Background: Transcription elongation is a rate-limiting step for inducible gene expression. BRD4 must be released from chromatin to regulate transcription elongation.
Results: Protein phosphatase 1α (PP1α) and histone deacetylases (HDAC) signaling pathways are required for this process.
Conclusion: Histone cross-talk in trans between H3S10ph and H4K5ac/K8ac connects PP1α and HDACs signaling pathways to control functional transition of BRD4.
Significance: BRD4 is regulated epigenetically for controlling stress-induced gene expression.
Keywords: Histone Deacetylase (HDAC), histone Modification, Phosphoprotein Phosphatase 1 (PP1), Stress Response, Transcription Elongation Factor, BRD4, P-TEFb, Histone Acetylation, Histone Phosphorylation, Inducible Gene Expression
Abstract
Transcription elongation has been recognized as a rate-limiting step for the expression of signal-inducible genes. Through recruitment of positive transcription elongation factor P-TEFb, the bromodomain-containing protein BRD4 plays critical roles in regulating the transcription elongation of a vast array of inducible genes that are important for multiple cellular processes. The diverse biological roles of BRD4 have been proposed to rely on its functional transition between chromatin targeting and transcription regulation. The signaling pathways and the molecular mechanism for regulating this transition process, however, are largely unknown. Here, we report a novel role of phosphorylated Ser10 of histone H3 (H3S10ph) in governing the functional transition of BRD4. We identified that the acetylated lysines 5 and 8 of nucleosomal histone H4 (H4K5ac/K8ac) is the BRD4 binding site, and the protein phosphatase PP1α and class I histone deacetylase (HDAC1/2/3) signaling pathways are essential for the stress-induced BRD4 release from chromatin. In the unstressed state, phosphorylated H3S10 prevents the deacetylation of nucleosomal H4K5ac/K8ac by HDAC1/2/3, thereby locking up the majority of BRD4 onto chromatin. Upon stress, PP1α-mediated dephosphorylation of H3S10ph allows the deacetylation of nucleosomal H4K5ac/K8ac by HDAC1/2/3, thereby leading to the release of chromatin-bound BRD4 for subsequent recruitment of P-TEFb to enhance the expression of inducible genes. Therefore, our study revealed a novel mechanism that the histone cross-talk between H3S10ph and H4K5ac/K8ac connects PP1α and HDACs to govern the functional transition of BRD4. Combined with previous studies on the regulation of P-TEFb activation, the intricate signaling network for the tight control of transcription elongation is established.
Introduction
In eukaryotic cells, the transcription of protein-coding genes is performed by RNA polymerase II (Pol II),4 which is tightly regulated in multiple stages. Although control of the promoter recruitment of RNA Pol II for transcription initiation has been a long held paradigm for transcription regulation, recent studies indicate that transcription elongation is another rate-limiting step for the rapid expression of signal-inducible genes in metazoans (1–4). Genome-wide surveys revealed that for a vast array of inducible genes, transcription initiation has already been accomplished even at the uninduced state, yet RNA Pol II is stalled at promoter-proximal regions (5–9). Upon stimulation, P-TEFb (positive transcription elongation factor b), a heterodimeric kinase consisting of CDK9 and mostly Cyclin T1, is recruited to these regions to convert the poised Pol II into a productive elongation mode, thereby triggering the synthesis of full-length mRNA (2, 10, 11). Hence, the recruitment of active P-TEFb emerges as a key point for modulating inducible gene expression. In cells, the activity of P-TEFb is tightly regulated, with the majority of P-TEFb being sequestrated in an inactive complex (termed 7SK snRNP) that also contains 7SK snRNA and nuclear proteins HEXIM1, MePCE, and LARP7 (12, 13). During stress response, P-TEFb is liberated from 7SK snRNP through the concerted actions of signal-activated protein phosphatase PP2B and PP1α, thereby making P-TEFb available for promoter recruitment (14).
Bromodomain-containing protein BRD4, a ubiquitously expressed nuclear protein, is a key recruitment factor for P-TEFb (15, 16). BRD4 belongs to the BET protein family, the members of which have a consensus structure of two N-terminal tandem bromodomains (BDI and BDII) followed by an extra terminal domain (17). Early studies indicated that the bromodomains of BRD4 are essential for its association with acetylated chromatin (18, 19). A recent study using systematic peptide array screening combined with crystallography revealed that the BDI of BRD4, which simultaneously binds to diverse diacetylated histones, is the major contributor for its interaction with chromatin (20). Consistent with its chromatin-targeting nature, BRD4 was found to be persistently associated with acetylated chromosomes during mitosis in many cell lines (18, 19). Later studies demonstrated that this association is crucial for the rapid expression of early G1 genes upon exiting mitosis (21–24). Although the mechanism is still unclear, BRD4 has been proposed to function in transmitting the epigenetic memory across cell division by marking the transcriptionally active genes (23, 25–27). Moreover, the association of BRD4 with mitotic chromosome was shown to be important for maintaining chromatin compaction (28), and facilitating the even segregation of viral DNAs into daughter cells during cell division (29).
Distinct from the other BET proteins, BRD4 contains a unique P-TEFb interaction domain (PID) at its extreme C terminus (30). In response to external signals, BRD4 associates with and recruits P-TEFb to gene promoters to stimulate transcription elongation of inducible genes (15, 16, 31–33). Consistent with its role in P-TEFb recruitment, BRD4 was shown to be crucial for cell cycle progression (21–23), HIV-1 transcription (15, 16, 34, 35), inflammatory response (33, 36, 37), and cardiac hypertrophy (38, 39). Moreover, BRD4 was identified by shRNA library screening as an essential factor for cancer development (40, 41). Studies with small molecule inhibitors of the BET proteins, such as JQ1 and I-BET, revealed the critical role of BRD4 in the development of several hematopoietic and somatic cancers, such as Burkitt's lymphoma, multiple myeloma (41–43), melanoma (44), colon (45), and breast cancer (46).
The diverse biological roles of BRD4 have been proposed to rely on its functional transition between chromatin targeting and transcriptional regulation (17, 25). In line with this notion, our recent study showed that, besides P-TEFb, the availability of BRD4 is also highly regulated (31). The survey of a variety of cell lines showed that almost all BRD4 is associated with chromatin in interphase cells and there is limited chromatin-free BRD4 available in the unstimulated state. Upon stress, BRD4 is released from chromatin, and subsequently it recruits P-TEFb to enhance expression of the HIV-1 gene (31). The signaling pathways and molecular mechanism for this process, however, was unknown.
Here, we report a novel role of phosphorylated serine 10 of nucleosomal histone H3 (H3S10ph) in governing the functional transition of BRD4 during stress response. By combining in vivo and in vitro experiments, we identified that both PP1α and histone deacetylase HDAC1/2/3 signaling pathways are essential for releasing chromatin-bound BRD4 for P-TEFb recruitment, which relies on histone cross-talk in trans between H3S10ph and H4K5ac/K8ac (acetylated lysine 5 and 8 of histone H4). In this context, the dephosphorylation of H3S10ph facilitates the expression of inducible genes. The function of the PP1α signaling pathway in coordinating BRD4 and P-TEFb activation for tight control of gene expression is discussed.
EXPERIMENTAL PROCEDURES
Chemicals
Trichostatin A and microcystin LR were from Santa Cruz Biotechnology. Doxorubicin (DOX), Entinostat (MS-275), and cyclosporin A were from LC Laboratories. Hexamethylene bisacetamide (HMBA) and nocodazole were from Sigma. Recombinant PP1α enzyme was from New England Biolabs. Micrococcal nuclease and the reverse transcriptase M-MLV Kit were from Takara Biotech (Dalian, China). DyNAmoTM ColorFlash Master Mix from Thermo. All other chemicals were from Amresco or Sigma.
Antibodies
Rabbit anti-HDAC1, -HDAC2, and -HDAC3 antibodies were from Proteintech. Rabbit anti-H3K14ac and H3K9ac from Cell Signaling. Rabbit anti-histone H4, H4K5ac, H4K8ac, H4K12ac, H4K16ac, H3K4me3, and H3K27me3 antibodies from Millipore. Rabbit anti-H3K36me3 was from Abcam. Rabbit anti-histone H3, H3S10ph, goat anti-histone H2A, H2B, and mouse anti-PP1 antibodies were from Santa Cruz Biotechnology. Mouse anti-β-ACTIN antibody, anti-HA-agarose beads, and anti-FLAG M2 affinity gel from Sigma. Rat anti-HA antibody was from Roche Applied Science. Rabbit anti-CDK9, Cyclin T1, HEXIM1, and BRD4 antibodies were raised in GeneScript (Nanjing, China) against the following peptides: RRKGSQITQQSTNQ (CDK9, amino acids 343–356), SGNTDKPRPPPLPS (Cyclin T1, amino acids 702-715), HRQQERAPLSKFGD (HEXIM1, amino acids 346–359), and SSQPQSMLDQQREL (BRD4, amino acids 1314–1327).
