Background: LPS-induced PDCD4 degradation leads to IL-10 induction.
Results: LPS-induced PDCD4 degradation results in release of Twist2, resulting in c-Maf induction and IL-10 production.
Conclusion: The PDCD4/Twist2 interaction has an important anti-inflammatory role in LPS signaling.
Significance: This study reports the mechanism of PDCD4/Twist2 interaction and provides a new insight of IL-10 production via suppression of PDCD4/Twist2 interaction.
Keywords: Inflammation, Interleukin, Lipopolysaccharide (LPS), mTOR Complex (mTORC), Transcription Factor, IL-10, PDCD4, Twist2, c-maf, Rapamycin
Abstract
Programmed cell death protein 4 (PDCD4) is a tumor suppressor and has also been shown to suppress production of the immunomodulatory cytokine IL-10. The precise role of PDCD4 in IL-10 induction in macrophages is still not fully understood. Incubation of macrophages with inhibitors of PI3K and mTOR blocked LPS-stimulated PDCD4 degradation and expression of c-Maf and IL-10 production. PDCD4 and the transcription factor Twist2 were shown to form a complex in untreated cells. LPS disrupted the complex allowing Twist2 to bind to the c-Maf promoter. PI3K and mTOR inhibitors prevented this disruption by stabilizing PDCD4 and thereby decreased Twist2 binding to the c-Maf promoter and induction of c-Maf mRNA. These results indicate a regulatory role for PDCD4 and Twist2 in LPS-induced IL-10 production in macrophages. LPS promotes PDCD4 degradation via a pathway involving PI3K and mTOR, releasing Twist2, which induces IL-10 via c-Maf.
Introduction
The Gram-negative bacterial product lipopolysaccharide (LPS) is a potent inducer of the proinflammatory response. LPS is also able to induce multiple negative feedback regulators of inflammation, notably the immunomodulatory cytokine interleukin-10 (IL-10). IL-10 limits the production of proinflammatory cytokines such as TNF, IL-6, and IL-12 (1). The induction of IL-10 by LPS requires activation of Toll-like receptor 4 (TLR4)2 signaling through p38 mitogen-activated protein kinase (p38); however, the mammalian target of rapamycin (mTOR) signaling pathway has also been shown to play an important role (2). The mTOR system comprises two different complexes, mTOR complex 1 (mTORC1) and mTORC2. mTORC1 comprises mTOR in a complex with mLST8 (mammalian lethal with SEC13 protein 8), PRAS40 (proline-rich AKT substrate of 40 kDa) and RAPTOR (regulatory-associated protein of mTOR). mTORC1 is sensitive to inhibition by rapamycin. mTORC2 comprises mTOR, RICTOR (rapamycin-insensitive companion of mTOR), mTOR-associated proteins Sin1 and mLST8, and is rapamycin-insensitive (3).
The activation of mTORC1 by LPS results in the activation of S6 kinase (S6K1) (4). S6K1 has multiple substrates, a notable example in the context of IL-10 being programmed cell death protein 4 (PDCD4). Following phosphorylation by S6K1, PDCD4 undergoes degradation by β-TRCP ubiquitin ligases (5, 6). PDCD4 was first described as a tumor suppressor (7). It inhibits translation, interacting via its MA3 domains with the eukaryotic translation-initiation factors eIF4a and eIF4G, targeting cap-dependent translation of mRNA, which include those encoding IL-10 and IL-4 (8–11). The induction of IL-10 by LPS has been shown to require degradation of PDCD4, allowing translation to occur (6), providing one mechanism whereby PDCD4 might suppress IL-10 production.
PDCD4 has been shown to interact with other proteins, a notable example being Twist1 (12). It has been shown that PDCD4 inhibits the transcriptional function of Twist1. In mammals, there are two Twist family members: Twist1 and Twist2 (also known as Dermo-1) (13, 14). Basic helix-loop-helix (bHLH) transcription factors such as Twist2 act as a homo- or heterodimer through their HLH motifs to form a DNA-binding domain. Twist2 has been shown to regulate c-Maf, via c-Maf IL-10 expression after LPS-induced stimulation (15).
In this study, we have addressed whether the inhibitory effect of PDCD4 on IL-10 production involves Twist2. We have found that PDCD4 interacts with Twist2 and that PDCD4 degradation results in the release of Twist2, allowing it to bind to the c-Maf E-box and resulting in c-Maf induction, leading to enhanced IL-10 production. Our study therefore provides a further function for PDCD4 in the regulation of IL-10, whereby it acts to limit c-Maf induction by sequestering Twist2.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
The HA-tagged PDCD4 expression vectors was kindly provided by Dr. Michele Pagano and subcloned into pc.DNA3.1. Dr. Toschi and Dr. Kohno kindly provided the HA-tagged PDCD4 S67A and the FLAG-tagged Twist1 expression vectors respectively. The Myc-DDK-tagged (FLAG tag) ORF clone of Twist2 was cloned into pCMV6 and was purchased from OriGene Technologies. LPS from Escherichia coli, serotype 0111:B4, was from Alexis. Rapamycin (R-5000) was from LC Laboratory. MG132 (M7449), wortmannin (W1628), LY294002 (L9908), PF-4708671 (PZ0143), and cycloheximide (C7698) were all purchased form Sigma. Oligonucleotides and SYBR primers were from Eurofins, and Taqman probes were from Applied Biosystems. SMARTpool siRNAs specific for mouse PDCD4 and S6K1 and negative control were from Dharmacon. Rabbit anti-Twist2 (ab66031) antibody was from Abcam or from Santa Cruz Biotechnology (H81 sc-15393), mouse anti-β-actin (AC-74), anti-HA (H6908), anti-FLAG (F7425), and rabbit IgG (I5006) antibodies were from Sigma. The antibodies PDCD4 (9535) and S6K (9202) were purchased from Cell Signaling. Protein A/G-plus agarose beads were from Santa Cruz Biotechnology. Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Lipofectamine 2000TM was from Invitrogen. IL-10 (DY-417) and IL-6 (DY-406) ELISA Duoset® kits were purchased from R&D Systems.
Cell Culture
The human embryonic kidney 293 TLR4-MD2-CD14 (HEK293-TLR4-MD2-CD14) cell line was obtained from Invivogen and cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% (v/v) heat-inactivated FCS, 50 mg/ml noromycin, 100 μg/ml blasticidin, and 100 μg/ml HygroGold obtained from Invivogen. RAW264.7 cells were obtained from the European Cell Culture Collection and cultured in DMEM containing 10% (v/v) FCS and 1% (v/v) penicillin/streptomycin. Bone marrow from wild type and PDCD4−/− were a kind gift from Dr. Derek Johnson and were differentiated for 7 days in DMEM supplemented with 10% (v/v) FCS, 1% (v/v) penicillin/streptomycin, and 20% (v/v) M-CSF (L929 mouse fibroblast supernatant). For experiments RAW264.7, BMDM, and HEK293-TLR4-MD2-CD14 cells were seeded at 5 × 105 cells/ml.
