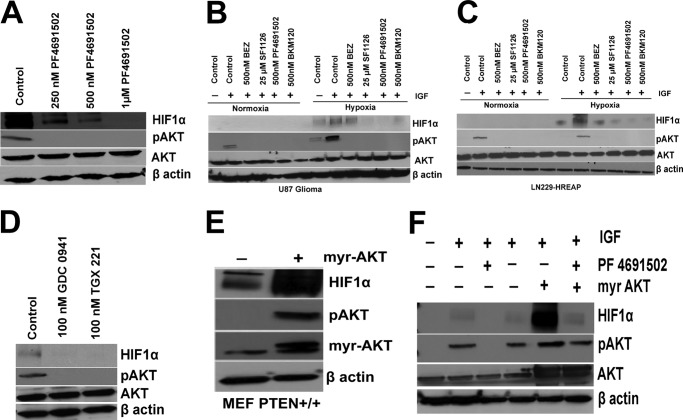
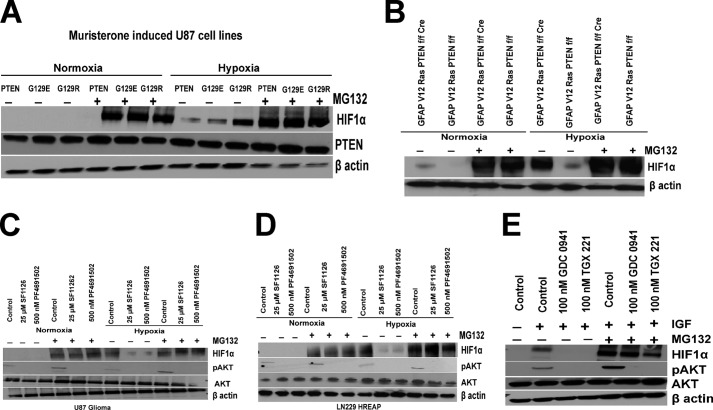
The observation that PTEN exerts control over HIF1α protein levels and its transcriptional activity (Figs. 1 and 2) led us to investigate the effects of different clinically relevant PI3K inhibitors (SF1126, BEZ235, PF4691502, BKM120, GDC0941, and TGX221) on the hypoxic induction of HIF1α in glioma models. SF1126 is a vascular RGDS/integrin targeted prodrug derivative of LY294002 that it has recently completed phase I clinical trials (40). SF1126 inhibits all isoforms of PI3K (pan-PI3K inhibitor), mTOR, ATM, PIM1, and PLK2. GDC-0941 is an orally bioavailable inhibitor of class I PI3K that is 100-fold more potent against class I compared with class II, III, and IV family members and is in clinical development for solid tumor indications including breast cancer (43). TGX-221 is a p110β-specific inhibitor, which is emerged as a new target for anti-thrombotic therapy (44). The other inhibitors are in early clinical development, including: BEZ235, a dual PI3K/mTOR inhibitor; PF4691502, a panPI3K inhibitor; and BKM120, a pan-PI3K inhibitor (45–47). We first determined whether these PI3K inhibitors are able to block accumulation of HIF1α under hypoxic conditions. Fig. 3A shows that PF4691502 blocks the hypoxic accumulation of HIF1α in a dose-dependent manner and blocks p-AKT in LN229-HRE-AP cells. The results shown in Fig. 3 (B and C) clearly demonstrate that the pan-PI3K inhibitors SF1126, BEZ, PF4691502, and BKM120 potently inhibit the hypoxic accumulation of HIF1α in LN229-HRE-AP and U87 glioma cell lines. SF1126, PF4691502, and BKM120 were equally effective in blocking HIF1α accumulation, whereas BEZ235 was less potent (Fig. 3, B and C). To determine the specificity of the PI3K signaling in HIF1α accumulation, we used isoform-specific PI3K inhibitors. Interestingly, both GDC0941 (p110α-specific inhibitor) and TGX-221 (p110β-specific inhibitor) block HIF1α accumulation in LN229-HRE-AP cells (Fig. 3D). To provide additional evidence that PTEN and PI3K control hypoxic HIF1α, we transfected Myr-AKT into MEF PTEN+/+ and LN229-HRE-AP cells to determine whether expression of constitutive active AKT would augment HIF1α accumulation under hypoxic conditions. We observed that the expression of Myr-AKT augmented the accumulation of HIF under hypoxia in MEF PTEN+/+ (Fig. 3E), as well as in LN229-HRE-AP cells (Fig. 3F). Of note, the PI3K inhibitor treatment blocked the accumulation of HIF1α induced by Myr-AKT transfection in LN229-HRE-AP cells (Fig. 3F). This observation is consistent with the literature, which has reported that Myr-AKT, although membrane-localized, requires phosphatidylinositol 1,4,5-trisphosphate-dependent phosphorylation on Thr308 by PDK1 for full activation (48).
FIGURE 3.
Inhibition of PI3K pathway blocks hypoxic accumulation of HIF1α. A, serum-starved LN229-HRE-AP cells (4 h) were treated with 250 nm, 500 nm, and 1 μm PF4691502 for 30 min followed by IGF stimulation and then placed under normoxic or hypoxic conditions (1% O2) for 4 h followed by the preparation of nuclear extracts for Western blot analysis of HIF1α and WCE for pAKT and AKT blots. B and C, serum-starved U87 and LN229-HRE-AP cells treated with 25 μm SF1126, 500 nm PF4691502, BEZ, and BKM120 inhibit HIF1α accumulation under hypoxia in U87 (B), LN229-HRE-AP (C) cells. Cells were serum-starved for 4 h pretreated with inhibitors for 30 min followed by IGF stimulation and then placed under normoxic or hypoxic conditions (1% O2) for 4 h followed by the preparation of nuclear and whole cell extracts for Western blot analysis. D, serum-starved LN229-HRE-AP cells were pretreated with 100 nm GDC0941 and TGX221 for 30 min followed by IGF stimulation and then placed under normoxic or hypoxic conditions (1% O2) for 4 h followed by the preparation of nuclear extracts for Western blot analysis of HIF1α and WCE for pAKT and AKT blots. E, MEF PTEN+/+ cells were transfected with 10 μg of Myr-AKT or empty vector using Lipofectamine Plus transfection reagent, and 48 h after transfection, cells were serum-starved for 4 h and subjected to hypoxia for 4 h followed by preparation of nuclear extracts and whole cell extracts. F, LN229-HRE-AP cells were transfected with 10 μg of Myr-AKT or empty vector using FuGENE transfection reagent and 48 h after transfection. The cells were serum-starved for 4 h pretreated with 500 nm of PF4691502 for 30 min followed by IGF stimulation and then placed under hypoxic conditions (1% O2) for 4 h followed by the preparation of nuclear extracts and WCE for Western blot analysis. The data are representative of two or three independent experiments.
