Background: Chaperonins like GroEL-GroES are required for the folding of many proteins.
Results: The GroEL C termini alter substrate protein conformation and accelerate folding.
Conclusion: Optimal protein folding requires the partial unfolding of misfolded states, a process that involves the GroEL C terminus.
Significance: Chaperonins can actively facilitate protein folding by altering the conformations of folding intermediates.
Keywords: Chaperone, Molecular Chaperone, Protein Dynamic, Protein Folding, Protein Misfolding, GroEL, Hsp60, Chaperonin
Abstract
Many essential cellular proteins fold only with the assistance of chaperonin machines like the GroEL-GroES system of Escherichia coli. However, the mechanistic details of assisted protein folding by GroEL-GroES remain the subject of ongoing debate. We previously demonstrated that GroEL-GroES enhances the productive folding of a kinetically trapped substrate protein through unfolding, where both binding energy and the energy of ATP hydrolysis are used to disrupt the inhibitory misfolded states. Here, we show that the intrinsically disordered yet highly conserved C-terminal sequence of the GroEL subunits directly contributes to substrate protein unfolding. Interactions between the C terminus and the non-native substrate protein alter the binding position of the substrate protein on the GroEL apical surface. The C-terminal tails also impact the conformational state of the substrate protein during capture and encapsulation on the GroEL ring. Importantly, removal of the C termini results in slower overall folding, reducing the fraction of the substrate protein that commits quickly to a productive folding pathway and slowing several kinetically distinct folding transitions that occur inside the GroEL-GroES cavity. The conserved C-terminal tails of GroEL are thus important for protein folding from the beginning to the end of the chaperonin reaction cycle.
Introduction
To function, most proteins must fold into specific three-dimensional structures. Although the native conformation of a protein is ultimately governed by the thermodynamics of its amino acid sequence in aqueous solution, protein folding is often prone to errors (1, 2). Side reactions, like misfolding and aggregation, frequently occur and can be especially serious for large and topologically complex proteins inside the concentrated interior of a living cell (3, 4). Fundamentally, the twin problems of misfolding and aggregation are kinetic in nature and biologically solved by the early evolution of several families of specialized machines known as molecular chaperones (5). In general, molecular chaperones prevent or correct folding and assembly errors and thereby permit proteins to attain the native states thermodynamically encoded in their sequences (6). Molecular chaperones are thus kinetic editors of protein folding reactions.
Within the network of molecular chaperones that maintain cellular protein homeostasis, the Hsp60s or chaperonins occupy a central and essential hub (6–8). The Hsp60s are ancient and widespread and are present in virtually every organism currently known. The chaperonin system of Escherichia coli, GroEL-GroES, is perhaps the best studied example of this molecular chaperone family (8–10). GroEL is a homotetradecamer composed of 57-kDa monomers arranged in two stacked, heptameric rings (11). Each GroEL ring contains a large solvent-filled cavity and the upper cavity-facing surface of each ring is lined with hydrophobic amino acids that capture incompletely folded substrate proteins (non-native proteins) (11, 12). Proteomic surveys suggest that ∼80–100 E. coli proteins possess an obligate dependence on GroEL for folding, with an additional larger number of proteins gaining an intermediate level of assistance from the GroEL-GroES system (13, 14).
Following capture, most substrate proteins are encapsulated within a sealed cavity formed between the GroEL ring and the smaller lid-like GroES co-chaperonin, a heptamer of 10-kDa subunits (15–19). The assembly of the GroEL-GroES folding cavity results in the initiation of protein folding by release and confinement of the substrate protein inside the privileged volume of the GroEL-GroES chamber (15). The formation of the GroEL-GroES folding cavity is a highly ordered process, in which binding of the non-native protein on the open trans ring of a GroEL-GroES complex is followed by the obligate binding of ATP and then GroES to the same ring (15, 20–23). The encapsulated protein can persist and fold within the GroEL-GroES cavity for a brief period of ∼5–25 s, depending on conditions (20, 21, 24). Hydrolysis of the ATP inside the GroEL-GroES cavity (the cis cavity) prepares the complex for disassembly upon binding of substrate protein and ATP to the second trans ring (20). Disassembly of the GroEL-GroES cavity results in the release of the full population of enclosed substrate protein, folded or not (25–27). Folding intermediates that do not commit to their native states within the lifetime of the GroEL-GroES cis cavity and that cannot complete folding in free solution must be recaptured for another round of processing. Thus, under the biologically relevant conditions of steady-state ATP turnover, the GroEL-GroES machine proceeds through a highly dynamic reaction cycle, the timing of which is ultimately set by the rate of ATP hydrolysis (20, 21, 24, 28).
Despite over 2 decades of effort, the precise manner in which GroEL facilitates protein folding remains controversial. Several mechanisms have been proposed, which cluster into two classes based upon whether GroEL is postulated to act passively or actively (6, 8–10). The prevailing passive mechanism, referred to as the Anfinsen cage model, assumes that the folding of GroEL-dependent proteins is, in general, only limited by the tendency of on-pathway folding intermediates to aggregate. In this view, GroEL facilitates folding by simply binding and sequestering aggregation-prone intermediates, blocking aggregation, and thereby allowing the inherent thermodynamic drive programmed into the protein sequence to express itself unencumbered (8, 10, 29). Active mechanisms, by contrast, accept the possibility that GroEL-dependent substrate proteins populate off-pathway states that have no direct access to the native state. Such misfolded conformations are also likely to be highly aggregation-prone but, because they cannot be rescued by simple sequestration, require an additional corrective action by the chaperonin (6, 9). The nature of this corrective mechanism remains poorly understood but has been suggested to come from either (i) repetitive unfolding and iterative annealing (30, 31) or (ii) smoothing of a substrate protein's free energy landscape as a result of confinement inside the GroEL-GroES cavity, where either spatial constraints or interactions between the substrate protein and the chaperonin cavity alter the ensemble of folding intermediates, eliminating inhibitory states or favoring productive ones (6, 9, 10).
One reason a coherent picture of GroEL-mediated protein folding has yet to emerge stems from the broad range of proteins upon which GroEL operates. Several proteins have been shown to satisfy the conditions required for a purely passive folding mechanism (32–35). However, other proteins appear to require more active participation of the GroEL-GroES system to fold (36–38). In examining active mechanisms of GroEL-mediated protein folding, we have focused on the CO2-fixing enzyme from Rhodospirillum rubrum, Rubisco,2 one of the most highly GroEL-dependent substrate proteins known. We previously demonstrated that Rubisco populates a kinetically trapped, misfolded monomer that is efficiently rescued by GroEL in the absence of aggregation (36). We also showed that GroEL assists Rubisco folding, at least in part, through two phases of multiple axis unfolding of the misfolded Rubisco monomer (21, 23, 36). In addition, we observed an encapsulation-dependent compaction of the Rubisco folding intermediate, hinting at the possibility of an active role for confinement inside the GroEL-GroES chamber (21, 23, 36). Work on Rubisco by others (37, 39) also suggested that confinement of the Rubisco monomer alters the folding landscape of the protein. In this case, confinement was suggested to be the dominant mechanism of folding assistance, and the nature of the effect was assigned to either the restricted volume or charge character of the GroEL-GroES cavity (37, 39–41). More recent observations have challenged some of these conclusions, however, calling into question confinement-based active mechanisms (29, 42, 43).
To probe the relative contribution of unfolding versus confinement, we have re-examined the folding of Rubisco using a variant of GroEL in which the character and volume of the GroEL cavity have been altered by removal of the unstructured C-terminal 23 amino acids of the GroEL subunits. Deletion of these C-terminal tails, which project upward from the equatorial plane of the GroEL ring into the bottom of the cavity, has no effect on the stability of the GroEL tetradecamer but has a notable impact of the folding of several proteins, as well as a modest in vivo phenotype (39, 40, 43–45). Additionally, although the C-terminal tails are not resolved in the x-ray crystal structures of the chaperonin, using high resolution cryo-EM we recently demonstrated that the C-terminal tails interact directly with the non-native Rubisco folding intermediate during and immediately after encapsulation by GroES (46). At the same time, other studies have suggested that the amphipathic character of the C-terminal tails is essential for efficient folding of several proteins (45). Still other work has suggested that the combined mass of the seven C termini at the base of the GroEL-GroES cavity is important for restricting the volume of the cavity (39, 40).