Plasmids
The ORF fragments of human histone H3 (NM_002107.4), H4 (NM_003545.3), HDAC1 (NM_004964), HDAC2 (NM_001527), and HDAC3 (NM_003883) were amplified by RT-PCR from RNA isolated from HeLa cells. The PCR fragments were inserted into BamHI/XbaI sites of a modified pLV-FLAG and pLV-HA lentiviral vectors (31). The nucleotide sequences of primers used in PCR are as following: 5′-CGC GGA TCC ATG GCT CGT ACA AAG CAG ACT G (forward) and 5′-GCC TCT AGA AGC ACG TTC TCC ACG TAT GC (reverse) for histone H3; 5′-CGC GGA TCC ATG TCT GGT CGC GGC AAA GGC (forward) and 5′-GCC TCT AGA GCC GCC GAA GCC GTA AAG AGT G (reverse) for histone H4; 5′-CGC GGA TCC ATGG CGC AGA CGC AGG GCA C (forward) and 5′-GCC TCT AGA GGC CAA CTT GAC CTC CTC CTT G (reverse) for HDAC1; 5′-CGC GGA TCC ATG GCG TAC AGT CAA GGA GGC (forward) and 5′-GCC TCT AGA GGG GTT GCT GAG CTG TTC TGAT T (reverse) for HDAC2; and 5′-CGC GGA TCC ATG GCC AAG ACC GTG GCC TAT TTC (forward) and 5′-GCC TCT AGA AAT CTC CAC ATC GCT TTC CTT GTC (reverse) for HDAC3. To generate mutant histone H3 without the Ser10 phosphorylation site, mutant histone H4 without K5- and/or K8-acetylation sites, and dominant-negative forms of HDACs, the following point mutations were introduced into corresponding plasmids by using COP-QuikChange (COP-QC) protocol (47): S10A for histone H3; K5R, K8R, or K5R/K8R for histone H4; H141A for HDAC1; H142A for HDAC2; and S423A for HDAC3. The primers used in COP-QC are as following: 5′-GCC CGC AAA GCC ACC GGT GGT AAA GCA CCC AG (forward) and 5′-ACC ACC GGT GGC TTT GCG GGC AGT CTG CTT TG (reverse) for S10A-H3, 5′-GGG CGA GGT CGC GGT GGC AAG GGG CTG (forward) and 5′-CTT GCC ACC GCG ACC TCG CCC AGA CAT (reverse) for K5R-H4; 5′-GGG CTG GGT CGC GGA GGC GCC AAG CGC CAC (forward) and 5′-GCG CCT CCG CGA CCC AGC CCC TTG CCA C (reverse) for K8R-H4; 5′-GGG CGA GGT CGC GGC GGA CGC GGA CTG GGT AAA GGA G (forward) and 5′-ACC CAG TCC GCG TCC GCC GCG ACC TCG CCC AGA C (reverse) for K5R/K8R-H4; 5′-GGC CTG CAC GCT GCA AAG AAG TCC GAG GC (forward) and 5′-GAC TTC TTT GCA GCG TGC AGG CCC CCA GC (reverse) for H141A-HDAC1; 5′-GAG GAT TAC ATG CTG CTA AGA AAT CAG AAG CAT (forward) and 5′-GAT TTC TTA GCA GCA TGT AAT CCT CCA GCC CAA (reverse) for H142A-HDAC2; and 5′-CCG GAA TTC ATG GCC AAG ACC GTG GCC TAT T (forward) and 5′-CGC GGA TCC AAT CTC CAC ATA GCT TCC TTG TCA T (reverse) for S423A-HDAC3. The previously described wild-type and H66N-mutant (inactive) PP1α (14) were subcloned into BamHI/XbaI sites of pLV-FLAG and pLV-HA lentiviral vector. All cDNAs were verified by sequencing.
shRNA Constructs
The short hairpin RNAs (shRNAs) targeting human HDAC1, HDAC2, and HDAC3 mRNA were cloned into modified pSicoR vector (31). The shRNAs in pSicoR vector targeting human PP1α and BRD4 mRNA were described previously (14, 31). The 19-nucleotide sequences of shRNAs are as following: shHDAC1, 5′-CTA TGG TCT CTA CCG AAA A; shHDAC2, 5′-AGC ATC AGG ATT CTG TTA C; shHDAC3, 5′-GCA TTG ATG ACC AGA GTT A; shBRD4, 5′-GAA CCT CCC TGA TTA CTA T; shPP1α#1, 5′-GAT CAA GTA CCC CGA GAA C; and shPP1α#2, 5′-TGC TGG CGC CAT GAT GAG T.
Cell lines, Transfection, and Infection
HEK293T, HeLa, and HeLa-based F1C2(CDK9-f) cells stably expressing FLAG-tagged CDK9 subunit of P-TEFb, and HeLa cells with an integrated HIV-LTR-luciferase reporter gene (HIV-LTR-Luc) were maintained as previously described (14, 31, 48, 49). Cells at ∼80% confluence were transfected with various cDNA constructs using a PEI transfection protocol as described previously (14). For puromycin selection, the constructs were co-transfected at a ratio of 5:1 with pBabe-puro vector that harbors a puromycin-resistant gene. Two days after transfection, the cells were selected in medium containing 1 μg/ml of puromycin for 36–48 h. The lentiviral infection was performed as described previously (31). For silencing PP1α, two shRNA lentiviral constructs were used at a 1:1 ratio.
Treatment of Cells with UV or Pharmacological Compounds
Cells at ∼50% confluence were preincubated with solvent, or the inhibitor trichostatin A (400 nm, 2 h), MS-275 (5 μm, 2 h), cyclosporin A (5 μm, 1 h), or microcystin LR (15 nm, 15 min), followed by treatment with 5 μg/ml of DOX (1 h), 10 mm HMBA (2 h). For UV irradiation, cells were exposed to 254 nm UV at 80 J/m2 without culture medium, and then incubated in the original medium for 1 h. Afterward, the cells were subjected to stepwise extraction with modified nuclear fractionation (MONF) protocol (31).
Preparation of LSF, HSF, NLF, and LSEN with MONF Protocol
The low salt fraction (LSF), high salt fraction (HSF), nuclear lysate (NLF), and low salt-extracted nuclei (LSEN) were prepared with MONF (31). Briefly, the buffer A-swollen cells were subjected to low salt extraction by incubating with 2× packed cell volume of buffer A (with 1% Nonidet P-40) to break the plasma membrane. After centrifugation, the supernatant was saved. The nuclei pellet was further extracted twice by resuspending in 1× packed cell volume of low salt buffer (75 mm), followed by centrifugation. The supernatants of the three extractions were combined and saved as the LSF. The LSEN were subsequently incubated with 6× packed cell volume of high salt buffer (0.3 m). After centrifugation, the supernatant was saved as the HSF. The high salt-extracted nuclei were lysed with SDS-loading buffer to yield NLF.
Immunopurification
FLAG-tagged proteins and their associated factors were isolated by anti-FLAG immunoprecipitation from LSF or HSF of F1C2 (CDK9-f) cells or transfected HeLa cells as described previously (31, 48). The levels of proteins were determined by Western blotting. To obtain stress-activated HDAC1, -2, or -3 enzymes, HEK293T cells transfected with FLAG- or HA-tagged HDAC expression vectors were treated with UV, DOX, or HMBA, followed by anti-FLAG or anti-HA affinity purification (48). To isolate intact nucleosomes, HeLa cells were infected with lentivirus expressing FLAG-tagged histone H3 or H4 for at least 96 h, and the LSEN prepared from ∼107 cells were digested with micrococcal nuclease (10 units in a total of 300 μl of low salt buffer) at 37 °C for 20 min to release mononucleosomes into supernatant. After centrifugation at 10,000 × g, 4 °C for 5 min, the supernatant was subjected to anti-FLAG affinity isolation of nucleosome. To obtain H3/H4 dimers, the LSEN were digested with 20 units of micrococcal nuclease at 37 °C for 1 h. The supernatant was adjusted to a salt concentration of 0.8 m NaCl to disrupt nucleosomes before anti-FLAG affinity isolation of H3/H4 dimers. To purify histone proteins, cells were lysed with high salt cell lysis buffer (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 0.5 mm EDTA, 0.5 m NaCl, 1% Triton X-100, 1 mm DTT, 0.5 mm PMSF, 1× protease inhibitor mixture), and sonicated before affinity isolation. The purity of isolated H3 was analyzed by Western blotting to verify that the H3 preparation is free of H4, and vice versa.
In Vitro Assay for BRD4 Release
To determine the effect of PP1α and/or HDAC enzymatic treatments on BRD4 release from chromatin or nucleosomes, the LSEN or nucleosomes were immobilized on anti-FLAG resin via H3-f or H4-f, incubated with 1.0 unit of recombinant PP1α and/or stress-activated f-HDAC1, -2, or -3 enzymes in 40 μl of 1× PP1 reaction buffer at 30 °C for 1 h. After centrifugation at 10,000 × g for 2 min at 4 °C, the supernatant and nuclear pellet or immobilized nucleosomes were saved for Western blotting. For two-round sequential incubation of LSEN, extensive washes with 1× PP1 reaction buffer was performed between the two incubations.
In Vitro Pulldown Assay with H3/H4 Dimers
The H3/H4 dimers were isolated as described above, and immobilized on anti-FLAG beads via H3-f or H4-f. After extensive washes with D0.8 M buffer and then D0.1 M buffer, the immobilized H3/H4 dimers were preincubated with 2 μg/μl of BSA (15 μl of beads in a 20-μl volume) for 15 min, followed by incubation with affinity-purified HA-tagged BRD4 at 30 °C for 15 min. The unbound proteins were washed away with D0.1 M buffer and the immobilized complex was eluted with FLAG peptide for Western blotting to determine the H3/H4 dimer-associated BRD4.
Luciferase Assay
HeLa cells with an integrated HIV-LTR-luciferase reporter gene (HIV-LTR-Luc) were infected with lentivirus expressing the desired proteins for 48–96 h, followed by incubation with 5 mm HMBA for 3–4 h as indicated. Cell lysates were prepared for measuring the luciferase activity (48). Data were represented as fold-enhancement compared with untreated cells. The error bars were standard deviations based on three independent experiments.
Quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from cells using TRIzol (Invitrogen) and treated with RQ1 DNase (Promega). Two micrograms of total RNA were used for reverse transcription, followed by qPCR as described previously (31, 48). Data were presented as fold-enhancement compared with untreated cells (31, 48). The qPCR primers are the following: 5′-CTA CCA CTC ACC CGC AGA CT (forward) and 5′-GTG GGA ATG AAG TT GG CAC T (reverse) for c-fos; 5′-GAC CTT ATG GCT ACA GTA AC (forward) and 5′-TGA GGA GGT CCG AGT TCT TG (reverse) for c-Jun; 5′-GAC GCA GAT CTT CAC CAC CT (forward) and 5′-GCC CCA ACA GAT TGT TGT CT (reverse) for HSP70; 5′-CAT TCT CTG TGG TAT CCA AG (forward) and 5′-ACC CTA CAA CAG ACC CAC AC (reverse) for IL-8; and 5′-ATC GTC CAC CGC AAA TGC TTC T (forward) and 5′-AGC CAT GCC AAT CTC ATC TTG T (reverse) for β-ACTIN. For measuring the initiation and elongation transcripts of HIV-LTR-Luc, the following PCR primers were used: (+1 ∼ +59, TAR region to represent initiation) 5′-GGG TCT CTC GAG TTA GAC CAG ATC TGA (forward) and 5′-GGG TTC CCT AGT TAG CCA GAG AGC (reverse), and (+468 ∼ +593, Luc region to represent elongation) 5′-CGC AGC CTA CCG TAG TGT TTG (forward) and 5′-ACT GAA ATC CCT GGT AAT CCG TT (reverse).