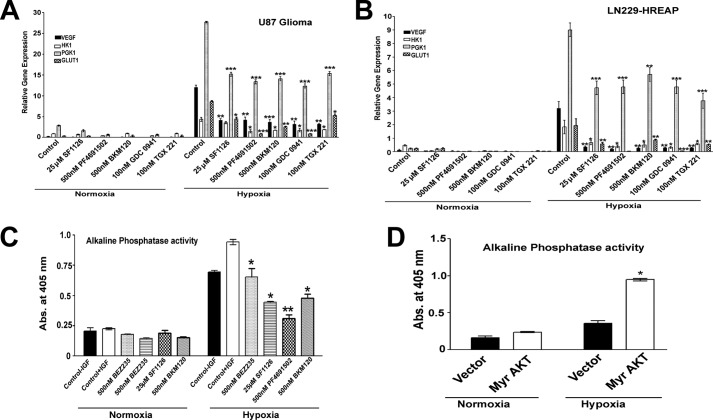
We next investigated whether PI3K inhibitors can block the transcriptional activation of HIF1α target genes. Fig. 4 (A and B) shows that PI3K inhibitors reduced VEGF, HK1, and GLUT1 mRNA levels in both PTEN containing and PTEN-deficient glioma cell lines. In agreement with these findings, PI3K inhibitors blocked the HIF1α transcriptional activity as measured by an alkaline phosphatase HRE reporter activity in the LN229-HRE-AP cell line (Fig. 4C). Interestingly, we found no change in the transcription of HIF1α mRNA under hypoxic conditions, as well as upon treatment with panPI3K inhibitor SF1126 in U87 glioma (data not shown) We next explored whether expression of AKT can increase the transcriptional activation of HIF1α target genes. Interestingly, expression of constitutively active Myr-AKT into LN229-HRE-AP cells increased HRE alkaline phosphatase reporter activity (Fig. 4D). Taken together, these results suggest that the PI3K-AKT signaling axis is necessary and sufficient for the hypoxic accumulation of HIF1α in glioma cells and MEFs (Figs. 3 and 4).
FIGURE 4.
PI3K/AKT pathway regulates HIF1α-induced transcription of target genes. A and B, serum-starved U87 and LN229-HRE-AP cells treated with 25 μm SF1126; 500 nm PF4691502, BEZ, and BKM120; and 100 nm GDC0941 and TGX221 were exposed to hypoxia for 24 h, followed by RNA isolation and real time PCR for VEGF, GLUT1, and PGK in U87 (A) and LN229-HRE-AP (B) cell lines. C, serum-starved LN229-HRE-AP cells treated with 25 μm SF1126 and 500 nm PF4691502, BEZ, and BKM120 for 30 min followed by IGF stimulation and then placed under normoxic (21% O2) or hypoxic conditions (1% O2) for 24 h. HRE-mediated AP activity was read at 405 nm using p-nitrophenyl phosphate as a substrate in the presence of different pan-PI3K inhibitors. D, LN229-HRE-AP cell lines transfected with 10 μg of Myr AKT or control plasmid were exposed to hypoxia (1% O2) for 24 h followed by alkaline phosphatase activity read at 405 nm as described before. Graphs present means ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control (A–D) in hypoxic conditions as determined using pair wise two-sided Student's t test. Please note that the expression values of all genes (A–D) in hypoxic conditions are significant, compared with normoxic conditions. Abs, absorbance.
PTEN and PI3K Inhibitors Induce the Hypoxic Degradation of HIF1α via the 26 S Proteasome Pathway
The above results (Figs. 1–4) clearly establish a key role for the PTEN-PI3K-AKT signaling pathway in hypoxia-induced HIF1α accumulation in glioma cells. To explore the mechanism by which PTEN-PI3K pathway affects HIF1α levels in glioma cells, we utilized the proteasome inhibitor MG132 to ask whether this process was related to proteasome-dependent degradation. Cells were serum-starved and treated with the proteasome inhibitor, MG132, for 5 min before being pulsed for 30 min with the pan-PI3K inhibitors, followed by IGF stimulation. Our data support the hypothesis that PTEN and pan-PI3K inhibitors affect HIF1α under hypoxic conditions by inducing the hypoxic degradation of HIF1α, because this effect can be reversed by pretreatment with the proteasome inhibitor, MG132 (Fig. 5). In Fig. 5A, we used a muristirone-inducible PTEN expression system to demonstrate that PTEN induced a marked reduction in HIF1α accumulation under hypoxic conditions. The effect of PTEN on hypoxic accumulation of HIF1α is completely reversed by MG132 treatment. Moreover, in our isogenic GFAP V12 Ras PTENfl/fl cre+ glioma cells, PTEN expression was associated with reduced hypoxia-induced accumulation of HIF1α, and this effect is completely reversed by treating these cells with MG132 (Fig. 5B). To further confirm that the disappearance of HIF1α is a proteasome-dependent event and that PI3K activation is required for the hypoxic stability of HIF1α, we treated glioma cell lines with a panel of PI3K inhibitors (Fig. 5, C–E). The rationale for selecting PI3K inhibitors SF1126 and PF4691502 was our observation that these inhibitors potently blocked HIF1α accumulation under hypoxia in U87 and LN229-HRE-AP glioma cells and blocked HIF1α target gene expression, e.g. VEGF, HK1, PGK1, and GLUT1 (Figs. 3, B and C, and 4, A and B). Moreover, the decreases in HIF1α accumulation observed with isoform-specific PI3K inhibitors GDC-0941 and TGX-221 were also reversed with treatment with MG132 (Fig. 5E). From these results, we conclude that both PTEN and PI3K control a heretofore unappreciated and potentially important physiologic process, the proteasome-dependent degradation of HIF1α under conditions of hypoxia.