Consistent with previous observations, we find that removal of the C-terminal tails results in a severalfold drop in Rubisco folding with actively cycling tetradecamer GroEL. However, when Rubisco folding is confined to the GroEL-GroES cavity by using a noncycling, single ring version of GroEL, removal of the C-terminal tails displays a much smaller impact on Rubisco folding. We show that, in part, the larger impact of tail removal on the cycling tetradecamer is due to a slowing of the GroEL reaction cycle and a large shift in a key allosteric transition of the GroEL machine. Removal of the tails also results in a modest slowing of several kinetically distinct Rubisco folding transitions inside the GroEL-GroES chamber. Surprisingly, however, the more substantial consequence of C-terminal tail removal is a large reduction in unfolding of the Rubisco monomer at the early stages of the GroEL reaction cycle. Reduced unfolding results in a decrease in the fraction of the Rubisco monomers that rapidly commits to the native state. The GroEL C-terminal tails thus not only assist in substrate protein encapsulation, but directly participate in protein unfolding, a process that is required for maximally efficient folding of the highly stringent substrate protein Rubisco.
EXPERIMENTAL PROCEDURES
Proteins
Wild-type and variants of GroEL (SR1, single-cysteine mutants and C-terminal truncation mutants), wild-type and E98C GroES, and wild-type and cysteine mutants of R. rubrum Rubisco were all expressed and purified as described previously (21–23, 36).
Labeling of Proteins for FRET
Rubisco variants were labeled as described previously (21–23, 36), using either 5-iodoacetamidofluorescein (fluorescein, F) and/or 5-(2-acetamidoethyl) aminonaphthalene 1-sulfonate (EDANS, ED). GroEL variants were labeled with fluorescein, and GroES-98C was labeled with EDANS as described (20, 47). All dyes were purchased from Molecular Probes (Eugene, OR) and prepared fresh in anhydrous N,N-dimethylformamide immediately before use. The extent of labeling was determined by protein quantification by the Bradford assay (Bio-Rad) and dye quantification under denaturing conditions using known molar extinction coefficients (20, 47). For Rubisco variants, site-specific labeling was verified through denaturing anion-exchange chromatography and analysis of proteolytic fragments (47). GroES-98ED was re-purified via anion exchange, and only protein labeled with 2 dyes/heptamer was used. All GroEL variants were labeled with 2–3 dyes/ring.
Stopped-flow Fluorescence
Stopped-flow experiments were conducted as described previously (23, 36, 47), using an SFM-400 rapid mixing unit (BioLogic) equipped with a custom-designed two-channel fluorescence detection system. Mixing was done using two syringes, one containing GroEL-Rubisco complexes and one containing GroES and ATP. The donor-side FRET efficiency was calculated from matched sets of donor-only and donor-acceptor experiments as described previously (20, 47).
Enzymatic Refolding
The refolding of wild-type Rubisco was assayed by enzymatic activity as described previously (15, 20). Acid-urea-denatured Rubisco was bound to an excess of full-length or truncated SR1. After a 10-min incubation at 25 °C, GroES and ATP were added. Intra-cavity folding was quenched by addition of EDTA and incubation at 4 °C, as described (21).
Steady-state and Time-resolved FRET
Steady-state fluorescence measurements were conducted with a PTI fluorometer, with temperature regulation through a jacketed cuvette holder (Neslab). Fluorescence lifetimes were measured in the time domain using a TimeMaster Fluorescence Lifetime Spectrometer (PTI), with sample excitation from a pulsed nitrogen laser coupled to an optical boxcar detector (48). The average distance between donor and acceptor probes was calculated from donor-side FRET efficiencies, extracted from both steady-state and time-resolved data, as described previously (23). The Förster distance, R0, was calculated independently for each donor-acceptor pair in each complex examined. The value of κ2 was assumed to be ⅔ for all distance measurements. In all cases, the average anisotropy of the donor and acceptor probes, at each labeling position, was the same when non-native Rubisco was bound to either full-length or truncated GroEL (data not shown). All protein complexes were equilibrated with a 10-min incubation at 25 °C prior to measurement. Note that in the absence of the C-terminal tails, and under active cycling conditions, 10–20% of the Rubisco initially bound to a GroEL ring can escape prior to GroES binding (46). However, under the experimental conditions employed here, in which Rubisco-GroEL binary complexes are incubated for several minutes prior to the addition of GroES and ATP, the amount of Rubisco that escapes from a Δ526 ring prior to encapsulation by GroES, compared with a wild-type GroEL ring, is negligible (<2%, data not shown).
Protease Protection
The protease sensitivity of non-native Rubisco bound to a GroEL ring was conducted as described previously (22, 23). Briefly, 58F-Rubisco was bound to asymmetric GroEL-GroES ADP bullets. Trypsin was added, and time points were taken, with the reaction stopped with PMSF. Samples were run on 10% SDS-PAGE and imaged using a Typhoon Trio (GE Healthcare).
RESULTS
Deletion of GroEL C Termini Slows Rubisco Folding and Perturbs the GroEL Reaction Cycle
To examine how a change in cavity character impacts the folding of a stringent substrate protein, we created a GroEL variant in which the C-terminal 23 amino acids of the GroEL subunits have been removed. This variant, GroELΔ526–548 (hereafter Δ526), displays no detectable alteration in tetradecamer assembly or stability at room temperature. The behavior of the Δ526 variant during expression, purification, and on gel filtration chromatography is identical to wild-type GroEL (data not shown). The Δ526 variant also supports a functional chaperonin cycle, in which GroES is bound and released in an ATP hydrolysis-dependent manner. However, key properties of the GroEL hydrolytic cycle change upon removal of the C termini. When folding of the stringent substrate protein ribulose-1,5-bisphosphate carboxylase oxygenase (Rubisco) from Rhodospirillum rubrum is examined by tryptophan fluorescence, we observe a greater than 2-fold decrease in the apparent folding rate constant with the Δ526 variant, compared with full-length GroEL (Fig. 1a). Notably, the yield of folded Rubisco is not significantly different between full-length GroEL and Δ526 (Fig. 1a, inset), indicating that the observed decrease in apparent folding rate is not due to an increase in aggregation in the presence of cycling Δ526. This result is consistent with observations made using other C-terminally perturbed tetradecameric GroELs, using different assay methods (39, 40, 43, 45).
FIGURE 1.