Chromatin Immunoprecipitation (ChIP)
ChIP was performed using HeLa cells containing an integrated HIV-LTR-Luc as described previously (31). Immunoprecipitated DNA was analyzed by real-time PCR with DyNAmoTM ColorFlash Master Mix (Thermo) and the primers for the promoter region and gene body of the following genes: HIV-LTR (promoter region: forward, 5′-GCT GAT ATC GAG CTT GCT AC and reverse, 5′-CCA ACA GTA CCG GAA TGC C, and gene body: forward, 5′-CCA TCT TCC AGG GAT ACG AC and reverse, 5′-GGC GTT GGT CGC TTC CGG), HSP70 (promoter region: forward, 5′-TGG ACA AGT GTC AAG AGG TC and reverse, 5′-CCT GGT ACA GTC CGC TGA TG, and gene body: forward, 5′-AAG GAC ATC AGC CAG AAC AAG CGA and reverse 5′-ACG TGT AGA AGT CGA TGC CCT CAA), c-fos (promoter region: forward, 5′-GGA ATT AAC CTG GTG CTG GAT ATT TTC and reverse, 5′-CAC CTC AAC AAT GCA TGA TCA GTA ACA, and gene body: forward, 5′-GCA CAA ATA ATG GCT GAT CGT and reverse, 5′-TCA GAG TCA TGT TGA CTT CTC C), c-jun (promoter region: forward, 5′-GCC AAC TCA TGC TAA CGC AG and reverse 5′-TTC TCT CCG TCG CAA CTT GT, and gene body: forward, 5′-GCC AAC TCA TGC TAA CGC AG and reverse, 5′-TTC TCT CCG TCG CAA CTT GT), and IL-8 (promoter region: forward, 5′-TGT CAT TGC CAG CTG TGT TG and reverse, 5′-AAC AAG TTT CAA CCA GCA AG). The PCR amplification was performed on Eppendorf Mastercycler ep realplex2 with the following program: 95 °C for 7 min followed by 40 cycles of 10 s at 95 °C, 30 s at 60 °C for annealing and extension. The results from two independent experiments were averaged and plotted as percentage of input.
Flow Cytometry Analysis
Cells were trypsinized, washed three times with PBS, and 1–2 × 106 cells were resuspended in 0.5 ml of PBS. The cells were dropped slowly into 2 ml of cold 87.5% ethanol with gentle mixing, and kept at 4 °C overnight. Before flow cytometry analysis, cells were centrifuged at 300 × g for 5 min, and the cell pellet was resuspended in 400 μl of PBS, digested with 100 μg/ml of RNaseA at 37 °C for 30 min, and incubated with 50 μg/ml of propidium iodine for 30 min in the dark. To synchronize cells to metaphase, the cells were incubated with 50 ng/ml of nocodazole for 16 h. For serum starvation, the cells were cultured without serum for 40 h.
RESULTS
Stress Induces H4K5ac/K8ac Deacetylation and H3S10ph Dephosphorylation
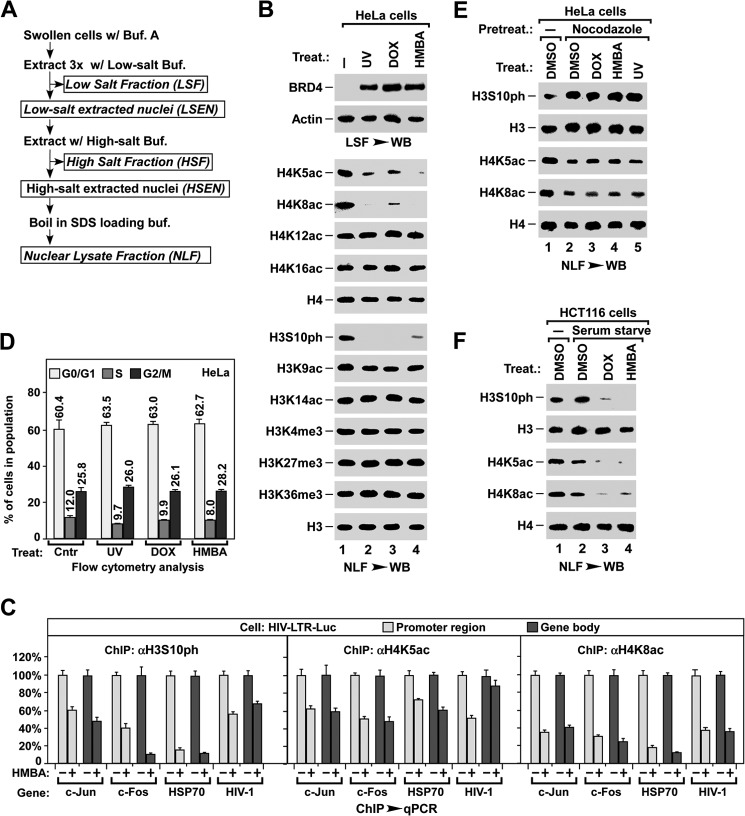
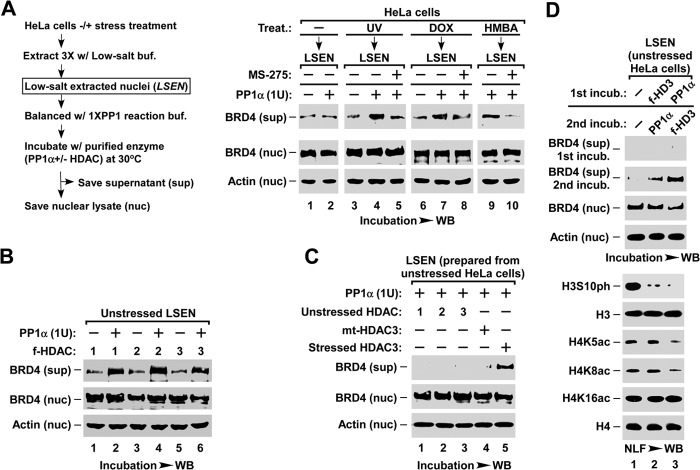
By using a MONF protocol (31) (outlined in Fig. 1A), our previous study showed that stress-induced release of chromatin-bound BRD4 is essential for the induction of HIV-1 expression (31). To identify the signal pathway(s) required for BRD4 release from chromatin, we first surveyed the changes in histone modifications after UV, DOX, or HMBA treatment (Fig. 1B). Intriguingly, concomitant with the release of chromatin-bound BRD4 as detected in the LSF (containing chromatin-free proteins), both the levels of acetylated lysine 5 and 8 of nucleosomal histone H4 (H4K5ac/K8ac) and phosphorylated serine 10 of nucleosomal histone H3 (H3S10ph) were reduced upon stress treatments (Fig. 1B). Consistently, the ChIP assay showed that HMBA treatment induced the reduction of H3S10ph and H4K5ac/K8ac at the promoter region or gene body of several endogenous genes as well as HIV-LTR-Luc reporter gene (Fig. 1C).
FIGURE 1.
Stress induces H4K5ac/K8ac deacetylation and H3S10ph dephosphorylation. A, outline for the MONF protocol. B, effect of stress treatment on the histone modifications of nucleosomal H3 and H4. NLF was prepared from HeLa cells with the indicated treatment and analyzed for the indicated modifications by Western blot (WB). C, ChIP-qPCR analysis of nucleosomal H3S10ph, H4K5ac, and H4K8ac levels in promoter and gene body regions of the indicated genes after HMBA treatments of HIV-LTR-luciferase cells. The levels in non-treated cells were set as 100%. D, flow cytometry analysis of cell cycle distribution after indicated stress treatments of non-synchronized HeLa cells. E, effect of stress on H3S10ph and H4K5ac/K8ac in nocodazole-synchronized HeLa cells. F, effect of stress on H3S10ph and H4K5ac/K8ac in serum-starved HCT116 cells.
As the level of H3S10ph was known to be greatly increased during metaphase (50), we examined whether stress treatment reduced the H3S10ph level by blocking cell cycle progression into metaphase, or by promoting cells to exit from metaphase. As shown in Fig. 1D, 1 h after UV treatment, or DOX or HMBA treatments for 2 h, did not change the cell cycle distribution of HeLa cells. When cells were synchronized to metaphase by nocodazole, the levels of nucleosomal H3S10ph or H4K5ac/K8ac were not affected by the stress treatments (Fig. 1E). In contrast, stress induced the H3S10ph dephosphorylation and H4K5ac/K8ac deacetylation in HCT116 cells that had been synchronized to the G0 phase by 40 h of serum starvation (Fig. 1F). The stress-induced H3S10ph dephosphorylation was also observed in both starved and non-starved HeLa cells (data not shown). Taken together, these observations suggest that stress-induced H3S10ph dephosphorylation and H4K5ac/K8ac deacetylation occur only in interphase and are not due to changes in cell cycle distribution.