FIGURE 5.
Hypoxic degradation of HIF1α by PTEN-PI3K pathway occurs via the 26 S proteosome. A and B, serum-starved (4 h) muristerone-induced U87 PTEN, G129E, or G129R cell lines (A) or GFAP V12 Ras PTENfl/fl and GFAP V12 Ras PTENfl/fl cre+ (B) cell lines were treated with the proteosome inhibitor 10 μm MG132 for 30 min before exposure to hypoxia (1% O2) for 4 h, followed by nuclear extract and WCE preparation. C and D, serum-starved U87 and LN229-HRE-AP cell lines were treated with the proteosome inhibitor 10 μm MG132 for 5 min before being pulsed for 30 min with, the pan-PI3K inhibitors, 25 μm SF1126, and 500 nm PF4691502, followed by IGF stimulation and hypoxia for 4 h. Nuclear extracts were prepared for HIF1α Western blots as described before. E, serum-starved LN229-HRE-AP cell lines were treated with 10 μm MG132 (as indicated) for 5 min before being pulsed for 30 min with 100 nm GDC0941 and TGX221, followed by IGF stimulation and hypoxia for 4 h.
PTEN and PI3K Regulate the MDM2 Induced Hypoxic Degradation of HIF1α
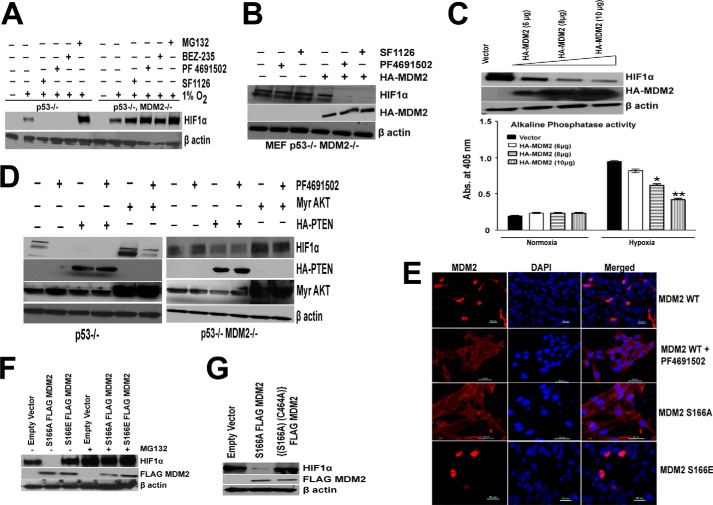
The observation that HIF1α can be degraded under hypoxic conditions in a PTEN- and PI3K-dependent manner suggested the involvement of a potential E3 ligase and proteasome-dependent degradation. We hypothesized that MDM2, an E3 ligase whose transport between the cytoplasm and nucleus is regulated by PI3K and AKT might be the E3 ligase controlling HIF1α degradation. We first biochemically confirmed the pattern of p53 and MDM2 expression in the p53−/− and MEF p53−/− MDM2−/− MEF cells (data not shown). To test our hypothesis, we compared HIF1α stability in p53−/− MDM2+/+ and p53−/− MDM2−/− cultured MEFs under conditions of PI3K inhibitor treatment. As expected, under normoxia, p53−/− MDM2−/− and p53−/− MDM2+/+ MEFs express no detectable HIF1α protein. When exposed to hypoxia for 4 h, p53−/− MDM2−/− MEFs express slightly higher basal levels of HIF1α protein than do p53−/− MDM2+/+ MEFs (Fig. 6A). Treatment of MEFs with the pan-PI3K inhibitors SF1126, PF4691502, and BEZ235 under hypoxic conditions resulted in abrogation of HIF1α protein levels in p53−/− MEFs, whereas the same treatment of p53−/− MDM2−/− MEFs was completely ineffective in the depletion of HIF1α under hypoxic conditions. From these results, we conclude that MDM2 is mediating the hypoxic degradation of HIF1α in the absence of signaling through PI3K. Our results support the hypothesis that inhibition of PI3K affects HIF1α under hypoxic conditions by inducing the hypoxic degradation of HIF1α and that this effect is dependent on the E3 ligase, MDM2. We further verified these results by reconstituting MDM2 in the p53−/− MDM2−/− cell line. Interestingly, reconstitution of MDM2 in this cell line blocked the hypoxic accumulation of HIF1α, and this effect is more pronounced in cells treated with PI3K inhibitor (Fig. 6B). To further confirm this observation, we transiently transfected PRK5-HA-MDM2 construct in LN229-HRE-AP cells. In Fig. 6C, we demonstrate a dose-dependent reduction of HIF1α stabilization and HIF1α HRE transcription reporter activity in LN229-HRE-AP cells. We next turned our attention to the molecular mechanism that underpins a role for PI3K/AKT signaling in the regulation of HIF1α stabilization in the presence or absence of MDM2; we overexpressed PTEN and AKT in both p53−/− and p53−/− MDM2−/− cell lines and treated them with PI3K inhibitor PF4691502. Fig. 6D shows that overexpression of PTEN drastically blocked HIF1α accumulation in p53−/−, and this effect is more pronounced when treated with PF4691502, whereas there was a modest decrease in HIF1α accumulation in p53−/− MDM2−/− cell lines upon expression of PTEN, and there is no drop in HIF1α protein levels when treated with PF4691502. Moreover, expression of Myr-AKT increased HIF1α accumulation in p53−/−, and this effect was inhibited upon treatment with PF4691502, whereas in p53−/−; MDM2−/− cells, PF4691502 was unable to block the increased HIF1α accumulation observed with Myr-AKT transfections (Fig. 6D). These data suggest that MDM2 is mediating the hypoxic degradation of HIF1α in the absence of signaling through PI3K. These results led us to investigate further the mechanism involved in the MDM2 regulated degradation of HIF1α. It is previously reported that there are two AKT phosphorylation sites (serine 166 and 186) in RXRXX(S/T) motifs within the human MDM2 protein (49) that mediate its translocation to the nucleus. This observation led to the hypothesis that MDM2 might serve as an E3 ligase for the degradation of HIF1α under hypoxic conditions.