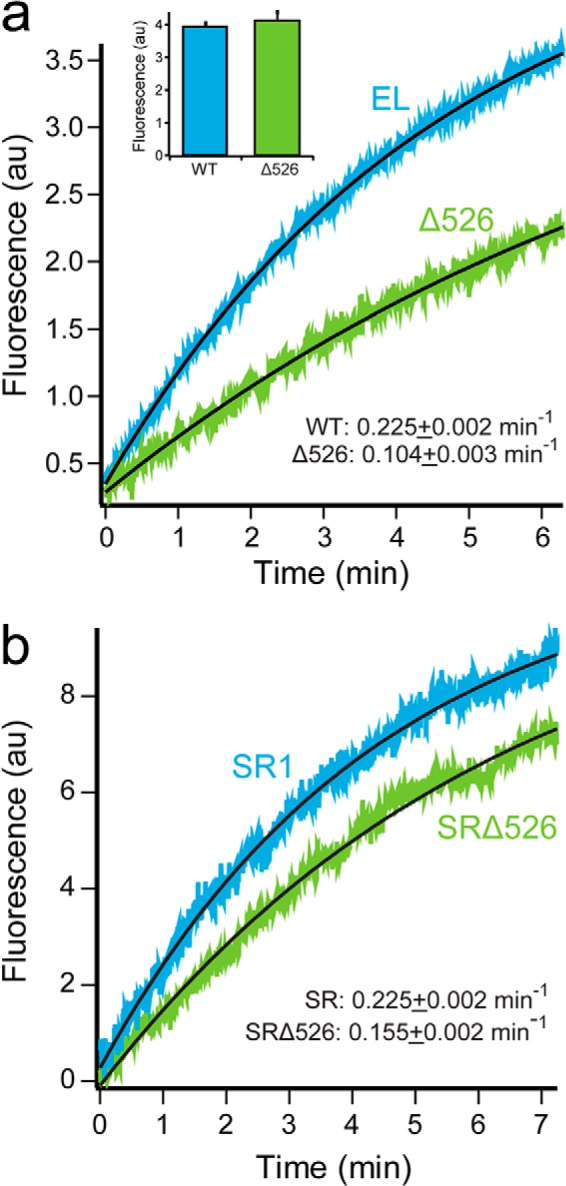
Presence of the GroEL C-terminal tails enhances protein folding. a, folding of Rubisco by cycling GroEL-GroES was monitored by an increase in tryptophan fluorescence. Chemically denatured, wild-type Rubisco (100 nm) was bound to GroEL (200 nm), wild-type (EL; blue), or C-terminal deletion (Δ526; green) and rapidly mixed with an equal volume of excess GroES (400 nm) and ATP (2 mm) in a stopped-flow apparatus. Curves were fit to a single-exponential rate law (black line), resulting in observed rate constants of 0.225 ± 0.002 min−1 for GroEL and 0.104 ± 0.003 min−1 for Δ526. In each case, the overall folding yield was examined by allowing each folding reaction to run to completion (∼30 min), followed by a measurement of the total tryptophan fluorescence (inset). b, folding of Rubisco inside stable single ring-GroES complexes monitored by an increase in tryptophan fluorescence. Chemically denatured wild-type Rubisco (100 nm) was bound to full-length SR1 (SR, blue) or Δ526 (SRΔ526, green) single-ring variants of GroEL (300 nm) and rapidly mixed with an equal volume of excess GroES (600 nm) and ATP (2 mm) in a stopped-flow apparatus. Curves were fit to single-exponential rate laws, yielding observed rate constants of 0.225 ± 0.002 min−1 for SR1 and 0.155 ± 0.002 min−1 for SRΔ526. n = 10 replicates, with a 100-ms sampling time. AU, arbitrary units.
We next considered whether slower Rubisco folding is due to a change in intra-cavity folding in the absence of the GroEL C termini. To prevent cycling and confine Rubisco folding to the interior of the GroEL-GroES cavity, we employed a previously described single-ring variant of GroEL, SR1 (18). For comparison, we constructed a Δ526 variant of SR1 (hereafter SRΔ526) that is the single ring analog of tetradecameric Δ526 (39, 43, 45). Binding of Rubisco and GroES to SRΔ526 was unchanged from SR1 (data not shown). However, Rubisco folding within the SRΔ526-ES cavity is 50–70% slower than folding inside the SR1-ES cavity (Fig. 1b). Thus, although we do observe a drop in intra-cavity folding rate, the magnitude of this decrease cannot fully explain the greater than 2-fold drop in folding rate observed with tetradecameric Δ526.
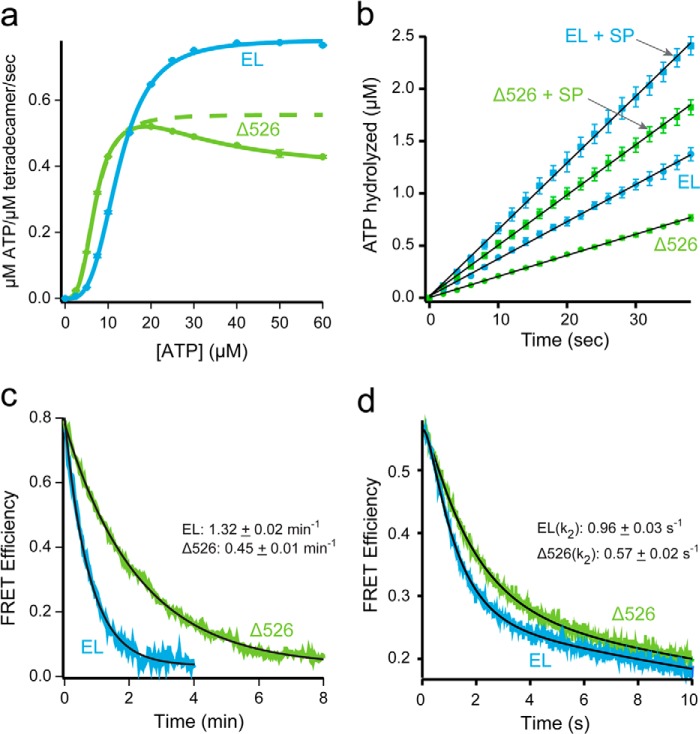
Because progression of the GroEL ATPase cycle is essential for facilitated folding, we next examined the hydrolytic cycle of Δ526 in greater detail. Previous studies demonstrated that C-terminal GroEL truncations like Δ526 can result in a reduction of the steady-state ATP hydrolysis rate (40, 43, 45, 49). However, the source of this reduced turnover, as well as the impact on the overall GroEL reaction cycle, has not been assessed. The decrease in ATPase rate caused by C-terminal truncations, as well as our previous observation of accelerated ATP turnover caused by dye-induced perturbations of the C termini (22), suggested that the C-terminal tails are coupled to the allosteric transitions of the GroEL machine. To test this, we examined the rate of ATP hydrolysis by both GroEL and Δ526 at different ATP concentrations. With full-length GroEL, we observe the well described, positively cooperative binding of ATP to the first GroEL ring (Fig. 2a) (50–52). Fitting of this transition to the Hill equation results in values for the turnover (kcat = 0.12 s−1 per active subunit), half-saturation point (K½ = 12.4 μm), and Hill coefficient (nH = 3.3) that are consistent with previous measurements. By contrast, ATP hydrolysis by Δ526 displays a substantially different response. Although Δ526 shows an initial, positively cooperative transition similar to full-length GroEL, the maximal ATPase rate attained is ∼ 40% lower than full-length (Fig. 2a).
FIGURE 2.
Stimulated folding of Rubisco is not the product of extended GroEL-ES cavity lifetime. a, full-length (EL; blue) or C-terminal deletion (Δ526; green) GroEL (200 nm) was mixed with various concentrations of ATP, and the steady-state rate of hydrolysis was measured. For GroEL, the observed change in initial rate was well fit by the Hill equation, yielding kcat = 0.12 ± <0.01 s−1 per active subunit, K½ = 12.4 ± 0.1 μm, nH = 3.3 ± 0.1. The data for Δ526 at low ATP concentrations was also well fit to a two-state Hill model (green dashed line), yielding kcat = 0.08 ± <0.01 s−1 per active subunit, K½ = 6.9 ± 0.1 μm, nH = 3.2 ± 0.1. The full Δ526 data set was also fit to the nested cooperativity model (52), yielding kcat, 1 = 0.09 ± <0.01 s−1 per active subunit; kcat 2 = 0.05 ± <0.01 s−1 per active subunit, K½ = 1.5 ± 0.1 μm, nH = 3.8 ± 0.1, L1 = 2.5 × 10−3 ± 0.1 × 10−3, L2 = 9.3 × 10−5 ± 2.7 × 10−5. b, steady-state ATP hydrolysis by GroEL (125 nm) and Δ526 (125 nm) was measured in the presence of excess GroES (250 nm), with and without non-native denatured Rubisco (dRub, 100 nm; SP). Addition of dRub to the GroEL-GroES system results in a hydrolysis rate enhancement of 1.8-fold (2.1 μm/min to 3.8 μm/min), consistent with previous observations using saturating levels of the substrate protein malate dehydrogenase (20). Addition of dRub to the Δ526-GroES system results in a rate enhancement of 2.4-fold (1.2 μm/min to 2.9 μm/min). c, lifetime of the GroEL-GroES complex in the absence of substrate protein was examined using a previously described FRET assay (20). ATP (2 mm) was added to GroES-98ED (100 nm) and either 315F-labeled full-length (EL; blue), or C-terminal deletion (Δ526; green) GroEL (125 nm). After a 1 min incubation, unlabeled GroES (2 μm) was introduced as a competitor. The change in FRET was calculated from matched donor-only and donor-acceptor traces. The curves were fit to a single-exponential rate law, yielding rates of 1.32 ± 0.02 min−1 (EL) and 0.45 ± 0.01 min−1 (Δ526). The average of three experiments is shown. d, lifetime of the GroEL-GroES complex upon addition of a stoichiometric amount of non-native Rubisco was examined using the same FRET assay as in c (20). Experiential conditions are similar; except that a stopped-flow apparatus was employed, and 100 nm denatured Rubisco was mixed with the cycling GroEL-GroES system simultaneously with excess, unlabeled GroES. The average of 10 experiments is shown. As reported previously, the observed change in FRET efficiency in the presence of non-native substrate protein requires a triple-exponential rate law for a good fit (20). The rate of the dominant decay component (k2), previously shown to reflect the substrate protein-induced acceleration of GroES release (20), is ∼1 s−1 for GroEL and ∼0.6 s−1 for Δ526.