RNA Isolation and Real Time PCR
Cells (primary bone marrow derived macrophages ((BMDM) or RAW264.7) were plated and serum-starved for 16 h prior stimulation. Cells were stimulated with LPS, rapamycin, MG132, wortmannin, or LY294002 as indicated in the figure legends. Total RNA was extracted using an RNeasy kit (Qiagen), modified to obtain small RNA species. cDNA for mRNA analysis was prepared for 5–100 ng/ml total RNA using the High Capacity cDNA archive kit (Applied Biosystems) according to the manufacturer's instructions, and incorporating TaqMan primers for mouse c-Maf, mouse IL-6, mouse IL-10, mouse GAPDH, and mouse 18S (Applied Biosystems). mRNA expression were measured on the 7900HT RT-PCR system (Applied Biosystems), and fold changes in expression were calculated by ΔΔCt method using mouse GAPDH and mouse 18S as an endogenous control for mRNA expression. All fold changes are expressed normalized to the non-stimulated control for each cell type.
Enzyme-linked Immunosorbent Assay
For cytokine measurements, BMDM and RAW264.7 cells were seeded at 5 × 105 cell/ml in a 12-well plate and stimulated in triplicate for each experiment. Supernatants were removed and analyzed for murine IL-10 and IL-6 (R&D Systems) using enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer's instructions.
Protein Expression
Differentiated BMDM or RAW264.7 cells were seeded at 5 × 105 in six-well plates and stimulated with LPS rapamycin, MG132, wortmannin, or LY294002 as indicated in the figure legends. Cells were lysed in low stringency lysis buffer complete with protease inhibitors. Protein concentration was then determined using the Coomassie Bradford reagent (Pierce). Lysates were resolved on 10% SDS-PAGE gels and transferred onto polyvinylidene difluoride membrane. Membranes were blocked in 5% (w/v) dried milk in TBS-T (50 mm Tris/HCl, pH 7.6, 150 mm NaCl, and 0.1% (v/v) Tween 20) before being immunoblotted with anti-PDCD4, anti-total S6K1, anti-HA, anti-FLAG, or anti-β-actin antibodies (1:1000 or 1:10,000 respectively) in 5% (w/v) dried milk in TBS-T at 4 °C overnight or at room temperature for at least 2 h. Membranes used for Twist2 determination were blocked in 5% (w/v) bovine serum albumin (Sigma) before being immunoblotted with anti-Twist2 antibody. Membranes were then incubated with the appropriate horseradish peroxidase-conjugated secondary antibody diluted 1:2000 in 5% (w/v) dried milk in TBS-T for 1 h. Blots were developed by enhanced chemiluminescence according to the manufacturer's instructions (Cell Signaling Technology, Inc.).
Co-immunoprecipitation Assay
HEK293-TLR4-MD2-CD14 were seeded at 4 × 105 cells/ml in 10-cm dishes. 24 h later, cells were transfected with a total of 4 μg of the indicated plasmids using GeneJuice® and serum-starved cultured for a further 24 h before 6 h stimulation with LPS rapamycin, MG132, wortmannin, or LY294002 as indicated in the figures. After treatment, cells were lysed in low stringency lysis buffer complete with protease inhibitors, and 50 μl of whole cell lysate was kept for analysis. Co-immunoprecipitations were performed with A/G-plus agarose beads and with an IgG or PDCD4 antibody. Cell lysates were centrifuged at 2.200 × g for 15 min before incubation with the beads and antibodies at 4 °C for 16 h. Following this, the lysate and beads were centrifuged at 80 × g for 2 min at 4 °C, the supernatant was removed, and the beads were washed three times in 1 ml of low stringency lysis buffer. Immune complexes were eluted by the addition of 50 μl of SDS-Laemmli buffer and boiling the samples. Co-immunoprecipitations were analyzed by SDS-PAGE and Western blotting.
Chromatin Immunoprecipitation
RAW264.7 cells were seeded at 4 × 105cell/ml in three 15-cm dishes per sample, 24 h later, cells were transfected with a total of 4 μg of the indicated plasmids using GeneJuice® and serum-starved cultured for a further 24 h before a 6-h stimulation with LPS rapamycin, MG132, wortmannin, or LY294002 as indicated in the figures. After treatment, cells were fixed by adding a final concentration of 1% formaldehyde to each culture dish and were incubated for 10 min at room temperature. A 1/20 volume of 2.5 m glycine was then added to each dish and allowed to set at room temperature for 5 min prior to washing in PBS and resuspension in 6 ml of ChIP lysis buffer (SDS lysis buffer with leupeptin, aprotinin, and PMSF) and immediately snap frozen in liquid nitrogen. The samples were thawed and resuspended in SDS:Triton buffer and then sonicated at 22% intensity, 10 × 30 s per sample, placing on ice in between pulses. 10 μl of whole cell lysate was kept for analysis. Lysates were incubated overnight with primary antibodies: anti-Twist2 (H81) (Santa Cruz Biotechnology, sc15393) and anti-IgG (Sigma, I5006). On the following day, cell lysates were incubated for 1 h at 4 °C with preblocked A/G-plus agarose beads. Next, the lysate and beads were centrifuged at 80 × g for 2 min at 4 °C, the supernatant was removed, and the beads were washed three times in 1 ml of ChIP lysis buffer. Quantitative RT-PCR was carried out using primers for either the c-Maf promoter consensus Twist2 binding site (forward, 5′-tgacgtcaggctcaaatgggaa-3′, reverse; 5′-agtcagctgattgggctgtgat-3′) or a non-Twist2 binding site (forward, 5′-ttcccagttcacattcagccct-3′; reverse, 5′-acgcatcatcattcctggct-3′). Data are presented as a percentage of input.