FIGURE 6.
MDM2 regulates PI3K induced hypoxic degradation of HIF1α. A, serum-starved p53−/− or p53−/− MDM2−/− MEFs (4 h) were treated with the proteosome inhibitor 10 μm MG132 for 5 min before being pulsed for 30 min with, the pan-PI3K inhibitors, 25 μm SF1126, and 500 nm PF4691502 and BEZ, followed by IGF stimulation and hypoxia for 4 h. Whole cell extracts were prepared for HIF1α blots. The results support a requirement for MDM2 for PI3K inhibitors effects on hypoxic HIF1α. In the absence of MDM2, inhibitor has no effect on HIF1α under hypoxic conditions. B, MEF p53−/− MDM2−/− cell lines were transfected with 10 μg of HA-MDM2 plasmid using Lipofectamine Plus transfection reagent. 48 h after transfection, cells were serum-starved, followed by treatment with, the pan-PI3K inhibitors, 25 μm SF1126, and 500 nm PF4691502 followed by IGF stimulation and hypoxia for 4 h and whole cell lysate preparation and Western blot analysis. C, LN229-HRE-AP cell lines transfected with PRK5-HA-MDM2 plasmid (6, 8, and 10 μg) or a control plasmid (10 μg) were serum-starved for 4 h and exposed to hypoxia for 4 h, followed by preparation of whole cell extracts (upper panel). LN229-HRE-AP cell lines transfected with PRK5-HA MDM2 plasmid (6, 8, and 10 μg) and control plasmid (10 μg) were exposed to normoxia or hypoxia for 24 h followed by lysate preparation and alkaline phosphatase activity as described before (lower panel). D, p53−/− (left panel) and p53−/− MDM2−/− (right panel) MEFs were transfected with 10 μg of HA-PTEN or Myr-AKT plasmids using Lipofectamine Plus transfection reagent. 48 h after transfection, cells were serum-starved for 4 h before being pulsed for 30 min with 500 nm of PF 4691502 followed by IGF stimulation and hypoxia for 4 h. Whole cell extracts were prepared for HIF1α, HA, and AKT blots. E, localization of MDM2 characterized by confocal microscopy. p53−/− MDM2−/− MEFs were transfected with 1 μg of FLAG-S166A/S166E-MDM2 or HA-WT MDM2 plasmid. Cells were serum-starved for 4 h after 48 h of transfection followed by treatment with 500 nm of PF4691502 for 30 min and were then processed for confocal microscopy as described under “Experimental Procedures.” F, p53−/− MDM2−/− cells were transfected with 10 μg of S166A or S166E MDM2 plasmids. 48 h after transfection, cells were serum-starved, followed by IGF treatment and pulse with 10 μm MG132 followed by hypoxia for 4 h and whole cell extract preparation. These lysates were run on SDS-PAGE, transferred on nitrocellulose membrane, and probed for HIF1α and FLAG tag. G, p53−/− MDM2−/− cells were transfected with 10 μg of S166A or C464A,S166A double mutant MDM2 plasmids. 48 h after transfection cells were serum-starved, followed by IGF treatment and hypoxia for 4 h and whole cell extract preparation. These lysates were run on SDS-PAGE, and proteins were transferred to a nitrocellulose membrane and probed for HIF1α and FLAG tag antibody.