H4K5ac/K8ac Are the Sites for BRD4 Binding to Nucleosome in Vivo
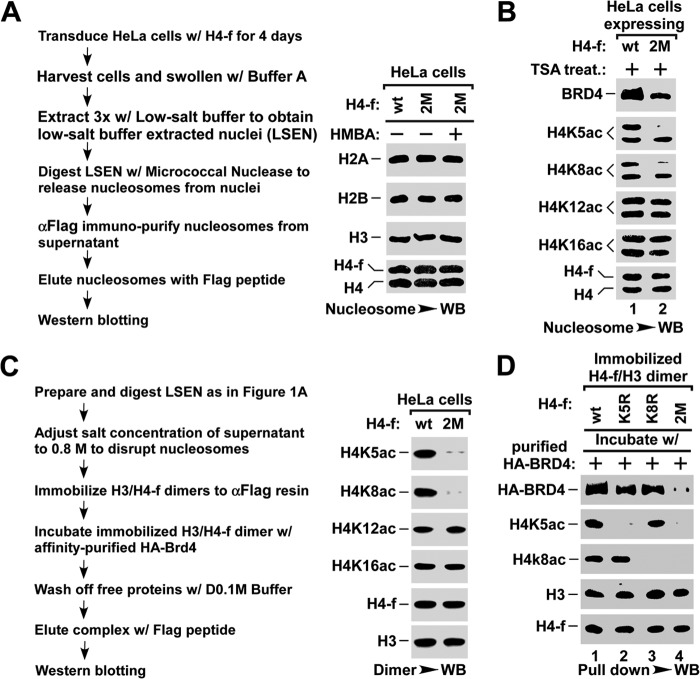
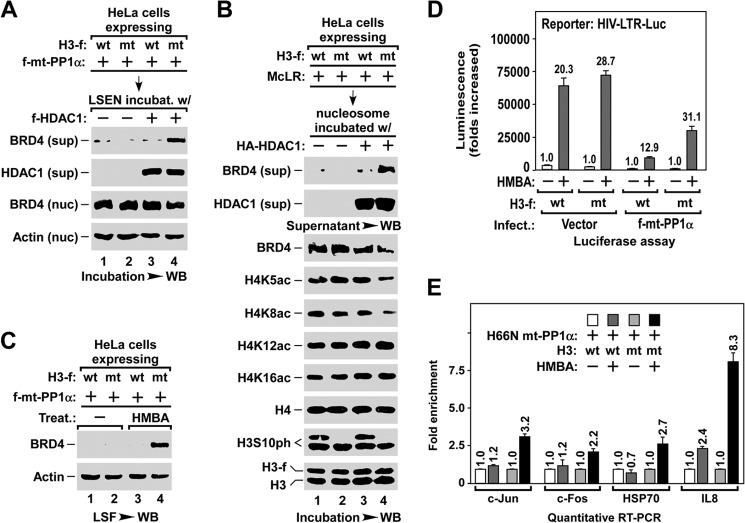
BRD4 has been shown to associate with H4K5ac/K8ac by an in vitro peptide binding assay (20). Here we tested whether these two sites are required for the chromatin association of BRD4 in vivo. To this end, we generated mutant histone H4 by substituting lysine 5 and/or 8 with arginine (K5R/K8R). HeLa cells were infected with lentivirus expressing FLAG-tagged wild type (wt-) or K5R/K8R double mutant (2M-) H4, followed by treatment with trichostatin A, a broad spectrum HDAC inhibitor, to ensure the maximal association of BRD4 with acetylated chromatin (31). The nucleosomes were immunoaffinity purified and the level of associated BRD4 was examined by Western blot (outlined in Fig. 2A). Notably, for the nucleosomes containing about half the amount of 2M-H4, the level of associated BRD4 was also reduced to half (Fig. 2B), suggesting that H4K5ac/K8ac are necessary for the binding of BRD4 to nucleosome. Moreover, we performed an in vitro pulldown assay to test the association of affinity purified HA-BRD4 with f-H4/H3 dimers that were immobilized via FLAG-tagged H4 (outlined in Fig. 2C). H4/H3 dimers containing wt, K5R, or K8R H4 retained a large amount of HA-BRD4, but those containing 2M-H4 retained little HA-BRD4 (Fig. 2D). These data indicate that H4K5ac/K8ac is essential for the in vivo binding of BRD4 to nucleosome.
FIGURE 2.
BRD4 binds to nucleosome via H4K5ac/K8ac sites in vivo. A, outline for preparing nucleosome (left panel). Nucleosomes were affinity purified from HeLa cells infected with lentivirus expressing FLAG-tagged wild-type (wt) or K5R/K8R-mutant (2M) H4 using anti-FLAG resin, and analyzed by Western blot (WB) for core histones (right panel). B, the association of BRD4 with nucleosomes containing wt- or K5R/K8R 2M-H4. The nucleosomes were affinity purified from HeLa cells with the indicated lentiviral infection and treatment, and were assayed for the associated BRD4 and the histone modifications by Western blot. C, outline for H3/H4 dimer pulldown assay (left panel) and Western blot analysis of the indicated acetyl-lysine of histone H4 (right panel). D, pulldown assay for the binding of BRD4 to H3/H4 dimers containing wt-, K5R-, K8R-, or K5R/K8R 2M-H4. The H3/H4 dimers containing the indicated H4-f were immobilized onto anti-FLAG resin and incubated with purified HA-BRD4. The amounts of BRD4 bound to H3/H4 dimers were assayed by Western blot.
HDAC1/2/3 Are Required for Stress-induced BRD4 Release and Inducible Gene Expression
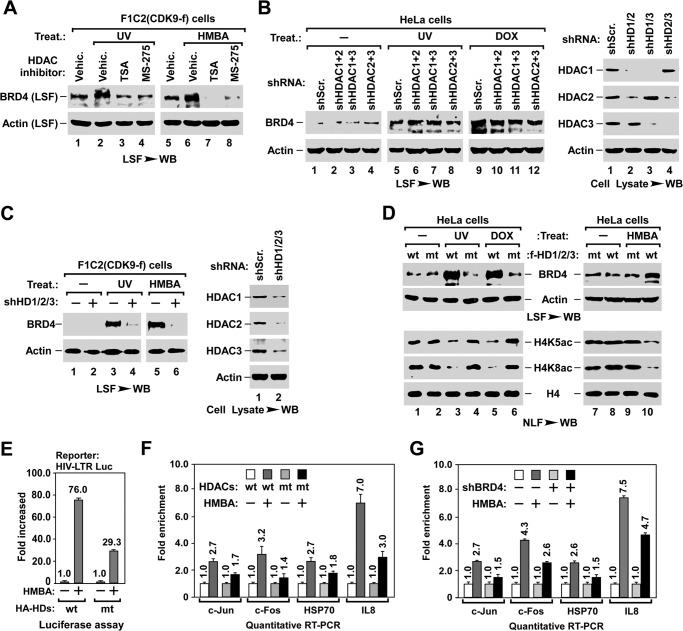
Next, we identified which HDAC was responsible for H4K5ac/K8ac deacetylation and BRD4 release during stress treatments. Humans have 11 HDACs belonging to four classes (51). We first focused on the ubiquitously expressed class I HDAC that includes HDAC1, -2, -3, and -8. Pre-treating cells with MS-275, which preferentially inhibits HDAC1/2/3 but not HDAC8 or other classes of HDACs (52), blocked stress-induced BRD4 release from chromatin with comparable efficiency to the wide spectrum HDAC inhibitor trichostatin A (Fig. 3A), suggesting that HDAC1/2/3 are required for this process. Unexpectedly, knocking down any one (data not shown) or any two of the three HDACs by shRNA (Fig. 3B) failed to block stress-induced release of chromatin-bound BRD4. Only when all three HDACs were knocked down was the stress-induced BRD4 release impaired (Fig. 3C), suggesting that HDAC1/2/3 are functionally redundant for stress-induced BRD4 release.
FIGURE 3.
HDAC1/2/3 are essential for stress-induced BRD4 release and inducible gene expression. A, effect of HDAC inhibitors on stress-induced BRD4 release. LSF was prepared from F1C2(CDK9-f) cells with the indicated HDAC inhibitor pre-treatment and stress treatment, and analyzed for the released BRD4 by Western blot (WB). B, effect of knocking down two of the three HDACs on stress-induced BRD4 release. LSF was prepared from F1C2(CDK9-f) cells with the indicated lentiviral co-infection and treatment, and assayed by Western blot. C, inhibitory effect of knocking down HDAC1/2/3 on stress-induced BRD4 release. D, inhibitory effect of co-expressing three inactive mutant (mt-) HDAC1/2/3 on stress-induced BRD4 release (top) and deacetylation of nucleosomal histone H4 (bottom). LSF and NLF prepared from F1C2(CDK9-f) cells with the indicated lentiviral co-infection and treatment were analyzed by Western blot. E and F, inhibitory effect of co-expressing mt-HDAC1/2/3 on HMBA-triggered expression of inducible genes. HIV-LTR-Luciferase cells with the indicated lentiviral co-infection and HMBA treatment were subjected to luciferase assay for the expression of HIV-LTR-Luc (E) and qRT-PCR analysis of the transcription of indicated endogenous genes (F). Data from three independent experiments were averaged and presented as fold-enhancement compared with untreated sample. G, knocking down BRD4 impairs inducible gene expression. HeLa cells with the indicated lentiviral infection and HMBA treatment were subjected to qRT-PCR analysis of the expression levels of the indicated genes.
In line with the HDAC knockdown experiments, ectopically expressing any one of the enzymatically inactive mutants of HDAC1/2/3 (mt-HDAC1/2/3) failed to block stress-induced BRD4 release (data not shown), whereas co-expressing all three mt-HDACs blocked both of the stress-induced deacetylation of H4K5ac/K8ac and the release of BRD4 (Fig. 3D), as well as the HMBA-induced expression of the HIV-LTR-luciferase reporter gene (HIV-LTR-Luc) (Fig. 3E) and endogenous genes (Fig. 3F) whose transcriptions are BRD4-dependent (Fig. 3G). Notably, without stress treatment, co-expressing wt-HDACs induced neither the release of BRD4, nor the deacetylation of H4K5ac/K8ac (Fig. 3D, lane 1), indicating that the HDAC pathway is necessary but not sufficient for mediating BRD4 release.