Strikingly, Δ526 has a marked roll-off in ATP turnover as the ATP concentration exceeds ∼20 μm. This decrease in ATP turnover is highly reminiscent of the negative cooperativity roll-off displayed by full-length GroEL, except in the case of Δ526, it occurs at a far lower ATP concentration. Positive cooperativity in ATP binding to the first GroEL ring, followed by negative cooperativity between the rings as the second ring fills with ATP is a defining feature of the nested cooperativity model of GroEL allostery (52). The ATPase response curve of Δ526 is well described by the nested cooperativity model, with parameters for the initial, positively cooperative event (nH = 3.8; L1 = 2.5 × 10−3, where L1 is the apparent allosteric constant for the first transition) shifted to a lower turnover rate (kcat = 0.09 per active subunit) and lower half-saturation concentration (K½ = 1.5 μm). Additionally, Δ526 displays a dramatic shift in the value of the second allosteric coupling parameter, which describes ATP binding to the second ring (L2 = 9.3 × 10−5), compared with full-length GroEL (L2 = 6.0 × 10−9, where L2 is the apparent allosteric constant for the second transition). These observations strongly suggest that both intra- and inter-ring allostery are altered by removal of the C termini in Δ526.
Because non-native substrate protein can accelerate the GroEL reaction cycle (20) and may also induce a shift from a cycle dominated by asymmetric bullet-shaped complexes to one dominated by symmetric football-shaped complexes (28, 53–55), we examined the effect of non-native substrate protein on both ATP turnover and GroES release. As with GroEL, non-native substrate protein accelerates the rate of ATP hydrolysis by Δ526 in the presence of GroES (Fig. 2b). However, although the magnitude of substrate-stimulated turnover is larger for Δ526 (1.8-fold for GroEL versus 2.6-fold for Δ526), the maximal turnover rate for Δ526 is still ∼60% slower than GroEL. These observations are consistent with steady-state ATPase stimulation seen with GroEL and a C-terminal truncation variant using other substrate proteins (49).
The changes in ATP turnover and allostery exhibited by Δ526 suggested that this C-terminal truncation variant progresses through the canonical GroEL reaction cycle more slowly than wild-type GroEL. If true, the lifetime of the Δ526-GroES complex should lengthen compared with full-length GroEL. To test this possibility, we exploited a previously described fluorescence resonance energy transfer assay (FRET) capable of tracking the dynamics of the GroEL-GroES complex (20). In this assay, an acceptor fluorescent probe was covalently attached to a nonperturbative cysteine substitution on the backside of the apical domain of either full-length GroEL or Δ526. A donor probe was attached to GroES, and the extent of complex formation between the GroEL and GroES was then observed by FRET. As shown in Fig. 2c, the lifetime of the Δ526-GroES complex in the absence of substrate protein is substantially longer than the GroEL-GroES complex (∼2.5 times) under conditions of steady-state ATP turnover. The rate of GroES release from a Δ526-GroES complex is also slower in the presence of non-native substrate protein, although the difference is not as dramatic (Fig. 2d). Interestingly, the differences between GroEL and Δ526 in substrate protein-stimulated GroES release and ATP turnover were essentially identical (Δ526 is 60% slower). This observation suggests that the Δ526 reaction cycle is limited by the same substrate-driven release of ADP from the trans ring as is GroEL (22, 28, 56), but in the absence of the C-terminal tails, the system responds more slowly. In combination, these results show that the progression of the GroEL reaction cycle is coupled to the dynamics of the GroEL C-terminal tails, such that removing the tails slows down the functional GroEL cycle.
It is important to note that the stimulated ATPase rate observed with near saturating levels of non-native substrate proteins (Fig. 2b) is unlikely to apply to the Rubisco folding experiments described here. To minimize aggregation, the GroEL-GroES or Δ526-GroES systems are typically present in a severalfold molar excess over the non-native Rubisco in refolding experiments. Consequently, the overall rate of ATP turnover will only be slightly affected by this amount of substrate protein, and the limiting cycling rate for both GroEL and Δ526 will approach the rate observed in the absence of substrate protein (Fig. 2, b and c).
GroEL C Termini Alter the Conformation of a Rubisco Folding Intermediate
We next considered whether, at different points in the GroEL reaction cycle, the conformation of the Rubisco monomer is altered by the presence of the C-terminal tails. We first examined whether the Rubisco monomer is bound in the same average position on a GroEL ring in the presence and absence of the C termini. For these FRET experiments, the donor was attached to the Rubisco monomer (Fig. 3a), and the acceptor was positioned either near the outer and upper edge of the ring (EL315-F) or near the bottom of the cavity (EL43-F). In all cases, the Rubisco labeling positions were homogeneously derivatized with the donor dye, although the GroEL variants were lightly modified at ∼2–3 dyes per ring. Acceptor-labeled, ADP-bullet complexes of both full-length GroEL and Δ526 were prepared and then mixed with one of four different, denatured, and donor-labeled Rubisco variants. Matched donor-only experiments were also performed with unlabeled full-length GroEL and Δ526. Following Rubisco binding, the extent of FRET between four different locations on the folding intermediate, relative to two positions on the GroEL tetradecamer, was determined. For all donor positions and both acceptor locations, a strong FRET signal is observed, ranging in efficiency from 0.42 to 0.75 (Fig. 3, b and c). Robust differences between the different donor-labeled positions suggest that the Rubisco monomer is asymmetrically bound to both the full-length GroEL and Δ526 trans rings. More striking, however, is the distinct pattern of differences that emerge when full-length GroEL and Δ526 are compared. For all Rubisco donor positions, full-length GroEL shows a consistently higher FRET efficiency than Δ526 when the acceptor dye is located at the base of the GroEL ring (Fig. 3b). By contrast, this pattern reverses when the acceptor dye is moved to the upper edge of the GroEL cavity, with Δ526 showing a consistently higher FRET efficiency (Fig. 3c). These observations suggest that the Rubisco monomer is, on average, shifted higher up the GroEL cavity when the C termini were removed.
FIGURE 3.