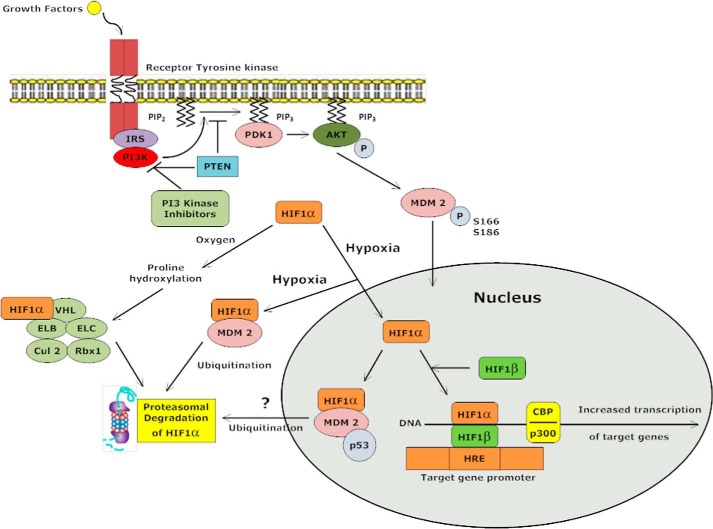
To test this hypothesis, wild type or phosphorylation site mutants of MDM2 that regulate subcellular localization of this protein were employed. S166A-MDM2 plasmid with a serine to alanine substitution cannot be phosphorylated by AKT; thus, it remains in the cytoplasm (49), whereas the S166E-MDM2 construct where serine 166 is mutated to glutamic acid mimics a phosphorylated MDM2 and is predominantly localized in the nucleus (49, 50). In this manner, we were able to test the effect of cytoplasmic versus a nuclear MDM2 on the hypoxic stability of HIF1α under conditions of PI3K and/or AKT inhibition. In these experiments, p53−/− MDM2−/− MEF cells were transduced with wild type HA-MDM2 plasmid and serum-starved for 4 h followed by treatment with 500 nm PF4691502 and hypoxia for 4 h. MDM2 under these conditions was localized to the cytoplasmic compartment as determined by confocal immunofluorescence microscopy (Fig. 6E). Under these conditions, HIF1α is degraded in a PI3K-dependent manner. In contrast, the S166E mutant of MDM2, which is obligately localized to nuclear compartment (Fig. 6E), does not induce the hypoxic degradation of HIF (Fig. 6F). Finally, the transfection of an obligate cytoplasmic MDM2 (S166A) mutant (Fig. 6E) results in the degradation of HIF1α in a PI3K-independent manner (Fig. 6F), confirming a model predicting that MDM2 in the cytoplasm mediates the degradation of HIF under hypoxia. Interestingly, we note that blocking the proteasome with MG132 reverses the effect caused by S166A MDM2 plasmid (Fig. 6F). Finally, we performed experiments to determine whether the E3 Ub-ligase activity of MDM2 is required for the degradation of HIF1α observed under conditions of hypoxia and PI3K inhibition. The Ub-protein ligase function of MDM2 is dependent on RING finger domain (residues 434–490) at the C terminus (51). It is reported that MDM2 mutant with a substitution of a cysteine residue at position 464 to alanine (C464A) is deficient in E3 Ub-protein ligase function (52). To validate our hypothesis, we generated an MDM2 double mutant that has mutations at the S166A and C464A sites. In contrast to S166A MDM2, the introduction of the S166A,C464A mutant into p53−/− MDM2−/− cells did not induce the degradation of HIF1α stability under hypoxic conditions (Fig. 6G). Based on these results, we propose a hypothetical model in which MDM2 degrades HIF1α under hypoxic conditions in the cytoplasm in a E3 ligase- and proteasome-dependent manner. The model would predict that the stimulation of PI3K and AKT via upstream signals will serve to promote MDM2 translocation into the nucleus and promote the stabilization of hypoxic HIF1α. The pathway will bring hypoxic HIF1α under the control of the PTEN-PI3K/AKT signaling axis to regulate an E3 ligase MDM2 to control hypoxic HIF1α levels, p53, and angiogenesis in vivo (Fig. 7). Consistent with this notion, we and other laboratories have reported the potent antiangiogenic activity of PTEN and PI3K inhibitors in vivo (53, 54). It is possible that potent antiangiogenic properties of PI3K inhibitors are to some degree dependent on this mechanism to suppress hypoxic HIF function.
FIGURE 7.
Mechanism by which the PTEN-PI3K-AKT-MDM2 signaling axis exerts control over HIF1α degradation in cytoplasm under hypoxic conditions via the MDM2 E3 ligase and 26 S proteasome. PI3K inhibitors block Ser166 phosphorylation of MDM2 resulting in cytoplasmic localization of MDM2; this new signaling pathway is therefore predicted to have therapeutic implications for the control of the HIF1α-VEGF axis in PI3K inhibitor cancer therapeutics.
DISCUSSION
Despite a number of reports that the PI3K/AKT pathway is involved in the regulation of HIF1α expression (20–22), the importance of the PI3K/AKT pathway in the hypoxic induction and/or stabilization of HIF1α remains controversial (23, 24). Hence, we designed experiments to investigate this issue in greater detail and made two important observations: 1) the PTEN-PI3K signaling axis regulates the hypoxic stability of HIF1α, and 2) we have identified the molecular basis for this effect of PI3K inhibitors and PTEN on HIF1α stability under hypoxic conditions and provide a new function for the E3 ligase, MDM2.
We have previously reported that PTEN regulates tumor-induced angiogenesis and the progression of gliomas to a malignant phenotype via the regulation of phosphoinositide-dependent signals (38). In the current study, using genetically engineered cell lines and different commercially available PI3K inhibitors, we demonstrate an important role of PTEN-PI3K signaling pathway in HIF1α stabilization and HIF1α-mediated transcription of VEGF, HK1, and GLUT1 (Figs. 1–4). Alvarez et al. (23) and Ashram et al. (24) investigated the role of PI3K in the regulation of HIF1α and concluded that PI3K/AKT signaling is neither required for hypoxic stabilization of HIF1α nor for HIF1α-dependent gene transcription. Our results clearly demonstrate that the loss of PTEN and activation of PI3K and AKT are required for HIF1α accumulation in glioma cells.