PP1α Pathway Is Involved in Stress-induced BRD4 Release and Inducible Gene Expression
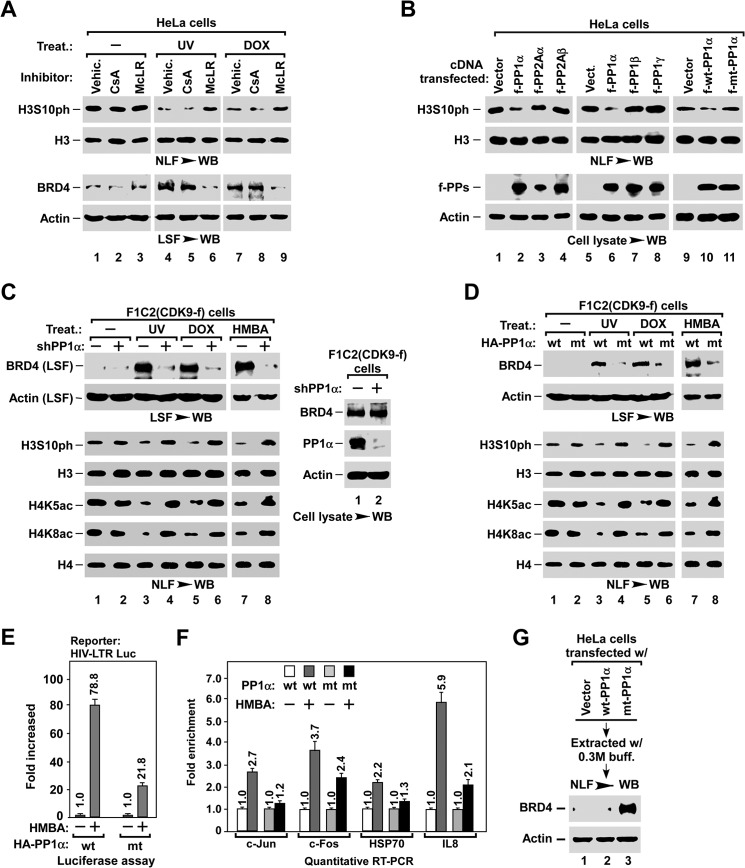
The observation that H3S10ph was dephosphorylated upon stress treatment (Fig. 1B) prompted us to study its functional relevance to BRD4 release. Remarkably, when stress-induced H3S10ph dephosphorylation was blocked by pre-treatment with the PP1/PP2A inhibitor microcystin LR (14), BRD4 release was also abolished, whereas the PP2B inhibitor cyclosporin A did not affect either process (Fig. 4A), suggesting that the PP1 or PP2A signaling pathways may be involved in this process. There are three isoforms of PP1 and two isoforms of PP2A in mammals (53). To identify which is responsible for H3S10ph dephosphorylation and BRD4 release, we examined whether ectopically expressing individual PP1 or PP2A isoforms could induce H3S10ph dephosphorylation. Significantly, overexpressing PP1α, but not the other phosphatases or the inactive form of H66N-mutant (mt-) PP1α, reduced the level of H3S10ph (Fig. 4B).
FIGURE 4.
PP1α is required for stress-induced BRD4 release and inducible gene expression. A, effect of phosphatase inhibitors on stress-induced H3S10ph dephosphorylation (top) and BRD4 release (bottom). LSF and NLF were prepared from HeLa cells with the indicated pre-treatment and treatment, and analyzed by Western blot (WB). McLR, PP1/PP2A inhibitor microcystin LR; CsA, PP2B inhibitor cyclosporin A. B, effect of exogenous expression of PP1 and PP2A isoforms on H3S10ph dephosphorylation in NLF (top). The levels of expressed protein phosphatases in cell lysates (bottom) were determined by Western blot (WB). C and D, inhibitory effect of knocking down PP1α (C) or overexpressing H66N mt-PP1α (D) on stress-induced BRD4 release and histone modifications. LSF and NLF were prepared from F1C2(CDK9-f) cells with the indicated lentiviral infection and treatment, and analyzed by Westen blot. E and F, inhibitory effect of overexpressing H66N mt-PP1α on HMBA-induced expression of inducible genes. HIV-LTR-Luc cells with the indicated lentiviral infection and HMBA treatment were subjected to luciferase assay (E) and qRT-PCR analysis (F) as described in the legend to Fig. 3, E and F. G, effect of overexpressing H66N mt-PP1α on the association of BRD4 with chromatin. NLF was prepared from D0.3 M high salt buffer-extracted nuclei of HeLa cells with the indicated lentiviral infection, and analyzed for the remaining BRD4 by Western blot.
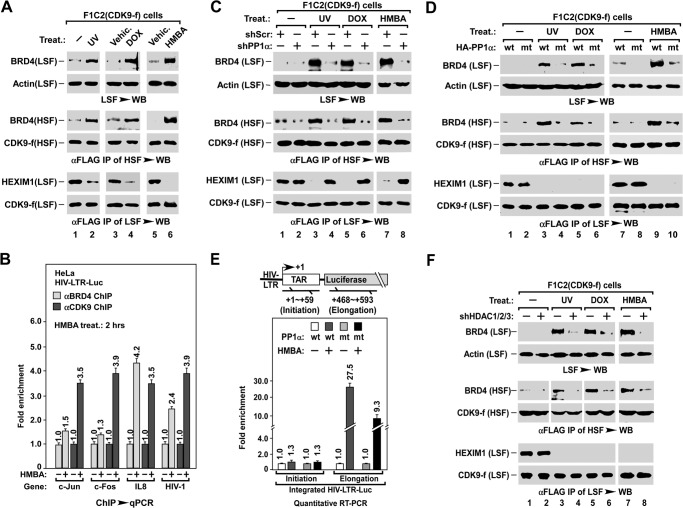
Next, we tested the role of PP1α in stress-induced BRD4 release. When PP1α was silenced with shPP1α, both the stress-induced H3S10ph dephosphorylation and BRD4 release were blocked (Fig. 4C). Moreover, ectopically expressing H66N mt-PP1α also abolished stress-induced H3S10ph dephosphorylation and BRD4 release (Fig. 4D), as well as HMBA-induced expression of HIV-LTR-Luc (Fig. 4E) and endogenous genes (Fig. 4F). Interestingly, impairing PP1α activity also abolished the stress-induced deacetylation of H4K5ac/K8ac (Fig. 4, C and D), suggesting that the PP1α pathway has an effect on the function of HDACs (see below). In line with these observations, overexpression of mt-PP1α enhanced the association of BRD4 with chromatin, as indicated by the high level of BRD4 remaining in nuclei after high salt extraction (Fig. 4G). Of note, neither knocking down PP1α nor overexpressing H66N mt-PP1α affected the expression level of BRD4 (data not shown). These data indicate that the PP1α pathway is also required for stress-induced BRD4 release.
PP1α and HDAC1/2/3 Act Sequentially to Induce BRD4 Release
Because both HDAC1/2/3 and PP1α pathways are involved in stress-induced BRD4 release from chromatin, we tested whether they are sufficient for this process. To this end, we employed an in vitro assay (outlined in Fig. 5A, left) by incubating the LSEN, which contains a significant amount of endogenous HDAC1/2/3 (data not shown), with recombinant PP1α enzyme. Interestingly, whereas PP1α failed to induce the release of chromatin-bound BRD4 in the LSEN that were prepared from unstressed HeLa cells (Fig. 5A, lane 2), it successfully induced BRD4 release in the LSEN prepared from stressed cells (lanes 4, 7, and 9). Remarkably, this release could be blocked by preincubating LSEN with HDAC1/2/3 inhibitor MS-275 (Fig. 5A, lanes 5, 8, and 10), suggesting that HDAC1/2/3 in the unstressed LSEN are functionally inactive, and only the stress-activated HDACs could induce BRD4 release. Hence, we affinity-purified HDACs from stress-treated HEK293T cells. When LSEN from unstressed cells were incubated with PP1α enzyme plus any one of the HDAC1/2/3 purified from stressed cells (Fig. 5B), BRD4 was released, whereas the HDACs purified from unstressed cells failed to do so (Fig. 5C). These data indicate that the stress-activated HDAC1/2/3 and PP1α pathways are sufficient for inducing BRD4 release from chromatin.
FIGURE 5.
Stress-activated HDAC1/2/3 and PP1α act cooperatively to induce BRD4 release. A, in vitro assay of PP1α-induced BRD4 release in LSEN. LSEN prepared from stressed or unstressed HeLa cells was preincubated with HDAC1/2/3 inhibitor MS-275, followed by incubation with recombinant PP1α enzyme as indicated. For A–D, the levels of BRD4 released into supernatant (sup) or those remaining in chromatin (nuc) were analyzed by Western blot (WB). B, in vitro assay of BRD4 release in LSEN induced by PP1α and HDAC co-incubation. LSEN prepared from unstressed HeLa cells was co-incubated with the indicated enzymes. FLAG-tagged HDAC1, -2, and -3 were affinity purified from HMBA-treated HEK293T cells that had been transfected with the corresponding cDNA constructs, and analyzed by Western blot (right). C, co-incubation with PP1α and non-activated HDAC1, -2, or -3 is unable to induce BRD4 release from the LSEN of the unstressed HeLa cells. For lanes 1-3, the FLAG-tagged wt-HDAC1, -2, and -3 were isolated from unstressed HEK293T cells with corresponding transfections. For lanes 4 and 5, FLAG-tagged mt-HDAC3 and wt-HDAC3 were isolated from stressed HEK293T cells transfected with the corresponding constructs. D, two-round enzymatic incubation assay of LSEN. LSEN of unstressed cells was sequentially incubated with HDAC3 and PP1α enzymes as indicated, with extensive washes between the two incubations. The changes in histone modifications were analyzed by Western blot.
As impairing PP1α blocked HDAC-mediated H4K5ac/K8ac deacetylation in vivo (see Fig. 4, C and D), we examined whether PP1α acts prior to HDACs when inducing BRD4 release in vitro. LSEN from unstressed cells were subjected to two-round incubations with stress-activated HDAC3 and recombinant PP1α enzyme, with extensive washes between the two incubations. Remarkably, BRD4 release and H4K5ac/K8ac deacetylation were much more efficient when PP1α was added before HDAC3 (Fig. 5D, lane 3), indicating that PP1α and HDACs act sequentially to induce BRD4 release.