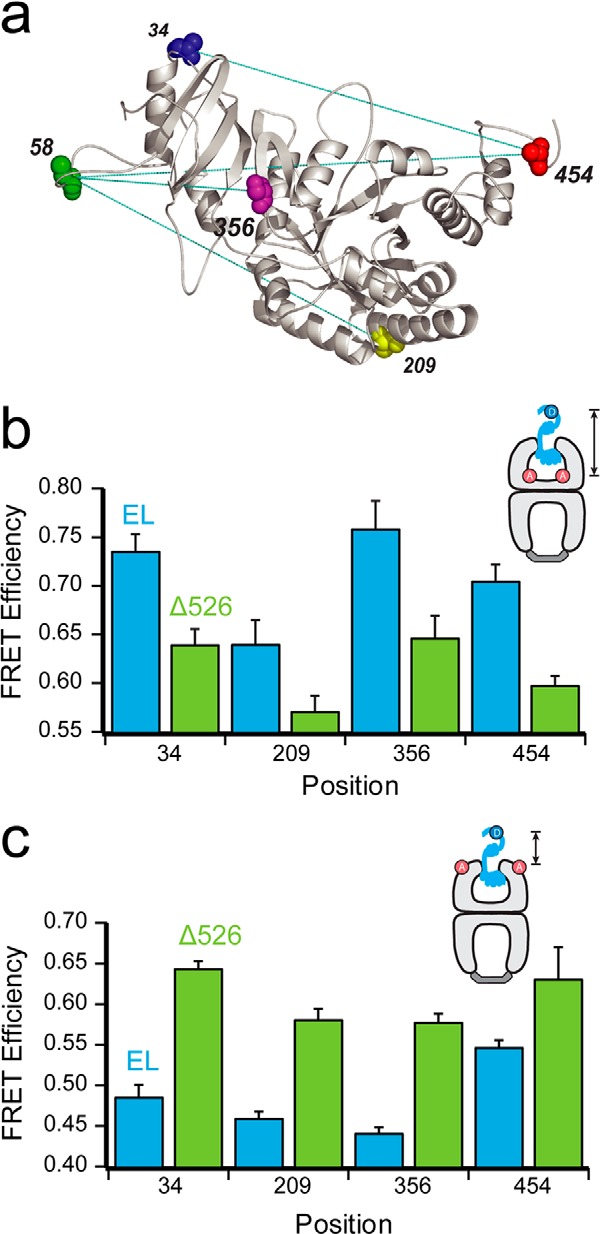
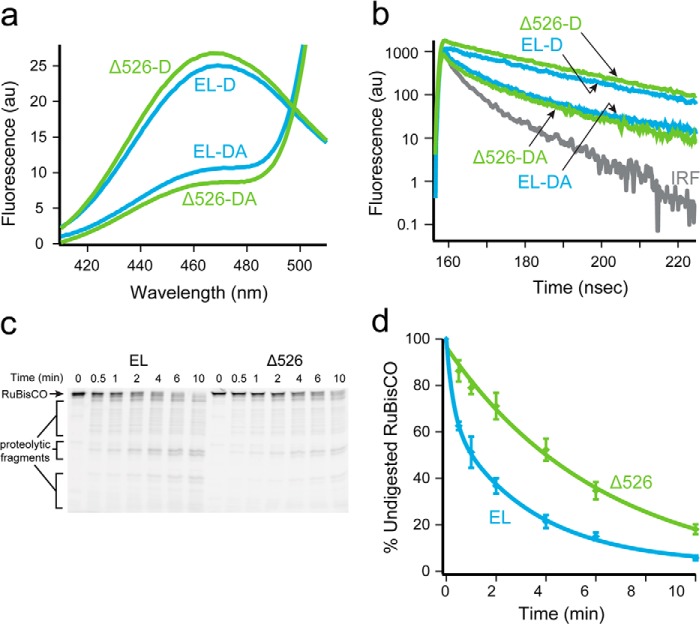
Contact with the C termini promotes deeper initial Rubisco binding within the GroEL cavity. a, structure of one monomer of the native Rubisco dimer (PDB ID: 9RUB) is shown. Positions of five amino acid locations employed for the attachment of exogenous fluorescent probes are indicated. Except for position 58, which is a naturally occurring surface-exposed Cys residue, these positions were mutated to encode Cys and labeled with thiol-alkylating fluorescent dyes as described previously (20, 47). Sites successfully paired for intramolecular FRET assays, in which donor and acceptor dyes are attached to the same Rubisco monomer, are indicated by dotted lines. b, steady-state FRET measurements between chemically denatured, donor-labeled Rubisco (100 nm) bound to the trans ring of acceptor-labeled, full-length (EL; blue) or C-terminal deletion (Δ526; green) GroEL-GroES ADP bullets (120 nm). For these measurements, Rubisco was labeled with EDANS at each of four sites (amino acids 34, 209, 356, and 454), and GroEL was labeled with fluorescein near the bottom of the cavity through a unique, introduced Cys at position 43 (65). Error bars represent the standard deviation for n = 3 experiments. c, steady-state FRET measurements as in b, but using GroEL labeled with fluorescein near the top of the cavity, through a unique introduced Cys at position 315 (20).
To investigate the effect of the C-terminal tails on Rubisco folding in greater detail, we next employed an intramolecular FRET assay. The assay is designed to follow the conformation of a Rubisco monomer as it transits the GroEL reaction cycle (21, 23, 36). A series of Rubisco variants were used, in which a donor probe was attached to one position and an acceptor probe attached to another (Fig. 3a). We used four distinct FRET pairs to measure the intramolecular FRET efficiency of the Rubisco monomer bound to the trans ring of full-length GroEL and Δ526 ADP bullets. In all cases, the donor-side FRET efficiency was determined by both steady-state and time-domain lifetime measurements. An example of experimental data from one FRET pair (209ED-58F) is shown in Fig. 4, a and b, and the FRET efficiencies and calculated distances for all four pairs are shown in Table 1. For each pair of sites, the measured intramolecular distance was shorter when the Rubisco monomer is bound to a Δ526 ring, compared with a full-length GroEL ring. These measurements suggest that the non-native Rubisco monomer is more compact and less unfolded when it is bound to a Δ526 ring. If true, this predicts that Rubisco bound to a Δ526 ring should be more resistant to protease digestion than a monomer bound to a full-length GroEL ring. As shown in Fig. 4, c and d, Rubisco bound to a full-length GroEL ring is digested by trypsin more rapidly (∼3 times) than Rubisco bound to a Δ526 ring.
FIGURE 4.
Rubisco adopts a more unfolded conformation on a GroEL ring in the presence of the C termini. a, steady-state fluorescence of chemically denatured, donor-only (209ED) or donor-acceptor (209ED-58F) labeled Rubisco (100 nm) bound to the trans ring of full-length (EL, blue) or C-terminal deletion (Δ526; green) GroEL-GroES ADP bullets (120 nm). The spectra shown are the average of n = 3 experiments. b, time-resolved fluorescence decay measurements of the same complexes in a. The instrument response function is also shown (IRF). c, fluorescently labeled Rubisco (58-F; 100 nm) was chemically denatured and bound to the trans ring of either GroEL or Δ526 ADP bullets (120 nm) and then treated with trypsin for the indicated times before quenching with PMSF (1 mm). The samples were analyzed by SDS-PAGE and laser-excited fluorescence gel scanning. An arrow shows the migration position of full-length Rubisco, and brackets indicate the position of three dominant groups of proteolytic fragments. d, amount of full-length Rubisco remaining at each time point in c was quantified and plotted as a function of time. The average half-time for the digestion of Rubisco bound to full-length ADP bullets was ∼1.5 min (EL, blue) and ∼4 min for C-terminal deletion ADP bullets (Δ526, green). AU, arbitrary units. Error bars represent the standard deviation of n = 3 experiments.
TABLE 1.
Intramolecular FRET measurements of Rubisco bound to wild-type and Δ526 GroEL
| FRET pair | Distancea |
||
|---|---|---|---|
| WTb | Δ526c | ||
| Å | |||
| Steady state | 34EDd-454Fe | 44.1 | 42.0 |
| 58F-209ED | 46.9 | 41.2 | |
| 58F-356ED | 43.9 | 40.1 | |
| 58F-454ED | 46.2 | 42.0 | |
| Time-resolved | 34ED-454F | 46.1 | 43.5 |
| 58F-209ED | 49.1 | 41.7 | |
| 58F-356ED | 43.2 | 39.3 | |
| 58F-454ED | 44.5 | 42.3 | |
a Steady-state and time-resolved intramolecular FRET measurements between different positions on chemically denatured, doubly-labeled RuBisCO bound to the trans ring of Δ526 GroEL/GroES ADP bullets.
b Full-length GroEL/GroES ADP bullets.
c Δ526 GroEL/GroES ADP bullets.
d Rubisco labeled with EDANS at amino acid 34.
e Rubisco labeled with fluorescein at amino acid 454.