Affinity Purification with Biotinylated Oligonucleotides
Oligonucleotides for the terminal Twist2 binding site on the c-Maf promoter were annealed at 90–95 °C for 3–5 min, and then the heat block was allowed to cool to room temperature (forward, 5′BIO-caggctcaaatgggaaattg-3′; reverse, 5′-caatttcccatttgagcctg-3′). RAW264.7 cells were seeded at 4 × 105 cell/ml and transfected with a total of 4 μg of Myc-DDK-tagged Twist2 plasmid using GeneJuice® and serum-starved and cultured for a further 24 h before a 6-h stimulation with LPS rapamycin. Cells were then lysed in 100 μl of oligonucleotide buffer (25 mm Tris, 5% glycerol, 50 mm EDTA, 5 mm NaF, Nonidet P-40 1%, 1 mm DTT, 150 mm NaCl, and protease and phosphatase inhibitors) and snap-frozen. Samples were then thawed on ice and diluted with a further 900 μl of oligonucleotide buffer containing no NaCl. A 10-μl sample of lysate was kept to which 40 μl of 5× SDS buffer was added. Remaining lysates were then precleared with 20 μl of prewashed streptavidin-agarose beads, rotating at 4 °C for 15 min before centrifuging at 80 × g for 5 min at 4 °C. Supernatants were removed to a fresh tube with 30 μl prewashed streptavidin-agarose beads and 30 μg of 5′-biotinylated oligonucleotide. Binding was performed for 2 h at 4 °C, and samples were rotated. Samples were centrifuged for 5 min at 4 °C to pellet the beads, which were washed three times before 50 μl of 5× SDS sample buffer was added to the beads. Samples were loaded on a 15% gel and Twist2 binding detected by Western blotting as described previously.
Site-directed Mutagenesis of Twist2
The QuikChange site-directed mutagenesis kit (Stratagene) was use to mutate or delete certain basis in the Twist2 gene. The manufacturer's instructions were followed using primers incorporating the desired mutations and deletions. To find a mechanistic function for Twist2, the amino acids located on 79 and 81 in the DNA binding domain were mutated. Previously, it has been shown that PDCD4 interacts with Twist1 (12) and eIF4A (7). It was found that the amino-terminal region of eIF4A possesses sequence homology with the bHLH of Twist1 (12) and Twist2 (data not shown), which is evolutionarily conserved. Therefore, amino acids located on 92, 93, 113, and 114 in the protein binding domain of Twist2 were mutated in the protein binding domain. Primers used were as follows: for Twist2 Gln-118-stop, 5′-catagacttcctctactaggtcctgcagagcga-3′ (forward) and 5′-tcgctctgcaggacctagtagaggaagtctatg-3′ (reverse), Twist2 Gln-66-stop, 5′-ggagctgcagagctagcgcatcctggc-3′ (forward) and 5′-gccaggatgcgctagctctgcagctcc-3′ (reverse); Twist2-FLAG T78A/S81A, 5′-agcgccagcgcgcccaggcgctcaacgag-3′ (forward) and 5′-ctcgttgagcgcctgggcgcgctggcgct-3′ (reverse); Twist2 Ile-92-stop, 5′-gcgctgcgcaagatctagcccacgctgccctct-3′ (forward) and 5′-agagggcagcgtgggctagatcttgcgcagcgc-3′ (reverse); Twist2-FLAG I92T/I93S/Y113C/I114S (I92T/I93S), 5′-cgcggcgctgcgcaagaccagccccacgct-3′ (forward) and 5′-agcgtggggctggtcttgcgcagcgccgcg-3′ (reverse); and Y113C/I114S, 5′-aagctggccgccaggtgcagagacttcctctacc-3′ (forward) and 5′-ggtagaggaagtctctgcacctggcggccagctt-3′ (reverse).
RESULTS
LPS-induced PDCD4 Degradation Requires mTOR Signaling
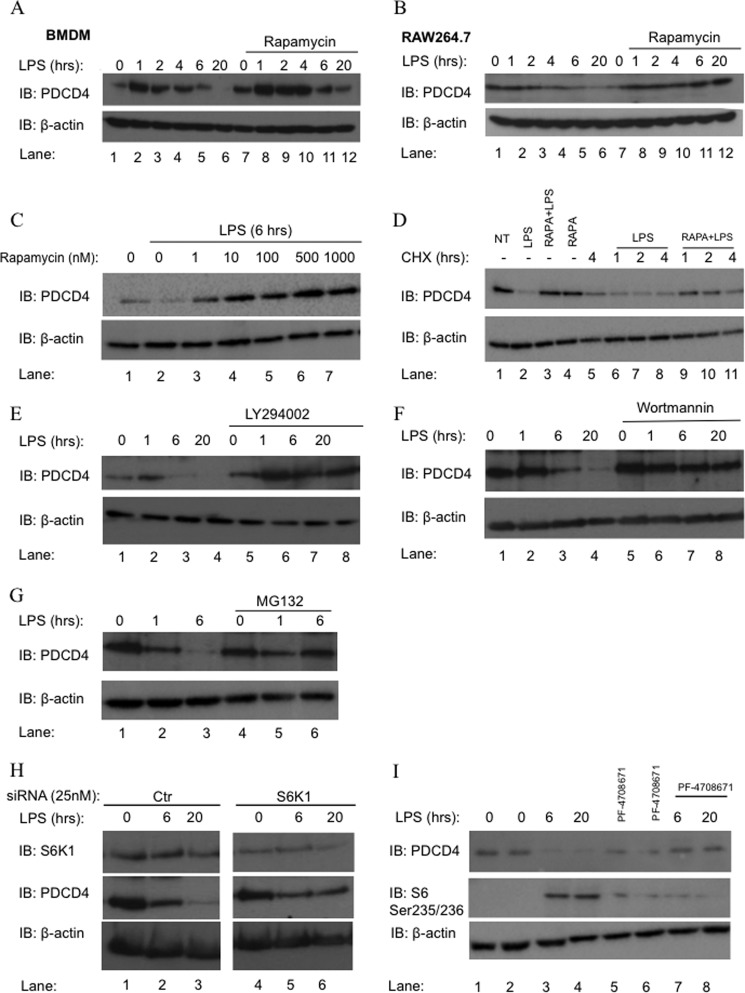
We first studied the effect of rapamycin on LPS induced PDCD4 degradation. As shown in Fig. 1A, in mouse primary BMDMs and in Fig. 1B in RAW264.7cells, LPS stimulation resulted in higher expression of PDCD4 protein after 1 h, followed by a gradual decrease after 6 and 20 h. In both BMDM and RAW264.7 cells, PDCD4 was almost completely degraded at 20 h (lane 6 in both Fig. 1, A and B). Pretreatment of cells with rapamycin inhibited LPS-induced PDCD4 protein degradation (compare lanes 4–6 and lanes 10–12 shown in Fig. 1, A and Fig. B). The residual degradation of PDCD4 occurring at 6 h and 20 h in BMDM in the presence of rapamycin is likely to be due to miR-21, as shown previously (6). The inhibitory effect of rapamycin was evident at 1 nm (Fig. 1C, lane 3 compared with lane 2) with higher doses increasing PDCD4 levels above basal level (Fig. 1C, lanes 4–7). These results confirmed those seen previously (16). We observed that pretreatment with rapamycin followed by 6 h of LPS stimulation almost completely prevented PDCD4 degradation in a dose-dependent manner (Fig. 1C). To verify that the increase in PDCD4 protein in response to inhibitors of the PI3K-mTOR pathway was a result of increased protein stability, we next determined the effect of cycloheximide on the rapamycin effect. We observed again a decrease in PDCD4 protein levels in RAW264.7 cells in the absence of rapamycin but stimulated with LPS (Fig. 1D, compare lane 2 with lane 1). The decrease in PDCD4 was still evident in cells pretreated with cycloheximide for various times followed by LPS (Fig. 1D, lanes 6–8). Rapamycin was still able to stabilize PDCD4 in LPS-treated cells that had been pretreated with cycloheximide (Fig. 1D, lanes 9–11). Although cycloheximide does cause a slight decrease in overall PDCD4 levels, as can be seen in Fig. 1D (lane 4). This result indicates that the increase in PDCD4 after rapamycin pretreatment was a result of protein stability. This has also been shown in MCF7 cells treated with conditioned medium and rapamycin pretreatment (17). The PI3K inhibitors LY294002 (Fig. 1E) and wortmannin (Fig. 1F) also inhibited the effect of LPS on PDCD4 (compare lanes 3 and 4 and 7 and 8 in