H3S10ph Dephosphorylation Is a Prerequisite for BRD4 Release and Inducible Gene Expression
To test whether the PP1α-mediated H3S10ph dephosphorylation or the other PP1α-dependent events was essential for BRD4 release, we generated a mutant histone H3 by substituting serine 10 with alanine (S10A mt-H3) to mimic the H3S10-dephosphorylated H3. If the dephosphorylation of H3S10ph is crucial for BRD4 release, it is expected that chromatin containing S10A mt-H3 might bypass requirement of the PP1α enzyme. Indeed, in the absence of PP1α, HDAC1 alone induced the release of BRD4 in the LSEN containing mt-H3, but not in that containing wt-H3 (Fig. 6A). Similarly, when affinity-purified nucleosomes were used as substrate, the deacetylation of H4K5ac/K8ac and the release of BRD4 were observed only from the nucleosomes containing mt-H3, but not those containing wt-H3 (Fig. 6B).
FIGURE 6.
Dephosphorylation of H3S10ph is a prerequisite for stress-induced BRD4 release for inducible gene expression. A, in vitro assay of HDAC1-induced BRD4 release in the LSEN containing either wt- or S10A mt-H3. LSEN of HeLa cells with the indicated lentiviral infection were incubated with HDAC1 enzyme, and the BRD4 released into supernatant (sup) and remained in nuclei (nuc) was analyzed by Western blot (WB). B, in vitro assay of HDAC1-induced BRD4 release from nucleosomes containing either wt- or S10A mt-H3. Nucleosome from HeLa cells with the indicated lentiviral infection and treatment was immobilized on anti-FLAG resin, and incubated with HA-HDAC1 enzyme. The levels of BRD4 released in the supernatant (sup) and remained in nucleosome, and the changes in histone modifications, were analyzed by Western blot. C, HMBA-induced BRD4 release in the cells expressing either wt- or S10A mt-H3. LSF prepared from HeLa cells with the indicated lentiviral infection and HMBA treatment was analyzed by Western blot. D and E, S10A mt-H3 allowing cells to bypass the requirement for PP1α during HMBA-induced expression of inducible genes. HIV-LTR-Luc cells with the indicated lentiviral infection and HMBA treatment were subjected to luciferase assay (D) and qRT-PCR analysis (E).
In vivo, H66N mt-PP1α inhibited HMBA-induced BRD4 release in cells expressing wt-H3 (Fig. 6C, lane 3), but not in cells expressing mt-H3 (lane 4). Accordingly, in the presence of H66N mt-PP1α, efficient induction of the expression of HIV-LTR-Luc (Fig. 6D) and endogenous genes (Fig. 6E) by HMBA were observed in cells expressing S10A mt-H3 but not wt-H3. Because the endogenous PP1α activity had been quenched either by dominant-negative H66N mt-PP1α (compare Fig. 6, A and C to E) or by pre-treatment with the PP1 inhibitor microcystin LR (Fig. 6B), these data ruled out other PP1α-dependent events, but indicated that H3S10ph dephosphorylation per se is a prerequisite for HDAC-mediated H4K5ac/K8ac deacetylation and BRD4 release for inducible gene expression.
Coordinated BRD4 Release and P-TEFb Liberation for Transcription Elongation
Our previous study showed that PP1α-mediated dephosphorylation of T186ph at the CDK9 T-loop is critical for stress-induced liberation of P-TEFb from the inactive 7SK complex (7SK snRNP) (14). Identification of the PP1α pathway in BRD4 release prompted us to test whether the two processes are concerted. F1C2(CDK9-f) cells, a cell line stably expressing FLAG-CDK9 (49), were stressed and extracted stepwise into LSF and HSF using a modified nuclear fractionation protocol (31) (outlined Fig. 1A). BRD4 release was examined by Western blot assay of the level of BRD4 in an aliquot of LSF (Fig. 7A, top 2 rows), the P-TEFb liberation was determined by analyzing the loss of P-TEFb-bound HEXIM1 in anti-FLAG immunoprecipitates of the same LSF (bottom 2 rows), and interaction of P-TEFb with BRD4 and recruitment of P-TEFb·BRD4 complex onto chromatin was assessed by measuring the level of P-TEFb-bound BRD4 in anti-FLAG immunoprecipitates of HSF (middle 2 rows). Interestingly, stress-triggered BRD4 release, P-TEFb liberation, as well as the recruitment of the BRD4·P-TEFb complex in parallel (Fig. 7A), without affecting the expressing levels of P-TEFb and its associated factors (data not shown). Consistently, the ChIP assay showed that HMBA treatment induced the enrichment of BRD4 and P-TEFb at the promoter region of inducible genes (Fig. 7B).
FIGURE 7.
PP1α coordinates BRD4 release and P-TEFb liberation for regulating transcription elongation. A, effect of stress on BRD4 release, P-TEFb liberation, and BRD4·P-TEFb recruitment. F1C2(CDK9-f) cells were treated as indicated, followed by stepwise fractionation to yield the LSF and HSF. BRD4 released into LSF (top), BRD4 in association with P-TEFb in the anti-FLAG immunoprecipitates of HSF (middle), and HEXIM1 in association with P-TEFb in the immunoprecipitates from the rest of LSF (bottom) were examined by Western blotting (WB). B, ChIP-qPCR analysis of BRD4 and CDK9/P-TEFb levels at promoter regions of the indicated genes after HMBA treatments of HIV-LTR-luciferase cells. The levels in non-treated cells were set as 1. C and D, effect of silencing PP1α (B) or expressing H66N mt-PP1α (C) on stress-induced BRD4 release, P-TEFb liberation, and BRD4/P-TEFb recruitment. shScr, scrambled shRNA control. LSF and HSF were prepared as described in the legend to Fig. 3, B and C, and assayed as in A. E, inhibitory effect of H66N mt-PP1α on HMBA-induced transcription elongation of the HIV-LTR-luciferase gene. HIV-LTR-Luc cells with the indicated lentiviral infection and HMBA treatment were subjected to qRT-PCR analysis of the levels of initiation and elongation transcripts as illustrated in top panel. Data were averaged and presented as fold-enhancement compared with untreated sample. F, knocking down HDAC1/2/3 blocks BRD4 release, but not P-TEFb liberation. LSF and HSF were prepared from F1C2(CDK9-f) cells with the indicated lentiviral co-infection and treatments, and assayed as in A.
Next, we tested the role of the PP1α pathway in these events by examining LSF and HSF, as shown in Fig. 4, C and D. When PP1α was knocked down by shRNA, in addition to the release of BRD4 (Fig. 7C, top), recruitment of the BRD4·P-TEFb complex (middle) and liberation of P-TEFb (bottom) were also blocked. However, overexpressing H66N mt-PP1α did not block P-TEFb liberation (Fig. 7D, bottom), but only abolished the stress-induced BRD4 release (top) and thereby BRD4·P-TEFb recruitment (middle), suggesting that H66N mt-PP1α only has a dominant-negative effect on BRD4 release. Significantly, overexpressing mt-PP1α attenuated HMBA-induced transcription elongation, but not initiation, as indicated by qRT-PCR analysis of the HIV-LTR-Luc transcripts representing initiation and elongation (Fig. 7E). Moreover, impairing cellular HDACs by either HDAC inhibitor (data not shown) or shRNA knockdown (Fig. 7F) blocked the release of BRD4 (top) and recruitment of the BRD4·P-TEFb complex (middle), but not P-TEFb liberation (bottom). Similar to the inhibitory effect of H66N mt-PP1α on HMBA-induced elongation of HIV-1 (Fig. 7E), overexpressing the dominant-negative mutant HDAC1/2/3 attenuated the HMBA-induced transcription elongation of HIV-LTR-Luc, but not transcription initiation (data not shown). Taken together, these observations support the notion that the coordinated BRD4 release and P-TEFb liberation are essential for the recruitment of the BRD4·P-TEFb complex and enhancement of transcription elongation.
DISCUSSION
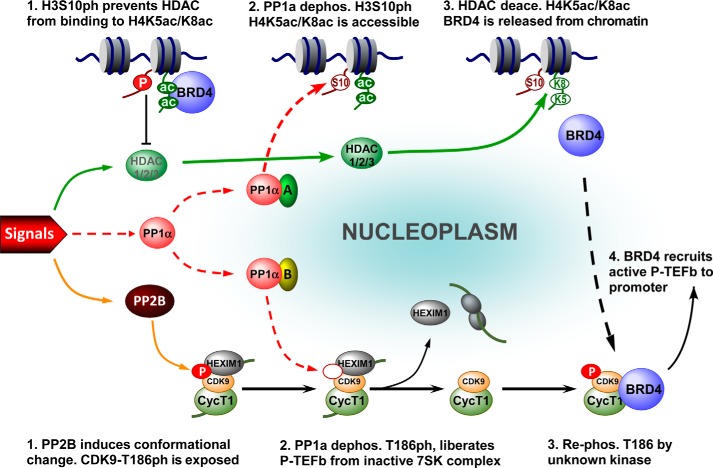
Combined with previous studies (14, 31), the data presented here are consistent with a model for the concerted regulation of BRD4 and P-TEFb during stress-induced gene expression (Fig. 8). In the unstressed state, HDAC1/2/3 are inactive, and the majority of BRD4 is associated with chromatin (Fig. 8, top). Meanwhile, most P-TEFb is sequestrated in the inactive 7SK complex (7SK snRNP, Fig. 8, bottom) (14). Upon stress treatment, HDAC1/2/3 are activated yet still not able to deacetylate nucleosomal H4K5ac/K8ac. PP1α-mediated dephosphorylation of H3S10ph is required to potentiate the nucleosomes to be deacetylated by HDAC1/2/3 at H4K5ac/K8ac, thereby releasing BRD4 from the nucleosome (Fig. 8, top). Concomitantly, stress-activated PP2B cooperates with PP1α to induce the dephosphorylation of CDK9 T-loop at Thr186, thereby leading to the liberation of P-TEFb from 7SK snRNP (14) (Fig. 8, bottom). Through selective association with P-TEFb that has its CDK9 re-phosphorylated at Thr186 by an unknown kinase (31) (Fig. 8, bottom), the released BRD4 recruits the active form of P-TEFb onto the promoter, where it modulates the processivity of RNA Pol II for productive transcription elongation. In this context, the dephosphorylation of H3S10ph controls the role of BRD4 switching from chromosomal targeting to transcription regulation of the expression of inducible genes.