On the same note, studies conducted by others have also suggested a role for PTEN and PI3K/AKT signaling in facilitating HIF1α-mediated gene expression or protein expression via mTOR (20–22). In our study, we address the molecular mechanism for how PI3K inhibitors and PTEN regulate proteasomal-dependent degradation of HIF1α. Recent studies have suggested that MDM2 binds to HIF1α under hypoxic conditions, and some investigators have suggested a regulatory role for MDM2 in HIF1α function (27, 30). Our results demonstrate that HIF1α stability is increased in p53−/− MDM2−/− MEFs and that PI3K inhibitors do not induce the hypoxic degradation of HIF1α in p53−/− MDM2−/− MEFs (Fig. 6A). In contrast, loss of p53 does not affect the hypoxic degradation of HIF1α caused by PI3K inhibition. These data suggest that MDM2 is capable of degrading HIF1α under conditions of hypoxia, under conditions where PI3K is inactivated or phosphatidylinositol 1,4,5-trisphosphate is regulated (PTEN). The observation that MDM2 can move from the cytoplasm into the nucleus in a PI3K-dependent manner (49) and that this cytoplasmic to nuclear shuttling is dependent on two AKT phosphorylation sites in MDM2 (Ser166 and Ser186) (49) led our group to hypothesize a role for MDM2 and MDM2 phosphorylation in the regulation of hypoxic HIF1α stability. Evidence to support this idea came from experiments examining the hypoxic stability of HIF under conditions of PI3K inhibition in p53−/− versus p53−/− MDM2−/− MEF cells. In the absence of MDM2 protein, HIF1α was not degraded under conditions where the PI3K signaling pathway was inhibited. To confirm this observation, we overexpressed PTEN or a constitutive active mutant of AKT in p53−/− or p53−/− MDM2−/− MEFs. These data demonstrate that MDM2 mediates hypoxic degradation of HIF1α in the absence of signaling through PI3K (Fig. 6, A and D). An observation that revealed a potential mechanism behind the hypoxic degradation of HIF1α emerged when we observed that the transfection of S166A MDM2 into p53−/− MDM2−/− cells resulted in the total degradation of HIF1α under hypoxia (Fig. 6F). This is an important finding suggesting that S166A MDM2, whose entry to nucleus is restricted, degrades HIF1α in cytoplasm. On the other hand, expression of S166E plasmid, which mimics phosphorylated MDM2 and localizes to the nucleus, is unable to degrade HIF1α (Fig. 6, E and F). The supporting evidence that MDM2 acts as an ubiquitin ligase for the degradation of MDM2 comes from the observation that the double mutant (S166A,C464A) MDM2 cytoplasmic localized, E3 ligase dead mutant is unable to degrade HIF1α under hypoxic conditions (Fig. 6G). Recent reports suggest that p53 serves as a potential bridge that links HIF1α and MDM2 into a ternary complex in nucleus (27, 30). Ravi et al. (30) reported that p53 promotes MDM2 mediated ubiquitination and proteasomal degradation of HIF1α in full serum and hypoxic conditions. The effects were modest (2-fold) in comparison with our results in Fig. 6 (F and G), in which 100% of HIF1α was lost/degraded upon the transfection of the cytoplasmic S166A MDM2 mutant. Other literature supports the association between nuclear MDM2 and HIF1α and between HIF1α and p53 (30), in which HIF1α accumulation under hypoxia is augmented by the loss of p53 leading to augmented tumor-induced angiogenesis. Our results are consistent with those of Ravi et al. (30) in that the MDM2 S166E mutant did not induce HIF1α degradation in p53 −/− MEFs. The localization of proteasome is well established in nucleus as well as in cytoplasm (55), but the factors involved in the inability of MDM2 S166E to degrade HIF1α in nucleus are not clear and will be a point of focus of future studies in our laboratory. For now, our results suggest a VHL-independent novel mechanism of HIF1α regulation where MDM2 induce degradation of hypoxic HIF1α in cytoplasm in a PTEN/PI3K-dependent manner (Fig. 7). Our results also suggest that AKT-dependent phosphorylation of MDM2 on Ser166 or is necessary and/or sufficient to regulate MDM2 capacity to induce the hypoxic degradation of HIF1α in vivo. In summary, we conclude that PTEN and PI3K inhibitors inhibit the hypoxic stabilization of HIF1α in a proteasome- and MDM2-dependent manner. The results reported here suggest that the PI3K-AKT signaling axis is required to maintain HIF1α stability under hypoxia. We suggest that the inhibition of this pathway could be important in the context of hypoxic tumor progression in that it supports the existence of another element of regulation of HIF1α other than the oxygen-dependent prolyl hydroxylation reaction as required for the proteasomal degradation of HIF1α in vivo. These findings provide new insight into the mechanisms controlling HIF1α degradation under hypoxic conditions and suggest new therapeutic considerations for PI3K and/or AKT for cancer therapeutics.
Acknowledgment
We thank Dr. David Schlaepfer for providing the PRK5-HA-MDM2 construct.
This work was supported, in whole or in part, by National Institutes of Health Grants HL091385 and CA94233 (to D. L. D.). This work was also supported by the Alex Lemonade Stand Foundation, a Hyundai Hope Grant, and the Cricket corporation (to D. L. D.).
- HIF
- hypoxia-inducible factor
- VEGF
- vascular endothelial growth factor
- HRE
- hypoxia response element
- IGF
- insulin-like growth factor
- AP
- alkaline phosphatase
- MEF
- mouse embryonic fibroblast
- WCE
- whole cell extract.