Fig. 1, E and F). The 26S proteasome inhibitor MG132 also inhibited the degradation of PDCD4 (Fig. 1G, compare lanes 3 and 6), indicating that the degradation is mediated by the proteasome. It has previously been shown that phosphorylation by S6K1 is critical for the proteasomal degradation of PDCD4 (5, 18). Knockdown of S6K1 using small interfering RNA (siRNA) resulted in a significant decrease in endogenous S6K1 expression (Fig. 1H, top panel, lanes 4–6). This led to an inhibition of LPS-induced degradation of PDCD4 protein (Fig. 1H, middle panel, compare lanes 3 and 6). Another method to implicate whether S6K1 is involved in this response was to pretreat RAW264.7 cells with S6K1 inhibitor PF-4708671. The cells pretreated with PF-478671 followed by 6 h of LPS stimulation, also led to an inhibition of LPS-induced PDCD4 protein degradation (Fig. 1I, compare lanes 3 and 4 with 7 and 8). These results show again that S6K1 is critical for PDCD4 degradation. It is therefore likely that the pathway stimulated by LPS here involves PI3K, mTORC1, and S6K1, leading to phosphorylation and degradation of PDCD4.
FIGURE 1.
LPS induces PDCD4 degradation requires mTOR signaling. BMDM (A–F) or RAW264.7 (B) cells were serum-starved for 16 h, followed by pretreatment for 30 min with DMSO or 500 nm rapamycin (RAPA) followed by 100 ng/ml LPS for the indicated times. C, cells were pretreated with various concentrations of rapamycin for 30 min (indicated above each lane) D, cells were pretreated with for 30 min with DMSO or 500 nm rapamycin followed by for 20 h, followed by post treatment of cycloheximide for the indicated times. E, cells were pretreated with 10 μm LY294002 followed by 100 ng/ml LPS for the indicated times (F), or 5 μm wortmannin (G), or 5 μm MG132 and then stimulated with 100 ng/ml LPS for 6 h. Protein expression was measured by Western blot with antibodies specific for PDCD4 or β-actin. Data are representative of three separate experiments. H, RAW264.7 cells were transfected with siRNA encoding for S6K1 or control (Ctr) and serum-starved for 24 h. The cells were stimulated with (100 ng/ml) LPS for indicated times. I, cells were serum-starved for 16 h followed by pretreatment for 30 min with DMSO or 10 μm PF-4708671 followed by 100 ng/ml LPS for the indicated times. Protein expression of S6K1 (70 kDa), PDCD4 (52 kDa), or β-actin (45 kDa) were measured by Western blot with specific antibodies. Results are representative of three independent experiments. IB, immunoblot.
Rapamycin Inhibits the Induction of IL-10 by LPS through PDCD4
We next examined the effect of rapamycin on the induction of IL-10. As shown in Fig. 2A, pretreatment of RAW264.7 cells with rapamycin followed by 6-h stimulation with LPS-inhibited IL-10 production with inhibition occurring from 1 nm and being optimal at 500 nm rapamycin (Fig. 2A). A similar decrease in Il10 mRNA was observed at the 500 nm dose of rapamycin (Fig. 2B). This effect on IL-10 was specific as IL-6 (both at the protein and mRNA level) was not affected by pretreatment with rapamycin followed by a 6-h stimulation of LPS (Fig. 2, C and D, respectively). Pretreatment with LY294002, wortmannin, or MG132 also decreased induction of IL-10 in response to LPS (Fig. 2E), as did knockdown of S6K1 and PF-408671 S6K1 inhibitor (Fig. 2, F and G).
FIGURE 2.
Rapamycin inhibits the induction of IL-10 by LPS through PDCD4. A, RAW264.7 cells were serum-starved for 16 h followed by pretreatment for 30 min with DMSO or various concentrations of rapamycin for 30 min (as indicated) and stimulated 6 h with 100 ng/ml LPS. B–D, RAW264.7 cells were serum-starved for 16 h, followed by pretreatment with DMSO (LPS) or 500 nm rapamycin (RAPA+LPS) for 30 min followed by 6 h of 100 ng/ml LPS stimulation. E, RAW2647 cells were serum-starved for 16 h, followed by pretreatment with DMSO or 500 nm rapamycin, 10 μm LY294002, 5 μm wortmannin, or 5 μm MG132 for 30 min, followed by 6 h of LPS (100 ng/ml). F, RAW264.7 cells were transfected with siRNA encoding for S6K1 or control (Ctr) and serum-starved for 24 h, followed by treatment with 100 ng/ml LPS for indicated times. G, RAW264.7 cells were serum-starved for 16 h, followed by pretreatment with DMSO (LPS) or 10 μm PF-4708671 (RPF-4708671+LPS) for 30 min followed by 6 and 20 h of 100 ng/ml LPS stimulation. H and I, RAW264.7 cells were transfected with siRNA specific for PDCD4 or control (Ctr) and serum-starved for 24 h. The cells were pretreated with DMSO or (500 nm) rapamycin for 30 min followed by 6 h of LPS stimulation. H, protein expression were measured by Western blot with antibodies specific for PDCD4 (52 kDa) or β-actin (45 kDa). Data are representative of three separate experiments. J, BMDMs from wild-type (WT) and PDCD4 deficient (PDCD4−/−) BMDMs were serum-starved for 16 h and pretreated with DMSO or 500 nm rapamycin, followed by 6 h LPS (100 ng/ml). After the indicated treatments, supernatants were analyzed by ELISA for IL-10 (A, E, F, G, I, and J) or IL-6 (C). RNA was extracted and the mouse IL-10 mRNA (B) and mouse IL-6 mRNA (D) were analyzed using TaqMan RT-PCR probes specific for murine IL-10 and IL-6. Data were normalized to GAPDH and 18S mRNA expression, and fold induction was calculated relative to untreated cells at 0 h. Data shown are representative of three separate experiments, with each point assayed in triplicate.*, p < 0.05; **, p < 0.01; ***, p < 0.001. IB, immunoblot.