FIGURE 8.
A model depicting the role of the PP1α signaling pathway in coordinating the activation of BRD4 and P-TEFb for inducible gene expression. Top: (1) in the absence of external signals, almost all BRD4 is locked up in nucleosomes containing histone modifications of H4K5ac/K8ac and H3S10ph. H3S10ph prevents HDAC from deacetylating H4K5ac/K8ac, thereby prohibiting the inappropriate activation of BRD4 (2). Upon stimulation, the external signals (left) trigger the activation of PP1α, which in association with partner A, dephosphorylates H3S10ph (3). Subsequently, the signal-activated HDAC1/2/3 deacetylate H4K5ac/K8ac, which leads to BRD4 release from chromatin. Bottom: (1) meanwhile, the signal-activated phosphatase PP2B induces a conformational change in the inactive 7SK complex to expose CDK9 T186ph (2). Consequently, PP1α with partner B is able to dephosphorylate CDK9 at Thr186, thereby liberating P-TEFb from the inactive 7SK complex (14) (3). Through selective association with the transcriptionally active form of P-TEFb that has its CDK9 T-loop re-phosphorylated by unknown kinase(s), (4) the released BRD4 mediates the recruitment of P-TEFb to the promoter-proximal region to enhance the processivity of RNA Pol II for productive elongation.
To form a functional BRD4·P-TEFb complex, BRD4 and P-TEFb must be available at the same time. Analysis of the release of chromatin-bound BRD4 and the liberation of sequestered P-TEFb using LSF and HSF prepared from stressed cells demonstrated that the two processes occur in parallel (Fig. 7A). How a cell coordinates these processes is intriguing. A recent report showed that overexpression of the PID domain of BRD4 could free P-TEFb from 7SK snRNP (54), suggesting that these processes may be coordinated by BRD4 itself. Our data showed, however, that when BRD4 release was blocked by either overexpressing H66N mt-PP1α (Fig. 7D) or impairing the function of HDAC (Fig. 7F), the stress-induced P-TEFb liberation was unaffected, indicating that BRD4 is not required for P-TEFb liberation. Interestingly, silencing of PP1α blocked both BRD4 release and P-TEFb liberation (Fig. 7C), indicating that the PP1α pathway serves as the coordinator of these processes. Of note, because H66N mt-PP1α inhibited only the release of BRD4, but not the liberation of P-TEFb (Fig. 7D), the functional PP1α complexes must be different for the two processes. In mammals, PP1 is estimated to have ∼650 interacting partners for diverse functions (55). Most likely, H66N mt-PP1α is able to associate with the partner (oval A in Fig. 8) that is essential for H3S10ph dephosphorylation and therefore has a dominant-negative effect on this process, but not with the partner (oval B) for P-TEFb liberation. In this way, the same core PP1α pathway could coordinate the regulation of BRD4 and the availability of P-TEFb for formation of the BRD4·P-TEFb complex.
Formation of the BRD4·P-TEFb complex relies on the liberation of P-TEFb and the release of BRD4, both of which are tightly controlled by more than one signaling pathway. For P-TEFb liberation, as shown previously (14), both PP2B and PP1α signaling pathways are required, with PP2B acting prior to PP1α (Fig. 8, bottom). For the release of BRD4 from nucleosomes, the PP1α and HDAC1/2/3 signaling pathways are employed (Fig. 8, top). In this case, PP1α must act prior to HDACs (Figs. 5D and 6). Of note, this order of actions is connected by trans-histone cross-talk between H3S10ph and H4K5ac/K8ac, in which PP1α-induced H3S10ph dephosphorylation enables HDAC-mediated H4K5ac/K8ac deacetylation. This was confirmed by the nucleosomes containing S10A mt-H3 that no longer require PP1α for HDAC-mediated deacetylation of H4K5ac/K8ac (Fig. 6). The molecular mechanism for the cross-talk, however, is unclear. Recently it was reported that HDAC1/2 was able to bind peptides corresponding to the unmodified H3 tail, but not those harboring H3S10ph (56). It is possible that the unmodified H3 peptide had some affinity for HDACs to facilitate the binding of HDACs to nucleosome. Alternatively, H3S10ph may pose steric hindrance in the context of a nucleosome for binding of HDAC.
For more than two decades, the signal-induced phosphorylation of H3S10 has been regarded as a positive epigenetic mark for transcriptional activation of several inducible genes (50, 57–61). Surprisingly, we found that dephosphorylation of H3S10ph is crucial for BRD4-mediated expression of inducible genes. Although apparently contradictory, these events are likely to occur at different regions of the genome. During heat shock of Drosophila cells, H3S10ph decreased globally, but increased at the promoters of the heat shock genes (62). In mammals, the signal-induced phosphorylation of H3S10 was detected only at some promoters (59), or at an intronic enhancer region (32). Moreover, the signal-induced phosphorylation of H3S10 usually correlates with the chromatin remodeling of promoter (50), which occurs prior to the transcription initiation, whereas the stress-induced dephosphorylation of H3S10ph enables BRD4 to augment transcription elongation (Fig. 7E and data not shown), an event that happened after initiation. Hence, one may envision that for a small fraction of nucleosomes located at promoters or enhancer regions, signal-induced H3S10 phosphorylation facilitates chromatin remodeling for transcription initiation, whereas the global decline in H3S10ph may have a distinct function in governing the release of chromatin-bound BRD4 for transcription elongation.
During stress response, the BRD4·P-TEFb complex must be recruited onto promoters to augment the expression of inducible genes (Fig. 7E and data not shown). Because H4K5ac/K8ac is essential for the association of BRD4 with nucleosomes but become deacetylated during stress treatment (Fig. 1, A and B), it is perplexing how the BRD4·P-TEFb complex is recruited to target genes. Several studies suggested that recruitment of the BRD4·P-TEFb complex to promoters may occur by its association with gene-specific transcription factors. For example, upon TNF-α stimulation, BRD4 was found to interact with acetylated NF-κB/RelA via its bromodomains, thereby bringing BRD4/P-TEFb to the promoters of NF-κB-dependent genes (37, 63). Moreover, P-TEFb could also be recruited by c-Myc to specific promoters (64). Furthermore, the acetylated K16 of nucleosomal histone H4 (H4K16ac) at the FOSL1 intronic enhancer was shown to play a role in tethering the BRD4·P-TEFb complex onto its promoter region after stimulating 293 cells with 50% serum (32). It is possible that different genes or regulatory elements could use diverse means for recruiting the BRD4·P-TEFb complex to promoter regions.
In summary, this study revealed the intricate network in which signal-induced histone cross-talk revises H3S10ph and H4K5ac/K8ac modifications, and thereby leads to BRD4 activation for augmenting inducible gene expression. The mechanism for the activation of PP1α and HDACs during stress response, as well as the molecular mechanism for the blocking effect of H3S10ph on the function of HDAC, await future investigation.
This work was supported by National Natural Science Foundation of China Grants 31270809, 81361120386, and 30930046 (to R. C.), 81171192 (to R. L.), and 31170752 and 81070307 (to F. D.), 973 program Grant 2013CB917802 (to R. C.), Natural Science Foundation of Fujian Province Grant 2010J01231 (to F. D.), China Postdoctoral Science Foundation project Grant 2013M530304 (to X. H.), and XMU Basic Training Program of Undergraduate Students Grant CXB2011019 (to Y. W).
- Pol II
- RNA polymerase II
- P-TEFb
- positive transcription elongation factor b
- BDI and BDII
- bromodomains I and II
- DOX
- doxorubicin
- HMBA
- hexamethylene bisacetamide
- MONF
- modified nuclear fractionation
- LSF
- low salt fraction
- HSF
- high salt fraction
- NLF
- nuclear lysate
- LSEN
- low salt-extracted nuclei
- qRT
- quantitative RT
- HDAC
- histone deacetylase
- PP1α
- protein phosphatase 1α.