REFERENCES
- 1. Semenza G. L. (2003) Targeting HIF-1 for cancer therapy. Nat Rev. Cancer 3, 721–732 [DOI] [PubMed] [Google Scholar]
- 2. Semenza G. L. (2004) Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda) 19, 176–182 [DOI] [PubMed] [Google Scholar]
- 3. Jiang B. H., Rue E., Wang G. L., Roe R., Semenza G. L. (1996) Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 271, 17771–17778 [DOI] [PubMed] [Google Scholar]
- 4. Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang L. E., Gu J., Schau M., Bunn H. F. (1998) Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 7. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., von Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 8. Mahon P. C., Hirota K., Semenza G. L. (2001) FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15, 2675–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Semenza G. L. (2001) HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 13, 167–171 [DOI] [PubMed] [Google Scholar]
- 10. Iyer N. V., Kotch L. E., Agani F., Leung S. W., Laughner E., Wenger R. H., Gassmann M., Gearhart J. D., Lawler A. M., Yu A. Y., Semenza G. L. (1998) Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12, 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maltepe E., Schmidt J. V., Baunoch D., Bradfield C. A., Simon M. C. (1997) Abnormal angiogenesis and responses to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature 386, 403–407 [DOI] [PubMed] [Google Scholar]
- 12. Ryan H. E., Lo J., Johnson R. S. (1998) HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 17, 3005–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kung A. L., Wang S., Klco J. M., Kaelin W. G., Livingston D. M. (2000) Suppression of tumor growth through disruption of hypoxia-inducible transcription. Nat. Med. 6, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 14. Ryan H. E., Poloni M., McNulty W., Elson D., Gassmann M., Arbeit J. M., Johnson R. S. (2000) Hypoxia-inducible factor-1α is a positive factor in solid tumor growth. Cancer Res. 60, 4010–4015 [PubMed] [Google Scholar]
- 15. Feldser D., Agani F., Iyer N. V., Pak B., Ferreira G., Semenza G. L. (1999) Reciprocal positive regulation of hypoxia-inducible factor 1α and insulin-like growth factor 2. Cancer Res. 59, 3915–3918 [PubMed] [Google Scholar]
- 16. Gao N., Shen L., Zhang Z., Leonard S. S., He H., Zhang X. G., Shi X., Jiang B. H. (2004) Arsenite induces HIF-1α and VEGF through PI3K, Akt and reactive oxygen species in DU145 human prostate carcinoma cells. Mol. Cell Biochem. 255, 33–45 [DOI] [PubMed] [Google Scholar]
- 17. Laughner E., Taghavi P., Chiles K., Mahon P. C., Semenza G. L. (2001) HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1α (HIF-1α) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol. Cell Biol. 21, 3995–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J., Davidson G., Huang Y., Jiang B. H., Shi X., Costa M., Huang C. (2004) Nickel compounds act through phosphatidylinositol-3-kinase/Akt-dependent, p70(S6k)-independent pathway to induce hypoxia inducible factor transactivation and Cap43 expression in mouse epidermal Cl41 cells. Cancer Res. 64, 94–101 [DOI] [PubMed] [Google Scholar]
- 19. Zelzer E., Levy Y., Kahana C., Shilo B. Z., Rubinstein M., Cohen B. (1998) Insulin induces transcription of target genes through the hypoxia-inducible factor HIF-1α/ARNT. EMBO J. 17, 5085–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang B. H., Jiang G., Zheng J. Z., Lu Z., Hunter T., Vogt P. K. (2001) Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 12, 363–369 [PubMed] [Google Scholar]
- 21. Zhong H., Chiles K., Feldser D., Laughner E., Hanrahan C., Georgescu M. M., Simons J. W., Semenza G. L. (2000) Modulation of hypoxia-inducible factor 1α expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 60, 1541–1545 [PubMed] [Google Scholar]
- 22. Zundel W., Schindler C., Haas-Kogan D., Koong A., Kaper F., Chen E., Gottschalk A. R., Ryan H. E., Johnson R. S., Jefferson A. B., Stokoe D., Giaccia A. J. (2000) Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 14, 391–396 [PMC free article] [PubMed] [Google Scholar]
- 23. Alvarez-Tejado M., Alfranca A., Aragonés J., Vara A., Landázuri M. O., del Peso L. (2002) Lack of evidence for the involvement of the phosphoinositide 3-kinase/Akt pathway in the activation of hypoxia-inducible factors by low oxygen tension. J. Biol. Chem. 277, 13508–13517 [DOI] [PubMed] [Google Scholar]
- 24. Arsham A. M., Plas D. R., Thompson C. B., Simon M. C. (2002) Phosphatidylinositol 3-kinase/Akt signaling is neither required for hypoxic stabilization of HIF-1α nor sufficient for HIF-1-dependent target gene transcription. J. Biol. Chem. 277, 15162–15170 [DOI] [PubMed] [Google Scholar]
- 25. An W. G., Kanekal M., Simon M. C., Maltepe E., Blagosklonny M. V., Neckers L. M. (1998) Stabilization of wild-type p53 by hypoxia-inducible factor 1α. Nature 392, 405–408 [DOI] [PubMed] [Google Scholar]
- 26. Blagosklonny M. V., An W. G., Romanova L. Y., Trepel J., Fojo T., Neckers L. (1998) p53 inhibits hypoxia-inducible factor-stimulated transcription. J. Biol. Chem. 273, 11995–11998 [DOI] [PubMed] [Google Scholar]
- 27. Chen D., Li M., Luo J., Gu W. (2003) Direct interactions between HIF-1α and Mdm2 modulate p53 function. J. Biol. Chem. 278, 13595–13598 [DOI] [PubMed] [Google Scholar]
- 28. Hansson L. O., Friedler A., Freund S., Rudiger S., Fersht A. R. (2002) Two sequence motifs from HIF-1α bind to the DNA-binding site of p53. Proc. Natl. Acad. Sci. U.S.A. 99, 10305–10309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xenaki G., Ontikatze T., Rajendran R., Stratford I. J., Dive C., Krstic-Demonacos M., Demonacos C. (2008) PCAF is an HIF-1α cofactor that regulates p53 transcriptional activity in hypoxia. Oncogene 27, 5785–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ravi R., Mookerjee B., Bhujwalla Z. M., Sutter C. H., Artemov D., Zeng Q., Dillehay L. E., Madan A., Semenza G. L., Bedi A. (2000) Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1α. Genes Dev. 14, 34–44 [PMC free article] [PubMed] [Google Scholar]