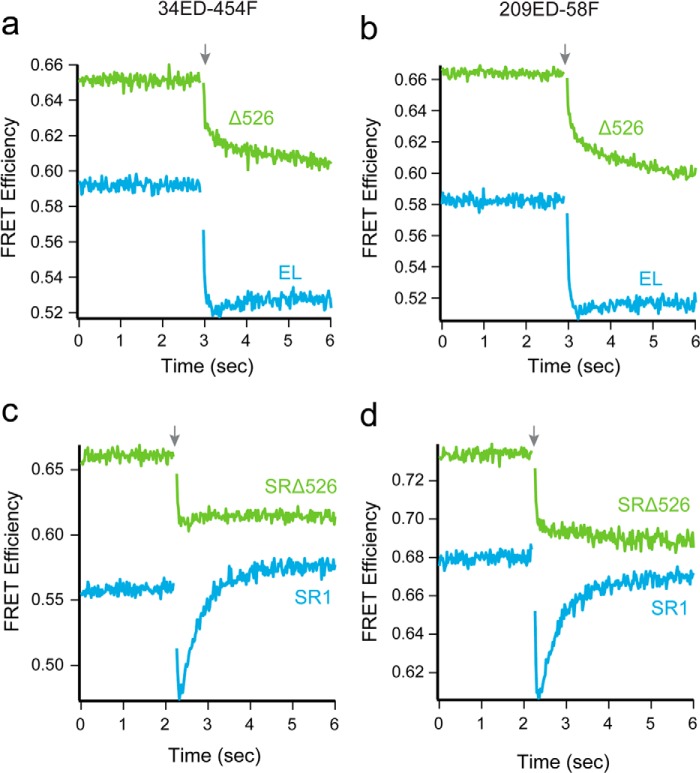
We previously demonstrated that the C-terminal tails help prevent premature substrate-protein escape during GroES binding (46). This finding suggests that the C termini maintain contact with the folding intermediate even as the apical domains, which bind different parts of the substrate protein, re-arrange in response to ATP binding (46). As the C-terminal tails appear to contribute to binding-induced unfolding, we reasoned they may also impact a later step, ATP-driven forced unfolding (21, 23). To test this possibility, we examined the conformation of the GroEL-bound Rubisco monomer during ATP and GroES binding. Using the same set of intramolecular FRET pairs described above, in combination with stopped-flow rapid mixing, we examined the time-resolved changes in conformation of a trans ring-bound Rubisco monomer as ATP and GroES bind. An example of the changes for two FRET pairs (34ED-454F and 209ED-58F) is shown in Fig. 5, a and b. All four pairs displayed similar overall behavior.
FIGURE 5.
Removal of the GroEL C termini diminishes both forced unfolding and compaction of Rubisco. a and b, change in FRET efficiency of labeled Rubisco (100 nm), bound to the trans ring of either a full-length GroEL-GroES (blue) or ELΔ526-GroES (green) complex (120 nm), during encapsulation by GroES. The change in FRET efficiency for 34ED-454F is shown in a and for 209ED-58F is shown in b. c and d, change in FRET efficiency of labeled Rubisco (100 nm) bound to the single ring GroEL variants SR1 (blue) or SRΔ526 (green) (500 nm), during encapsulation by GroES (1 μm). The change in FRET efficiency for 34ED-454F labeled Rubisco is shown in c and 209ED-58F is shown in d. In each case, the starting FRET efficiency of non-native Rubisco bound to each GroEL variant is shown, beginning at t= 0. At t = 2.25 s (arrow), the complex was rapidly mixed with ATP and GroES; n = 21 replicates, with sampling times of 20 ms. The small gap in each plot is a consequence of the removal of data points collected during the mixing dead time.
The kinetic behavior observed with full-length GroEL is consistent with our previous observations of forced unfolding upon ATP binding, followed by compaction of the Rubisco monomer as GroES binds (21, 23, 36). We observe a very rapid drop in FRET efficiency over the first 200–500 ms, which is followed by a slower increase in FRET efficiency (Fig. 5, a and b). With Δ526, we also observe the early and rapid drop in the FRET efficiency with all four site pairs, although the amplitude of the rapid FRET decrease is smaller with Δ526 compared with full-length GroEL (Fig. 5, a and b). More striking is the change in the GroES-dependent compaction phase, which is absent when the Rubisco intermediate is encapsulated on a Δ526 trans ring.
The change in compaction of the Rubisco monomer was also observed using SR1 and SRΔ526. With SR1, we readily observed forced expansion of the Rubisco monomer, followed by compaction of the folding intermediate. Because each FRET pair again displayed similar overall behavior, we only show the data for the 34ED-454F and 209ED-58F pairs in Fig. 5, c and d. Interestingly, we observe a larger amplitude compaction phase upon GroES binding to SR1 than is observed with the trans ring of the GroEL-GroES complex (21). The reason for this difference is not known, but is likely due, as least in part, to the fact that the SR1 sample does not cycle. By contrast, for tetradecamer GroEL, cycling was initiated within the first several seconds of the experiment, resulting in asynchronous reaction phases for the Rubisco monomers and an apparent difference in average FRET efficiency. When Rubisco is bound to SRΔ526, we observed a reduced forced expansion phase, as well as the complete disappearance of the GroES-dependent compaction phase. The extent of GroES binding and Rubisco encapsulation was similar for all of the GroEL variants employed in these experiments (both Δ526 and full-length) under the conditions used (data not shown; see “Experimental Procedures”), ruling out trivial explanations for this change. Taken together, these observations indicate that the C termini directly contribute to unfolding by binding and holding segments of a folding intermediate near the bottom of the GroEL cavity.
GroEL C Termini Enhance Productive Rubisco Folding
In addition to their involvement in protein unfolding, the C-terminal tails appear to affect protein folding within the GroEL-GroES cavity. We and others observe a modest but clear difference in intra-cavity folding rate in the presence and absence of the C termini (Fig. 1) (40, 43). To gain greater insight into the impact of the C termini on intra-cavity folding, we examined the changes in intramolecular FRET efficiency of labeled Rubisco monomers during folding inside the SR-GroES and SRΔ526-GroES cavities. Because the four FRET vectors span several different regions and length scales of the Rubisco monomer, we anticipated that this assay would provide a higher resolution examination of intra-cavity folding and potentially illuminate how the C termini enhance the process.
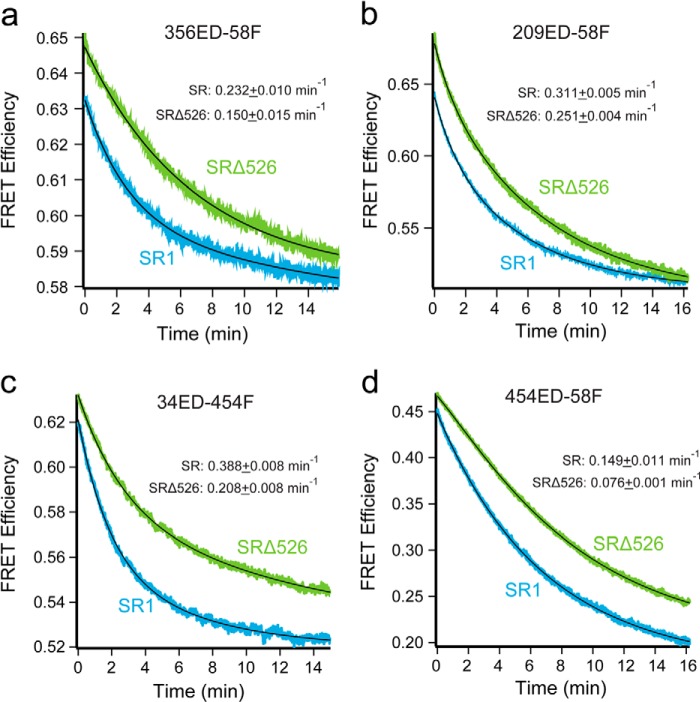
All four Rubisco FRET pairs demonstrate substantial decreases in FRET efficiency upon the initiation of folding inside both SR1-GroES and SRΔ526-GroES cavities (Fig. 6). The observed decrease in FRET efficiency shows that as the monomer folds, the labeled sites move apart, indicating that, on average, the kinetically trapped Rubisco folding intermediate is more structurally collapsed than the committed monomer. In most cases, the observed FRET transients were not well fit by single exponential rate laws and required double exponentials for good fits (Table 2). The precise reason for this kinetic complexity is unknown. However, one plausible explanation is the appearance of two kinetically resolvable Rubisco subpopulations in the GroEL-GroES cavity that possess distinct conformational transition rates in the regions probed by the different FRET pair. Despite this kinetic complexity, the four FRET pairs detect three average temporal regimes in the folding process (Table 2). Although the 356ED-58F pair appears to track the same committed step in folding that is observed by enzymatic activity, the other pairs report on steps both slower (454ED-58F) and faster (209ED-58F and 34ED-454F). Interestingly, each of the three kinetic phases identified during folding within the SR1-GroES cavity are also present when Rubisco folds in the absence of the C termini inside the SRΔ526-GroES cavity. However, the average rate of each kinetic phase is slower by 20–50% when folding proceeds in the SRΔ526-GroES cavity (Table 2).