It has been shown previously that there is more IL-10 produced in response to LPS in cells lacking PDCD4 (6). We predicted that pretreatment with rapamycin in cells lacking PDCD4 would not affect IL-10 production if rapamycin mediates its effect through PDCD4 stabilization. Therefore, we tested the effect of rapamycin on cells in which we knocked down PDCD4 (Fig. 2H). As can be seen in Fig. 2I, there was a 3-fold increase in IL-10 production in PDCD4 knockdown cells after LPS stimulation in comparison with the cells with control siRNA (Fig. 2I, 6-h LPS-treated samples, compare gray and white bars). Rapamycin had no effect on the induction of IL-10 by LPS in the cells in which PDCD4 was knocked down (Fig. 2I, 6-h LPS-treated samples, compare gray hatched bar with gray bar). In cells treated with control siRNA, rapamycin inhibited this IL-10 response (Fig. 2I, 6-h treated sample, compare black bar with white bar). Similar results were observed when PDCD4-deficient BMDM and wild-type BMDM were used as shown in Fig. 2J. IL-10 production in the PDCD4-deficient BMDM was increased compared with the wild-type BMDM (Fig. 2J, 6-h LPS treated samples, compare gray bar to white bar), but its induction was insensitive to rapamycin (Fig. 2J, 6-h LPS-treated samples, compare gray hatched bar with gray bar), unlike wild-type cells that showed inhibition by rapamycin (Fig. 2J, 6-h LPS-treated samples, compare black bar with white bar). These data confirm the importance of limiting PDCD4 via mTOR to induce IL-10 in response to LPS.
PDCD4 and Twist-2 Form a Complex
Next, we tested whether Twist2 and PDCD4 can interact. As shown in Fig. 3A, overexpressed HA-tagged PDCD4 interacted with overexpressed FLAG-tagged Twist2 (top panel, lane 4). Stimulation with LPS reduced this interaction (Fig. 3A, top panel, lane 5). Only a fraction of the overexpressed PDCD4 interacted with Twist2 because LPS did not cause a detectable decrease in the levels of total PDCD4 in the transfected cells (Fig. 3A, middle panel, lane 5). When cells were pretreated with rapamycin, the dissociation between PDCD4 and Twist2 following LPS stimulation was prevented (Fig. 3A, lane 7). Pretreatment of cells with wortmannin also inhibited the dissociation (Fig. 3B, compare lane 7 with lane 5). We next tested whether deficiency of S6K-mediated phosphorylation of PDCD4 stabilized its interaction with Twist2. As shown in Fig. 3C, overexpressed HA-tagged PDCD4 interacted with overexpressed FLAG-tagged Twist2 (top panel, lane 6). Stimulation with LPS reduced this interaction (Fig. 3C, top panel, lane 8). The interaction can also be determined in cells with overexpressed HA-tagged PDCD4 with a S6K phosphorylation site-deficient Ser-to-Ala mutant (Fig. 3C, lane 7) (5). However, after stimulation with LPS the decrease in the interaction between this mutant and the wild-type PDCD4 was attenuated (Fig. 3C compare lane 9 with lane 8). A previous study has shown PDCD4 can interact with Twist1; therefore, we also tested Twist1 as a control (12). As shown in Fig. 3D, overexpressed HA-tagged PDCD4 interacted with overexpressed and overexpressed FLAG-tagged Twist1 (top panel, lane 7). Stimulation with LPS also reduced the interaction with Twist1 (Fig. 3D, top panel, lane 9). These results show that PDCD4 and Twist2 can form a complex, which can be disrupted by LPS in a rapamycin-sensitive manner.
FIGURE 3.
PDCD4 interacts with Twist2. A and B, HEK293/TLR4-MD2-CD14 cells were transfected with plasmids encoding Twist2-FLAG, HA-PDCD4, or empty vector (EV) as indicated. A, following 24-h post transfection and serum starvation, cells were left untreated or pretreated with DMSO (L) or 500 nm rapamycin (R/L) for 30 min followed with 6-h of LPS (100 ng/ml) stimulation. B, cells were pretreated with DMSO or 500 nm rapamycin (R) or 5 μm wortmannin (W) for 30 min, followed by 6 h LPS (100 ng/ml). C and D, HEK293/TLR4-MD2-CD14 cells were transfected with plasmids encoding Twist2-FLAG, HA-PDCD4, HA-PDCD4 S67A (C), FLAG-Twist1 (D), or empty vector (EV) as indicated. C and D, following 24 h post transfection and serum starvation, cells were left untreated stimulated 6 h with 100 ng/ml LPS. After treatment, cells were lysed, and 50 μl of whole cell lysate was kept for analysis. The remainder was immunoprecipitated (IP) with IgG or PDCD4 for 16 h at 4 °C. Whole cell lysates and immunoprecipitated samples were analyzed by Western blotting for Twist2-FLAG (18 kDa) or HA-PDCD4 (52 kDa) were measured with antibodies specific for FLAG and HA. Results are representative of three independent experiments.