- 31. Bárdos J. I., Chau N. M., Ashcroft M. (2004) Growth factor-mediated induction of HDM2 positively regulates hypoxia-inducible factor 1α expression. Mol. Cell Biol. 24, 2905–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu L., Simon M. C. (2004) Regulation of transcription and translation by hypoxia. Cancer Biol. Ther 3, 492–497 [DOI] [PubMed] [Google Scholar]
- 33. Semenza G. L. (2009) Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology (Bethesda) 24, 97–106 [DOI] [PubMed] [Google Scholar]
- 34. Montagner M., Enzo E., Forcato M., Zanconato F., Parenti A., Rampazzo E., Basso G., Leo G., Rosato A., Bicciato S., Cordenonsi M., Piccolo S. (2012) SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature 487, 380–384 [DOI] [PubMed] [Google Scholar]
- 35. Liu Y. V., Baek J. H., Zhang H., Diez R., Cole R. N., Semenza G. L. (2007) RACK1 competes with HSP90 for binding to HIF-1α and is required for O2-independent and HSP90 inhibitor-induced degradation of HIF-1α. Mol. Cell 25, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Narita T., Yin S., Gelin C. F., Moreno C. S., Yepes M., Nicolaou K. C., Van Meir E. G. (2009) Identification of a novel small molecule HIF-1α translation inhibitor. Clin. Cancer Res. 15, 6128–6136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rong Y., Post D. E., Pieper R. O., Durden D. L., Van Meir E. G., Brat D. J. (2005) PTEN and hypoxia regulate tissue factor expression and plasma coagulation by glioblastoma. Cancer Res. 65, 1406–1413 [DOI] [PubMed] [Google Scholar]
- 38. Wen S., Stolarov J., Myers M. P., Su J. D., Wigler M. H., Tonks N. K., Durden D. L. (2001) PTEN controls tumor-induced angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 4622–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ding H., Roncari L., Shannon P., Wu X., Lau N., Karaskova J., Gutmann D. H., Squire J. A., Nagy A., Guha A. (2001) Astrocyte-specific expression of activated p21-ras results in malignant astrocytoma formation in a transgenic mouse model of human gliomas. Cancer Res. 61, 3826–3836 [PubMed] [Google Scholar]
- 40. Mahadevan D., Chiorean E. G., Harris W. B., Von Hoff D. D., Stejskal-Barnett A., Qi W., Anthony S. P., Younger A. E., Rensvold D. M., Cordova F., Shelton C. F., Becker M. D., Garlich J. R., Durden D. L., Ramanathan R. K. (2012) Phase I pharmacokinetic and pharmacodynamic study of the pan-PI3K/mTORC vascular targeted pro-drug SF1126 in patients with advanced solid tumours and B-cell malignancies. Eur. J. Cancer 48, 3319–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andrews N. C., Faller D. V. (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmittgen T. D., Livak K. J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 43. Folkes A. J., Ahmadi K., Alderton W. K., Alix S., Baker S. J., Box G., Chuckowree I. S., Clarke P. A., Depledge P., Eccles S. A., Friedman L. S., Hayes A., Hancox T. C., Kugendradas A., Lensun L., Moore P., Olivero A. G., Pang J., Patel S., Pergl-Wilson G. H., Raynaud F. I., Robson A., Saghir N., Salphati L., Sohal S., Ultsch M. H., Valenti M., Wallweber H. J., Wan N. C., Wiesmann C., Workman P., Zhyvoloup A., Zvelebil M. J., Shuttleworth S. J. (2008) The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-t hieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J. Med. Chem. 51, 5522–5532 [DOI] [PubMed] [Google Scholar]
- 44. Jackson S. P., Schoenwaelder S. M., Goncalves I., Nesbitt W. S., Yap C. L., Wright C. E., Kenche V., Anderson K. E., Dopheide S. M., Yuan Y., Sturgeon S. A., Prabaharan H., Thompson P. E., Smith G. D., Shepherd P. R., Daniele N., Kulkarni S., Abbott B., Saylik D., Jones C., Lu L., Giuliano S., Hughan S. C., Angus J. A., Robertson A. D., Salem H. H. (2005) PI 3-kinase p110β: a new target for antithrombotic therapy. Nat. Med. 11, 507–514 [DOI] [PubMed] [Google Scholar]
- 45. Yuan J., Mehta P. P., Yin M. J., Sun S., Zou A., Chen J., Rafidi K., Feng Z., Nickel J., Engebretsen J., Hallin J., Blasina A., Zhang E., Nguyen L., Sun M., Vogt P. K., McHarg A., Cheng H., Christensen J. G., Kan J. L., Bagrodia S. (2011) PF-04691502, a potent and selective oral inhibitor of PI3K and mTOR kinases with antitumor activity. Mol. Cancer Ther. 10, 2189–2199 [DOI] [PubMed] [Google Scholar]
- 46. Maira S. M., Stauffer F., Brueggen J., Furet P., Schnell C., Fritsch C., Brachmann S., Chène P., De Pover A., Schoemaker K., Fabbro D., Gabriel D., Simonen M., Murphy L., Finan P., Sellers W., García-Echeverría C. (2008) Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol. Cancer Ther. 7, 1851–1863 [DOI] [PubMed] [Google Scholar]
- 47. Bendell J. C., Rodon J., Burris H. A., de Jonge M., Verweij J., Birle D., Demanse D., De Buck S. S., Ru Q. C., Peters M., Goldbrunner M., Baselga J. (2012) Phase I, dose-escalation study of BKM120, an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. 30, 282–290 [DOI] [PubMed] [Google Scholar]
- 48. Hart J. R., Vogt P. K. (2011) Phosphorylation of AKT: a mutational analysis. Oncotarget 2, 467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mayo L. D., Donner D. B. (2001) A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. U.S.A. 98, 11598–11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gu L., Zhu N., Zhang H., Durden D. L., Feng Y., Zhou M. (2009) Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell 15, 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Honda R., Tanaka H., Yasuda H. (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 52. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Regulation of p53 stability by Mdm2. Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 53. Su J. D., Mayo L. D., Donner D. B., Durden D. L. (2003) PTEN and phosphatidylinositol 3′-kinase inhibitors up-regulate p53 and block tumor-induced angiogenesis: evidence for an effect on the tumor and endothelial compartment. Cancer Res. 63, 3585–3592 [PubMed] [Google Scholar]
- 54. Garlich J. R., De P., Dey N., Su J. D., Peng X., Miller A., Murali R., Lu Y., Mills G. B., Kundra V., Shu H. K., Peng Q., Durden D. L. (2008) A vascular targeted pan phosphoinositide 3-kinase inhibitor prodrug, SF1126, with antitumor and antiangiogenic activity. Cancer Res. 68, 206–215 [DOI] [PubMed] [Google Scholar]
- 55. Wójcik C., DeMartino G. N. (2003) Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35, 579–589 [DOI] [PubMed] [Google Scholar]