FIGURE 6.
Intra-cavity folding of Rubisco with and without the GroEL C termini monitored by intramolecular FRET. Rubisco folding inside full-length and truncated single ring GroEL-GroES complexes monitored by intramolecular FRET with four different site pairs as follows: a, 356ED-58F; b, 209ED-58F; c, 34ED-454F, and d, 454ED-58F. Chemically denatured, fluorescently labeled Rubisco (100 nm) was bound to full-length SR1 or SRΔ526 (500 nm). This complex was rapidly mixed with an equal volume of excess GroES (1 μm) and ATP (2 mm) in a stopped-flow apparatus. Shown is the average of n = 12 replicates of matched experimental pairs, calculated from donor-only and donor-acceptor samples. Sampling time was 150 ms. In all cases, the change in FRET efficiency was well fit by a bi-exponential rate law (black line), and the average folding rate constant is shown for each case. Table 2 contains the rate constants for each fit.
TABLE 2.
Intra-cavity folding rates of Rubisco with and without the GroEL C-termini
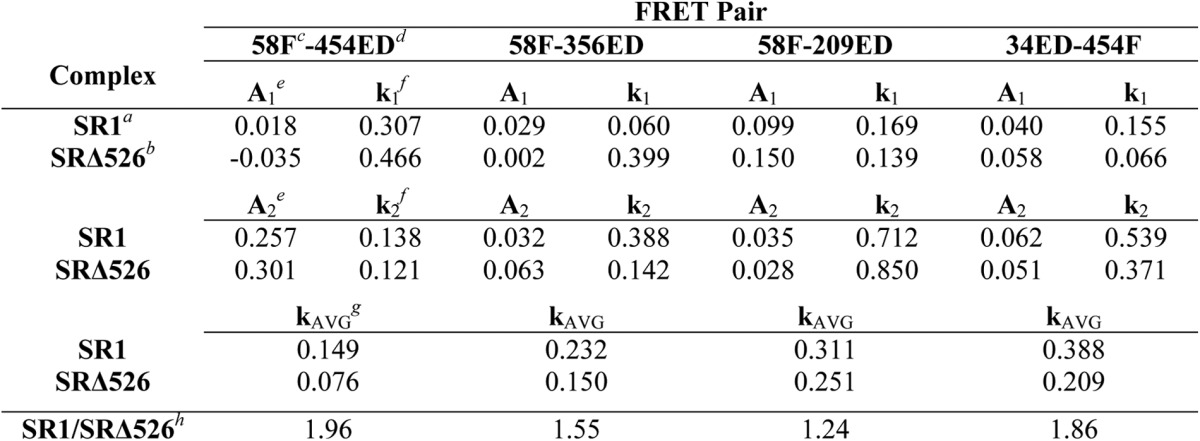
a Folding was inside full-length SR1-GroES cavity.
b Folding was inside truncated SRΔ526-GroES cavity.
c Rubisco was labeled with fluorescein at amino acid 58.
d Rubisco was labeled with EDANS at amino acid 454.
e Pre-exponential amplitude terms were from double exponential fits of folding transients.
f Rate constant terms were from double exponential fits of folding transients.
g Calculated average rate constant was for the double exponential fit.
h Ratio of average folding rate constant was for wild-type and Δ526 GroEL.
As a complement to the stopped-flow FRET measurements, we employed changes in tryptophan fluorescence to report on the folding of the Rubisco monomer at early times inside the SR1-GroES and SRΔ526-GroES cavities. The tryptophan emission spectra of non-native Rubisco bound to SR1 and SRΔ526 are very similar (Fig. 7a, inset), suggesting that the average exposure and environment of the Rubisco tryptophan residues are also similar in both starting complexes. Consistent with our earlier measurements, a rapid decrease in fluorescence intensity was followed by a much slower rise in fluorescence that matches the rate of the committed folding step (Fig. 7b), when ATP and GroES are added to the SR1 complex (15). The early drop in tryptophan fluorescence intensity likely reports on the process of conformational expansion, GroES binding, and release of the Rubisco monomer into the SR1-GroES cavity. However, the early folding behavior with SRΔ526 is quite different. Although we observe a rapid decrease in fluorescence intensity upon the addition of ATP and GroES to the SRΔ526 complex, the decrease is slower, biphasic, and shows a greater amplitude compared with SR1 (Fig. 7, a and b). In addition, the rising phase that reports on formation of the committed Rubisco monomer is dramatically delayed, relative to SR1.
FIGURE 7.
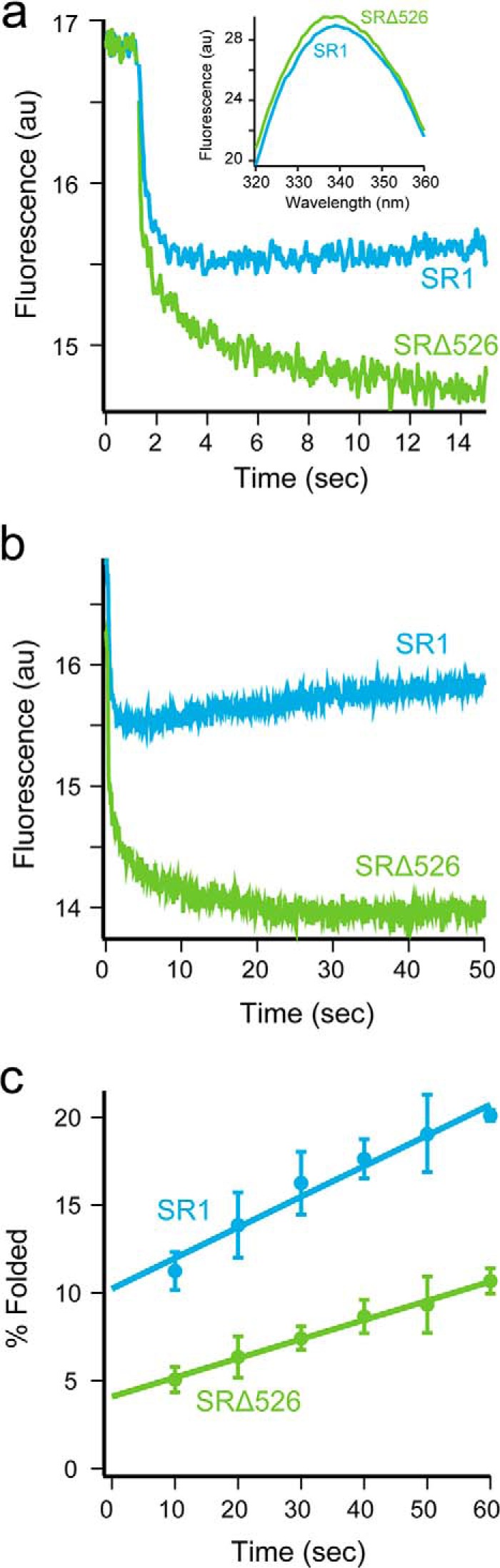
GroEL C termini enhance the fraction of Rubisco that folds rapidly upon encapsulation beneath GroES. a, intra-cavity folding of Rubisco at early times monitored by changes in tryptophan fluorescence following addition of GroES and ATP to complexes of wild-type Rubisco bound to SR1 (blue) or SRΔ526 (green). Conditions are as in Fig. 1b, except with sampling time of 20 ms. The steady-state, tryptophan fluorescence emission spectra of wild-type Rubisco bound to SR1 (blue) or SRΔ526 (green) are shown (inset). b, same as in a but with the time scale expanded to show the full 50 s of folding monitored in this experiment. c, Rubisco folding at early times with SR1 and SRΔ526 monitored by regain of enzymatic activity. Chemically denatured, wild-type Rubisco (100 nm) was bound to either 500 nm SR1 (blue) or SRΔ526 (green), to which excess GroES (1 μm) and ATP (2 mm) were added. Folding was quenched by adding EDTA and simultaneously lowering the temperature to nonpermissive folding conditions. AU, arbitrary units. Error bars represent the standard deviation from n = 3 experiments.