Defining the Domains Responsible for the Interaction
Having confirmed an interaction between PDCD4 and Twist2, we next investigated which domains of Twist2 mediated the interaction. Five Twist2 constructs in addition to full-length Twist2 were generated and are depicted in Fig. 4A. Fig. 4B illustrates the results from the interaction analysis with the constructs. Lane 4 (top panel) shows the interaction between PDCD4 and Twist2 as demonstrated previously in Fig. 3A. The higher percentage gel (18%) revealed a nonspecific band running faster than Twist2. Our results suggest that both the Twist Box and the bHLH domain are required for the interaction as removal of either domains abolished the interaction (Fig. 4B, lanes 5, 6, and 8). The mutations generated in the DNA binding domain (Fig. 4B, lane 7) did not affect the interaction between PDCD4 and Twist2; however, mutations in the protein binding domain appeared to reduce the interaction (Fig. 4B, lane 9). These results led us to investigate the importance of the different domains for IL-10 production. As shown in Fig. 4C, in RAW264.7 cells transfected with constructs lacking a functional Twist-Box and protein-binding domain (Twist2 Ile-92-stop) or mutated protein-binding domain (Twist2-FLAG I92T/I93S/T113C/I114S), there was an induction of IL-10 production (white bars). LPS was able to induce IL-10 in cells transfected with a plasmid encoding Twist2-FLAG WT (black bar, Twist2-FLAG WT). Transfection with a plasmid encoding Twist2 with the mutated DNA binding domain (black bar, Twist2-FLAG T78A/S81A) decreased its response, whereas transfection with a plasmid encoding the construct lacking the Twist-Box and protein binding domain (Twist2 Ile-92-stop) or mutated protein-binding domain (Black bar, Twist2-FLAG I92T/I93S/T113C/I114S) both showed a potentiated response. None of the constructs affected the induction of IL-6 by LPS (Fig. 4D, black bars). These results suggest that the Twist box and protein-binding domain in the bHLH domain are involved in the interaction with PDCD4, their absence allowing Twist2 to increase IL-10 production most likely due to an inability to be sequestered by PDCD4. The DNA-binding domain in Twist2 is needed for induction of IL-10.
FIGURE 4.
Defining the domains responsible for the interaction of Twist2 with PDCD4. A, schematic representation of Twist2-FLAG deletion mutants and Twist2-FLAG with mutations in the DNA binding domain (DBD) and protein binding domain (PBD) in the bHLH. B, HEK293T cells were transfected with equal amounts of various encoding plasmids Twist2-FLAG construct as described in A and HA-PDCD4 or empty vector (EV) plasmids as indicated. 24-h post transfection and serum starvation, cells were lysed, and 50 μl of whole cell lysate was kept for analysis. The remainder was immunoprecipitated (IP) with IgG or PDCD4 for 16 h at 4 °C. Whole lysates and immunoprecipitated samples were analyzed by Western blotting Twist2 (18 kDa). Results are representative of three independent experiments. C and D, RAW264.7 cells were transfected with equal amounts of various plasmids encoding Twist2-FLAG constructs and empty vector as described in A and HA-PDCD4. After 24 h, cells were left untreated or stimulated for 6 h with LPS (100 ng/ml). The supernatants were analyzed by ELISA for IL-10 (C) or IL-6 (D). Data shown are the mean ± S.D. from three separate experiments all carried out in triplicate *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Twist2 Binds to a Highly Conserved Site on the c-Maf Promoter after LPS Stimulation
Our data suggested that Twist2 might be released from PDCD4 to bind to the c-Maf promoter in response to LPS. We next examined the binding of Twist2 to the c-Maf promoter using a number of approaches. First, oligonucleotide pulldown assays were employed to investigate binding of Twist2 to the E-Box on the c-Maf promoter. As shown in Fig. 5A, lane 2, increased Twist2 binding to this specific and highly conserved site on the c-Maf promoter was induced by LPS. Pretreatment with 500 nm rapamycin inhibited this response (Fig. 5A, lane 4). To investigate this finding further, and in an endogenous context, ChIP at the Twist2 binding site in the c-Maf promoter was carried out. As shown in Fig. 5B, black bars, Twist2 bound to the c-Maf promoter basally, with binding being increased after 6 h LPS treatment. Pretreatment with rapamycin, wortmannin, LY294002, and MG132 inhibited the binding of Twist2, decreasing it below basal level. Twist2 did not bind to a non-Twist2 binding site in the c-Maf promoter (Fig. 5C, black bars) or the β-actin promoter (data not shown). These findings confirmed that LPS induces Twist2 binding to the Twist2 binding site at the E-Box on the c-Maf promoter. Rapamycin, wortmannin, LY294002, and MG132 all inhibited the induction of c-Maf mRNA by LPS (Fig. 5D). Induction of c-Maf mRNA by LPS was enhanced in PDCD4-deficient BMDM (Fig. 5E, 6-h LPS-treated samples, compare gray bar with white bar) but unlike in wild-type BMDM, this induction was not inhibited by rapamycin (Fig. 5E, 6-h LPS-treated samples, compare gray hatched bar with gray bar). These data indicate that PDCD4 targeting is more important for c-Maf induction by LPS.
FIGURE 5.
Twist2 binds to a highly conserved site on the c-Maf promoter after LPS stimulation. A and B, RAW264.7 cells were serum-starved for 16 h. Cells were then left untreated or pretreatment for 30 min with DMSO or 500 nm rapamycin and stimulated with 100 ng/ml LPS (L) for 6 h. Cells were also treated with 500 nm rapamycin alone for 6.5 h (R) (A, left panel). Cells were then lysed and an oligonucleotide (oligo) pulldown (OPD) assay was carried out with the E-Box Twist2-binding site oligonucleotide sequence on the c-Maf promoter. Samples were probed for Twist2 (18 kDa) by Western blot (A, right panel). Densitometry was carried are relative to non-treated controls. Bar graph represents the mean of two independent experiments. B and C, RAW264.7 cells were transfected with plasmids encoding Twist2-FLAG or HA-PDCD4. 24-h post transfection and serum starvation, cells were left untreated, or cells were pretreated for 30 min with DMSO or 500 nm rapamycin (R/L), 5 μm wortmannin (W/L), 10 μm LY294002 (LY/L), or 5 μm MG132 (M/L), and stimulated with 100 ng/ml LPS (L) for 6 h. After stimulation a ChIP assay was performed. Primers specific for the Twist2-binding site (∼1141 base pairs upstream the c-Maf promoter of the genomic DNA) (B) and, additionally, the non-Twist2- binding site (∼20 kb upstream the c-Maf promoter (C)) were designed, and binding events were measured as percent of input by real time PCR using antibodies against IgG (control antibody, white bars) or Twist2 (black bars). D, RAW264.7 cells were serum-starved for 16 h followed by pretreatment for 30 min with DMSO or 500 nm rapamycin, 5 μm wortmannin, 10 μm LY294002, or 5 μm MG132. E, BMDMs from wild-type (WT) and PDCD4-deficient (PDCD4−/−) BMDMs were serum-starved for 16 h and pretreated with DMSO or 500 nm rapamycin, followed by 6 h LPS (100 ng/ml). D and E, after the indicated treatments, RNA was extracted, and the mouse c-Maf mRNA was analyzed using TaqMan RT-PCR probes specific for murine c-Maf. Data were normalized to GAPDH and 18S mRNA expression, and fold induction was calculated relative to untreated cells (E) or relative to untreated WT BMDM. Data shown are the mean ± S.D. from three separate experiments all carried out in triplicate. *, p < 0.05; **, p < 0.01. F, LPS activates PI3K/mTOR signaling inducing the expression of c-Maf and IL-10 by degradation of PDCD4. The degradation of PDCD4 allows Twist2 protein to bind on the c-Maf promoter, inducing c-Maf expression, which then induces IL-10 gene expression. The inhibitors shown maintain PDCD4-Twist2 complex inhibiting IL-10 induction.