REFERENCES
- 1. Core L. J., Lis J. T. (2008) Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319, 1791–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Price D. H. (2008) Poised polymerases: on your mark.get set.go!. Mol. Cell 30, 7–10 [DOI] [PubMed] [Google Scholar]
- 3. Fujita T., Schlegel W. (2010) Promoter-proximal pausing of RNA polymerase II: an opportunity to regulate gene transcription. J. Recept. Signal Transduct. Res. 30, 31–42 [DOI] [PubMed] [Google Scholar]
- 4. Nechaev S., Adelman K. (2011) Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta 1809, 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muse G. W., Gilchrist D. A., Nechaev S., Shah R., Parker J. S., Grissom S. F., Zeitlinger J., Adelman K. (2007) RNA polymerase is poised for activation across the genome. Nat. Genet. 39, 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zeitlinger J., Stark A., Kellis M., Hong J. W., Nechaev S., Adelman K., Levine M., Young R. A. (2007) RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39, 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Core L. J., Waterfall J. J., Lis J. T. (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koch F., Jourquin F., Ferrier P., Andrau J. C. (2008) Genome-wide RNA polymerase II: not genes only!. Trends Biochem. Sci. 33, 265–273 [DOI] [PubMed] [Google Scholar]
- 10. Peterlin B. M. (2010) Transcription elongation takes central stage: the P-TEFb connection. Cell Cycle 9, 2933–2934 [DOI] [PubMed] [Google Scholar]
- 11. Lenasi T., Barboric M. (2010) P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Biol. 7, 145–150 [DOI] [PubMed] [Google Scholar]
- 12. Zhou Q., Yik J. H. (2006) The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 70, 646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diribarne G., Bensaude O. (2009) 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 6, 122–128 [DOI] [PubMed] [Google Scholar]
- 14. Chen R., Liu M., Li H., Xue Y., Ramey W. N., He N., Ai N., Luo H., Zhu Y., Zhou N., Zhou Q. (2008) PP2B and PP1α cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 22, 1356–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jang M. K., Mochizuki K., Zhou M., Jeong H. S., Brady J. N., Ozato K. (2005) The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19, 523–534 [DOI] [PubMed] [Google Scholar]
- 16. Yang Z., Yik J. H., Chen R., He N., Jang M. K., Ozato K., Zhou Q. (2005) Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19, 535–545 [DOI] [PubMed] [Google Scholar]
- 17. Chiang C. M. (2009) Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000 Biol. Rep. 1, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K. (2003) The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. U.S.A. 100, 8758–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dey A., Ellenberg J., Farina A., Coleman A. E., Maruyama T., Sciortino S., Lippincott-Schwartz J., Ozato K. (2000) A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 20, 6537–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J. P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Müller S., Pawson T., Gingras A. C., Arrowsmith C. H., Knapp S. (2012) Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mochizuki K., Nishiyama A., Jang M. K., Dey A., Ghosh A., Tamura T., Natsume H., Yao H., Ozato K. (2008) The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem. 283, 9040–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang Z., He N., Zhou Q. (2008) Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell. Biol. 28, 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dey A., Nishiyama A., Karpova T., McNally J., Ozato K. (2009) Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol. Biol. Cell 20, 4899–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao R., Nakamura T., Fu Y., Lazar Z., Spector D. L. (2011) Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell. Biol. 13, 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Devaiah B. N., Singer D. S. (2013) Two faces of BRD4: mitotic bookmark and transcriptional lynchpin. Transcription 4, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Voigt P., Reinberg D. (2011) BRD4 jump-starts transcription after mitotic silencing. Genome Biol. 12, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang W., Prakash C., Sum C., Gong Y., Li Y., Kwok J. J., Thiessen N., Pettersson S., Jones S. J., Knapp S., Yang H., Chin K. C. (2012) Bromodomain-containing Protein 4 (BRD4) Regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J. Biol. Chem. 287, 43137–43155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang R., Li Q., Helfer C. M., Jiao J., You J. (2012) Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J. Biol. Chem. 287, 10738–10752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. You J., Croyle J. L., Nishimura A., Ozato K., Howley P. M. (2004) Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117, 349–360 [DOI] [PubMed] [Google Scholar]
- 30. Bisgrove D. A., Mahmoudi T., Henklein P., Verdin E. (2007) Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. U.S.A. 104, 13690–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ai N., Hu X., Ding F., Yu B., Wang H., Lu X., Zhang K., Li Y., Han A., Lin W., Liu R., Chen R. (2011) Signal-induced Brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation. Nucleic Acids Res. 39, 9592–9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zippo A., Serafini R., Rocchigiani M., Pennacchini S., Krepelova A., Oliviero S. (2009) Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138, 1122–1136 [DOI] [PubMed] [Google Scholar]
- 33. Hargreaves D. C., Horng T., Medzhitov R. (2009) Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138, 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karn J. (2013) A new BET on the control of HIV latency. Cell Cycle 12, 545–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou M., Huang K., Jung K. J., Cho W. K., Klase Z., Kashanchi F., Pise-Masison C. A., Brady J. N. (2009) Bromodomain protein Brd4 regulates human immunodeficiency virus transcription through phosphorylation of CDK9 at threonine 29. J. Virol. 83, 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang G., Liu R., Zhong Y., Plotnikov A. N., Zhang W., Zeng L., Rusinova E., Gerona-Nevarro G., Moshkina N., Joshua J., Chuang P. Y., Ohlmeyer M., He J. C., Zhou M. M. (2012) Down-regulation of NF-κB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J. Biol. Chem. 287, 28840–28851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang B., Yang X. D., Zhou M. M., Ozato K., Chen L. F. (2009) Brd4 coactivates transcriptional activation of NF-κB via specific binding to acetylated RelA. Mol. Cell. Biol. 29, 1375–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anand P., Brown J. D., Lin C. Y., Qi J., Zhang R., Artero P. C., Alaiti M. A., Bullard J., Alazem K., Margulies K. B., Cappola T. P., Lemieux M., Plutzky J., Bradner J. E., Haldar S. M. (2013) BET bromodomains mediate transcriptional pause release in heart failure. Cell 154, 569–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spiltoir J. I., Stratton M. S., Cavasin M. A., Demos-Davies K., Reid B. G., Qi J., Bradner J. E., McKinsey T. A. (2013) BET acetyl-lysine binding proteins control pathological cardiac hypertrophy. J. Mol. Cell. Cardiol. 63, 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zuber J., Shi J., Wang E., Rappaport A. R., Herrmann H., Sison E. A., Magoon D., Qi J., Blatt K., Wunderlich M., Taylor M. J., Johns C., Chicas A., Mulloy J. C., Kogan S. C., Brown P., Valent P., Bradner J. E., Lowe S. W., Vakoc C. R. (2011) RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blobel G. A., Kalota A., Sanchez P. V., Carroll M. (2011) Short hairpin RNA screen reveals bromodomain proteins as novel targets in acute myeloid leukemia. Cancer Cell 20, 287–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Delmore J. E., Issa G. C., Lemieux M. E., Rahl P. B., Shi J., Jacobs H. M., Kastritis E., Gilpatrick T., Paranal R. M., Qi J., Chesi M., Schinzel A. C., McKeown M. R., Heffernan T. P., Vakoc C. R., Bergsagel P. L., Ghobrial I. M., Richardson P. G., Young R. A., Hahn W. C., Anderson K. C., Kung A. L., Bradner J. E., Mitsiades C. S. (2011) BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dawson M. A., Prinjha R. K., Dittmann A., Giotopoulos G., Bantscheff M., Chan W. I., Robson S. C., Chung C. W., Hopf C., Savitski M. M., Huthmacher C., Gudgin E., Lugo D., Beinke S., Chapman T. D., Roberts E. J., Soden P. E., Auger K. R., Mirguet O., Doehner K., Delwel R., Burnett A. K., Jeffrey P., Drewes G., Lee K., Huntly B. J., Kouzarides T. (2011) Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Segura M. F., Fontanals-Cirera B., Gaziel-Sovran A., Guijarro M. V., Hanniford D., Zhang G., González-Gomez P., Morante M., Jubierre L., Zhang W., Darvishian F., Ohlmeyer M., Osman I., Zhou M. M., Hernando E. (2013) BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res. 73, 6264–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rodriguez R. M., Huidobro C., Urdinguio R. G., Mangas C., Soldevilla B., Domínguez G., Bonilla F., Fernandez A. F., Fraga M. F. (2012) Aberrant epigenetic regulation of bromodomain Brd4 in human colon cancer. J. Mol. Med. 90, 587–595 [DOI] [PubMed] [Google Scholar]
- 46. Alsarraj J., Hunter K. W. (2012) Bromodomain-containing protein 4: a dynamic regulator of breast cancer metastasis through modulation of the extracellular matrix. Int. J. Breast Cancer 2012, 670632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang H., Zhou N., Ding F., Li Z., Chen R., Han A., Liu R. (2011) An efficient approach for site-directed mutagenesis using central overlapping primers. Anal. Biochem. 418, 304–306 [DOI] [PubMed] [Google Scholar]
- 48. Chen R., Liu M., Zhang K., Zhou Q. (2011) Isolation and functional characterization of P-TEFb-associated factors that control general and HIV-1 transcriptional elongation. Methods 53, 85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yik J. H., Chen R., Nishimura R., Jennings J. L., Link A. J., Zhou Q. (2003) Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12, 971–982 [DOI] [PubMed] [Google Scholar]
- 50. Nowak S. J., Corces V. G. (2004) Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 20, 214–220 [DOI] [PubMed] [Google Scholar]
- 51. Haberland M., Montgomery R. L., Olson E. N. (2009) The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Rev. Genet. 10, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Beckers T., Burkhardt C., Wieland H., Gimmnich P., Ciossek T., Maier T., Sanders K. (2007) Distinct pharmacological properties of second generation HDAC inhibitors with the benzamide or hydroxamate head group. Int. J. Cancer 121, 1138–1148 [DOI] [PubMed] [Google Scholar]
- 53. Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 54. Schröder S., Cho S., Zeng L., Zhang Q., Kaehlcke K., Mak L., Lau J., Bisgrove D., Schnölzer M., Verdin E., Zhou M. M., Ott M. (2012) Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J. Biol. Chem. 287, 1090–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heroes E., Lesage B., Görnemann J., Beullens M., Van Meervelt L., Bollen M. (2013) The PP1 binding code: a molecular-lego strategy that governs specificity. FEBS J. 280, 584–595 [DOI] [PubMed] [Google Scholar]
- 56. He S., Khan D. H., Winter S., Seiser C., Davie J. R. (2013) Dynamic distribution of HDAC1 and HDAC2 during mitosis: association with F-actin. J. Cell. Physiol. 228, 1525–1535 [DOI] [PubMed] [Google Scholar]
- 57. Banerjee T., Chakravarti D. (2011) A peek into the complex realm of histone phosphorylation. Mol. Cell. Biol. 31, 4858–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rossetto D., Avvakumov N., Côté J. (2012) Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics 7, 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sawicka A., Seiser C. (2012) Histone H3 phosphorylation: a versatile chromatin modification for different occasions. Biochimie 94, 2193–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mahadevan L. C., Willis A. C., Barratt M. J. (1991) Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 65, 775–783 [DOI] [PubMed] [Google Scholar]
- 61. Chadee D. N., Hendzel M. J., Tylipski C. P., Allis C. D., Bazett-Jones D. P., Wright J. A., Davie J. R. (1999) Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J. Biol. Chem. 274, 24914–24920 [DOI] [PubMed] [Google Scholar]
- 62. Nowak S. J., Pai C. Y., Corces V. G. (2003) Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Mol. Cell. Biol. 23, 6129–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang W., Yao X., Huang Y., Hu X., Liu R., Hou D., Chen R., Wang G. (2013) Mediator MED23 regulates basal transcription in vivo via an interaction with P-TEFb. Transcription 4, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kanazawa S., Soucek L., Evan G., Okamoto T., Peterlin B. M. (2003) c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene 22, 5707–5711 [DOI] [PubMed] [Google Scholar]