DISCUSSION
Here, we demonstrate a previously undescribed aspect of IL-10 regulation by LPS. The stimulation of macrophages with LPS leads to the dissociation of PDCD4 from Twist2 with PDCD4 being degraded. Twist2 is thereby released and binds to the c-Maf promoter. The increase in c-Maf would lead to induction of IL-10. PI3K, mTOR, and S6K are all required for PDCD4 degradation, indicating another aspect of mTOR involvement in IL-10 induction (Fig. 5F). It has been shown previously that PDCD4 is a negative regulator of IL-10 production. PDCD4 is known to be degraded following LPS stimulation, resulting in increased IL-10 production (6). Our results are in agreement with these previous studies; however, we have also shown that pretreatment with rapamycin inhibits LPS-induced PDCD4 degradation. We have also shown that the increase of PDCD4 after rapamycin pretreatment was a result of protein stability, as has been shown in a previous study in MCF7 cells treated with conditioned medium and rapamycin pretreatment (17). It has also previously been shown that rapamycin, LY294002, or MG132 prevented the loss of PDCD4 protein after 8 and 24 h in macrophages exposed to conditioned medium (17). When the mTOR pathway is activated, mTOR can phosphorylate S6K1. S6K1 has previously been shown to be critical for the phosphorylation and proteasomal degradation of PDCD4 in T98G glioblastoma cells after serum activation and in HEK293 cells activated with TPA (5, 18). We have also shown that S6K1 is involved in PDCD4 degradation in response to LPS using RAW264.7 cells transfected with S6K1-specific siRNA and the S6K1 inhibitor PF-4708671. However, S6K1 depletion with siRNA appears to only partially block LPS-induced PDCD4 degradation. This is most likely due to the incomplete knockdown of S6K1 as demonstrated by Western blot in Fig. 1H. The residual S6K1 present could still lead to partial PDCD4 degradation. In total, our data add to our understanding of processes whereby LPS promotes PDCD4 degradation.
Our findings that PI3K inhibitors and rapamycin reduce LPS induction of IL-10 in macrophages are in agreement with previous studies (19, 20). It has previously been demonstrated that rapamycin has the ability to decrease IL-10 mRNA and protein in monocytes and dendritic cells treated with LPS (16, 20–22). However, a mechanism behind this was not completely understood.
PDCD4 has been described as an inhibitor of cap-dependent translation of mRNA, including IL-10 mRNA, through the interaction via MA3 domains with eukaryotic initiation factors eIF4a and eIF4G (8, 9). We have identified Twist2 as another binding partner of PDCD4. The interaction was shown to occur via the bHLH and Twist box. LPS caused a decrease in this interaction most likely through PDCD4 degradation. We also demonstrated that the DNA-binding domain in Twist2 was required for IL-10 induction.
Sharabi et al. (15) demonstrated that in Twist2-deficient mice, there was a decrease in IL-10 cytokine secretion and c-Maf mRNA after LPS stimulation (15). Furthermore, a study with c-Maf-deficient cells showed that c-Maf is an essential transcription factor for IL-10 gene expression in macrophages activated with LPS (23). Sharabi et al. (15) demonstrated that Twist2 could bind to the c-Maf promoter, suggesting that Twist2 is able to promote the expression of IL-10 via direct activation of c-Maf transcription (15). We confirmed these findings, demonstrating in addition that LPS increases Twist2 binding to the E-box of the c-Maf promoter in a PI3K and rapamycin-sensitive manner.
The mechanism for IL-10 production through the PDCD4-Twist2 complex as demonstrated in this study might be common to more TLR pathways and not only found in TLR4 pathway signaling, as it has been shown in innate immune cells that TLRs can recruit and activate PI3K (24–26). Activation of the PI3K pathway by TLR2 increases the production of IL-10, and inhibition of the PI3K pathway leads to a decrease of IL-10 production (19). This may imply that PDCD4/Twist2 could be involved.
Given the influence of IL-10 as a potent anti-inflammatory cytokine, these findings may have important therapeutic implications. IL-10 plays a critical role in preventing excessive inflammation and autoimmune diseases as it has been shown that IL-10 deficiency can lead to the development of inflammatory bowel disease and other auto-immune diseases (27). Therefore, the mechanisms that regulate IL-10 expression are critical to developing therapeutic strategies. In our study, we have shown that IL-10 expression can be regulated with the PDCD4-Twist2 complex through the PI3K-mTOR pathway, uncovering an important mechanism of IL-10 regulation.
Dysregulation of this process might lead to increased inflammation, or the specific activation of the pathways might have anti-inflammatory effects. Therefore, it will be important to define in further studies the role of the PDCD4-Twist2 complex in disease.
Taken together, the data indicate that rapamycin, by inhibiting mTORC1 activity, can prevent LPS-induced PDCD4 degradation. Rapamycin allows the PDCD4-Twist2 complex to remain stable, thereby preventing Twist2 from binding to the c-Maf promoter, limiting c-Maf expression. This in turn will limit IL-10 induction by LPS. Overall, we provide a novel insight into the important anti-inflammatory role of the mTOR/PDCD4/Twist2/c-Maf pathway in LPS-induced signaling, with this signaling process leading to IL-10 induction.
Acknowledgments
We acknowledge Dr. Pagano for kindly providing the PDCD4 expression vector, Dr. Toschi for providing the S6K phosphorylation site-deficient PDCD4 mutant, and Dr. Kohno for providing the FLAG-Twist1 construct.
This work was supported by the Science Foundation Ireland, Health Research Board/Marie Curie CoFund, and the European Research Council (based in Ireland).
- TLR4
- Toll-like receptor 4
- mTOR
- mammalian target of rapamycin
- mTORC1
- mTOR complex 1
- PDCD4
- programmed cell death protein 4
- bHLH
- basic helix-loop-helix
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Saraiva M., O'Garra A. (2010) The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10, 170–181 [DOI] [PubMed] [Google Scholar]
- 2. Pengal R. A., Ganesan L. P., Wei G., Fang H., Ostrowski M. C., Tridandapani S. (2006) Lipopolysaccharide-induced production of interleukin-10 is promoted by the serine/threonine kinase Akt. Mol. Immunol. 43, 1557–1564 [DOI] [PubMed] [Google Scholar]
- 3. Benjamin D., Colombi M., Moroni C., Hall M. N. (2011) Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 10, 868–880 [DOI] [PubMed] [Google Scholar]
- 4. Lim H. K., Choi Y. A., Park W., Lee T., Ryu S. H., Kim S. Y., Kim J. R., Kim J. H., Baek S. H. (2003) Phosphatidic acid regulates systemic inflammatory responses by modulating the Akt-mammalian target of rapamycin-p70 S6 kinase 1 pathway. J. Biol. Chem. 278, 45117–45127 [DOI] [PubMed] [Google Scholar]
- 5. Dorrello N. V., Peschiaroli A., Guardavaccaro D., Colburn N. H., Sherman N. E., Pagano M. (2006) S6K1- and βTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314, 467–471 [DOI] [PubMed] [Google Scholar]
- 6. Sheedy F. J., Palsson-McDermott E., Hennessy E. J., Martin C., O'Leary J. J., Ruan Q., Johnson D. S., Chen Y., O'Neill L. A. (2010) Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 11, 141–147 [DOI] [PubMed] [Google Scholar]
- 7. Yang H. S., Knies J. L., Stark C., Colburn N. H. (2003) Pdcd4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene 22, 3712–3720 [DOI] [PubMed] [Google Scholar]
- 8. Yang H. S., Jansen A. P., Komar A. A., Zheng X., Merrick W. C., Costes S., Lockett S. J., Sonenberg N., Colburn N. H. (2003) The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 23, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loh P. G., Yang H. S., Walsh M. A., Wang Q., Wang X., Cheng Z., Liu D., Song H. (2009) Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J. 28, 274–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hilliard A., Hilliard B., Zheng S. J., Sun H., Miwa T., Song W., Göke R., Chen Y. H. (2006) Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J. Immunol. 177, 8095–8102 [DOI] [PubMed] [Google Scholar]
- 11. Chang J. H., Cho Y. H., Sohn S. Y., Choi J. M., Kim A., Kim Y. C., Jang S. K., Cho Y. (2009) Crystal structure of the eIF4A-PDCD4 complex. Proc. Natl. Acad. Sci. U.S.A. 106, 3148–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shiota M., Izumi H., Tanimoto A., Takahashi M., Miyamoto N., Kashiwagi E., Kidani A., Hirano G., Masubuchi D., Fukunaka Y., Yasuniwa Y., Naito S., Nishizawa S., Sasaguri Y., Kohno K. (2009) Programmed cell death protein 4 down-regulates Y-box binding protein-1 expression via a direct interaction with Twist1 to suppress cancer cell growth. Cancer Res. 69, 3148–3156 [DOI] [PubMed] [Google Scholar]
- 13. Wolf C., Thisse C., Stoetzel C., Thisse B., Gerlinger P., Perrin-Schmitt F. (1991) The M-twist gene of Mus is expressed in subsets of mesodermal cells and is closely related to the Xenopus X-twi and the Drosophila twist genes. Dev. Biol. 143, 363–373 [DOI] [PubMed] [Google Scholar]
- 14. Li L., Cserjesi P., Olson E. N. (1995) Dermo-1: a novel twist-related bHLH protein expressed in the developing dermis. Dev. Biol. 172, 280–292 [DOI] [PubMed] [Google Scholar]
- 15. Sharabi A. B., Aldrich M., Sosic D., Olson E. N., Friedman A. D., Lee S. H., Chen S. Y. (2008) Twist-2 controls myeloid lineage development and function. PLoS Biol. 6, e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haidinger M., Poglitsch M., Geyeregger R., Kasturi S., Zeyda M., Zlabinger G. J., Pulendran B., Hörl W. H., Säemann M. D., Weichhart T. (2010) A versatile role of mammalian target of rapamycin in human dendritic cell function and differentiation. J. Immunol. 185, 3919–3931 [DOI] [PubMed] [Google Scholar]
- 17. Schmid T., Bajer M. M., Blees J. S., Eifler L. K., Milke L., Rübsamen D., Schulz K., Weigert A., Baker A. R., Colburn N. H., Brüne B. (2011) Inflammation-induced loss of Pdcd4 is mediated by phosphorylation-dependent degradation. Carcinogenesis 32, 1427–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmid T., Jansen A. P., Baker A. R., Hegamyer G., Hagan J. P., Colburn N. H. (2008) Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res. 68, 1254–1260 [DOI] [PubMed] [Google Scholar]
- 19. Martin M., Schifferle R. E., Cuesta N., Vogel S. N., Katz J., Michalek S. M. (2003) Role of the phosphatidylinositol 3 kinase-Akt pathway in the regulation of IL-10 and IL-12 by Porphyromonas gingivalis lipopolysaccharide. J. Immunol. 171, 717–725 [DOI] [PubMed] [Google Scholar]
- 20. Baker A. K., Wang R., Mackman N., Luyendyk J. P. (2009) Rapamycin enhances LPS induction of tissue factor and tumor necrosis factor-α expression in macrophages by reducing IL-10 expression. Mol. Immunol. 46, 2249–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weichhart T., Costantino G., Poglitsch M., Rosner M., Zeyda M., Stuhlmeier K. M., Kolbe T., Stulnig T. M., Hörl W. H., Hengstschläger M., Müller M., Säemann M. D. (2008) The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity 29, 565–577 [DOI] [PubMed] [Google Scholar]
- 22. Wang H., Brown J., Gu Z., Garcia C. A., Liang R., Alard P., Beurel E., Jope R. S., Greenway T., Martin M. (2011) Convergence of the mammalian target of rapamycin complex 1- and glycogen synthase kinase 3-β-signaling pathways regulates the innate inflammatory response. J. Immunol. 186, 5217–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cao S., Liu J., Song L., Ma X. (2005) The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J. Immunol. 174, 3484–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arbibe L., Mira J. P., Teusch N., Kline L., Guha M., Mackman N., Godowski P. J., Ulevitch R. J., Knaus U. G. (2000) Toll-like receptor 2-mediated NF-κB activation requires a Rac1-dependent pathway. Nat. Immunol. 1, 533–540 [DOI] [PubMed] [Google Scholar]
- 25. Sarkar S. N., Peters K. L., Elco C. P., Sakamoto S., Pal S., Sen G. C. (2004) Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat. Struct. Mol. Biol. 11, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 26. Santos-Sierra S., Deshmukh S. D., Kalnitski J., Küenzi P., Wymann M. P., Golenbock D. T., Henneke P. (2009) Mal connects TLR2 to PI3Kinase activation and phagocyte polarization. EMBO J. 28, 2018–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sellon R. K., Tonkonogy S., Schultz M., Dieleman L. A., Grenther W., Balish E., Rennick D. M., Sartor R. B. (1998) Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infection and immunity 66, 5224–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]