Given the more extensive unfolding and compaction of the Rubisco monomer seen with SR1 (Figs. 4 and 5), the reduced perturbation in tryptophan fluorescence over the same time period is surprising. A simple explanation is suggested by the increase in tryptophan fluorescence that accompanies the committed folding step, which follows release of the less fluorescent, non-native monomer into the cavity (Figs. 1 and 7, a and b) (15). In SR1, the committed step is completed much sooner than in SRΔ526 (Fig. 7, a and b). If a subpopulation of the Rubisco monomers achieves the committed monomer conformation very rapidly, then formation of small amounts of this more fluorescent, committed conformation would partially compensate for the drop in fluorescence of the more slowly progressing subpopulation, thereby accounting for the smaller amplitude of the tryptophan fluorescence drop within SR1. The larger apparent drop in tryptophan fluorescence within SRΔ526 would then be the result of less extensive unfolding and a correspondingly smaller subpopulation of a rapidly folding monomer. This explanation predicts that folding within SRΔ526 should result in a substantially reduced fraction of Rubisco monomers that commit rapidly upon addition of ATP and GroES. Rapid commitment is detected as a dead-time burst in native enzyme activity in single turnover folding experiments (23). As shown in Fig. 7c, we observe a greater than 2-fold drop in the early fraction of rapidly committed Rubisco monomers in comparison with SR1. The fact that this prediction is met suggests strongly that the C-terminal tails help stimulate Rubisco folding through structural disruption of kinetically trapped Rubisco monomers.
DISCUSSION
The results we present here strongly support an active role for GroEL in assisted protein folding (Fig. 8). We have shown that the unstructured C-terminal tails of GroEL bind and pull a partially structured Rubisco folding intermediate toward the bottom of the GroEL cavity. In the absence of the C termini, several distinct folding transitions of the Rubisco monomer slow inside the GroEL-GroES cavity, suggesting that the conformational search executed by the Rubisco folding intermediate is more efficient in the presence of the C-terminal tails. In addition, the C termini participate in protein unfolding, both during the initial capture of a folding intermediate on an open GroEL ring, as well as during the forced expansion of the intermediate upon ATP binding. When unfolding is diminished by removal of the C termini, the fraction of the Rubisco monomers that populate the most rapid folding pathways is substantially reduced.
FIGURE 8.
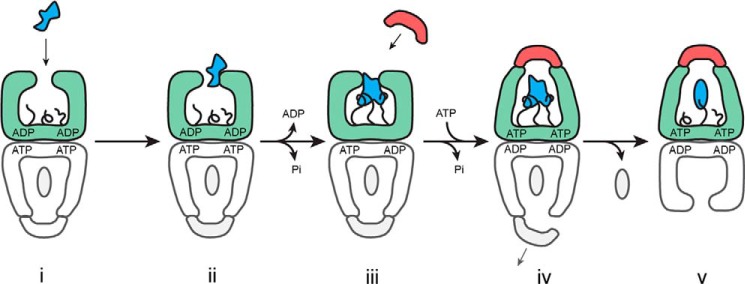
Model for the role of the GroEL C termini in substrate protein unfolding. A schematic is shown for the steps involved in substrate protein loading and the initiation of folding by GroEL. Step i, a non-native substrate protein (irregular blue shape) enters the GroEL reaction cycle on the open trans ring (green) of the ATP bullet complex (21). Step ii, substrate protein binding accelerates both the release of ADP from the trans ring and ATP hydrolysis in the opposite, cis ring (gray) (22, 28, 56, 66). Step iii, binding of the non-native substrate protein by the C-terminal tails (black), helps pull the substrate protein into the GroEL cavity and, in combination with additional binding by multiple apical domains, results in substrate protein unfolding. Step iv, ongoing association of the C termini with the substrate protein during ATP-driven encapsulation by GroES both retards pre-mature protein release (46) and provides an anchor point for forced expansion of the substrate protein as the apical domains shift to accommodate GroES binding. Assembly of the new folding cavity on the trans ring is directly coupled to the disassembly of the folding cavity on the opposite ring, potentially through a transient, symmetric intermediate (28, 53–55). Step v, a subsequent allosteric shift of the GroEL-GroES complex results in full ejection of the substrate protein in the enclosed GroEL-GroES cavity and the initiation of folding (46). The C-terminal tails may continue to interact with the folding intermediate, influencing the spectrum of states populated during folding.
Our observations are consistent with a key role for substrate protein unfolding in GroEL-stimulated folding but inconsistent with an exclusive role for substrate protein confinement. Removal of the C-terminal tails in the Δ526 GroEL variant not only slows the rate of Rubisco folding but also simultaneously increases the lifetime of the GroEL-GroES complex. If the stimulation of folding occurs mainly through substrate protein confinement within the GroEL-GroES cavity, then the longer lifetime of the Δ526-GroES complex presents a paradox; even taking into account the modest reduction in intra-cavity folding observed with SRΔ526 (Figs. 1 and 6), the considerably longer Δ526-GroES cavity lifetime (Fig. 2) should result in better Rubisco folding compared with full-length GroEL, not worse. This points to some other property of the GroEL machine that must contribute to the stimulation of Rubisco folding. We previously demonstrated that a Rubisco monomer is subjected to two phases of multiple axis unfolding prior to the initiation of productive folding inside the GroEL-GroES cis cavity as follows: 1) binding-driven unfolding upon capture of a folding intermediate by the GroEL ring, and 2) forced unfolding upon ATP binding to a Rubisco-occupied GroEL ring (21, 23, 36). The fraction of the Rubisco population that populates efficient folding pathways is proportional to the magnitude of this unfolding (23). Furthermore, the rapidly cycling GroEL-GroES system can achieve assisted folding rates that are substantially faster than what confined folding in the GroEL-GroES cavity can achieve alone (21). These observations are consistent with predictions of the iterative annealing model of stimulated protein folding by GroEL (30, 31).
Importantly, the behavior of the Δ526 GroEL variant also satisfies predictions of the iterative annealing model. If GroEL acts, at least in part, as an iterative annealing machine, disrupting inhibitory and kinetically trapped states through partial unfolding, then any modification to the machine that reduces the extent of unfolding and slows the rate of turnover should result in a reduced rate of stimulated folding (31). As we have shown with observations of the Δ526 GroEL variant, these predictions are met. Not only does removal of the C-terminal tails reduce both binding-induced and forced unfolding, resulting in a substantial decrease in the fraction of Rubisco that rapidly commits to the native state, but removal of the tails also slows the rate at which the GroEL-GroES system cycles, reducing the frequency with which a given population of Rubisco folding intermediates can be subjected to unfolding.
Although our observations with Δ526 GroEL are consistent with key elements of the iterative annealing model, they do not exclude a role for protein confinement. Indeed, we previously demonstrated that encapsulation of the Rubisco monomer beneath GroES is associated with compaction of the folding intermediate, a conformational restriction that is, in principle, consistent with confinement-based models (21, 36). Notably, the magnitude and kinetics of this compaction event are compromised by removal of the GroEL C termini (Fig. 5). At the same time, intra-cavity folding of Rubisco is measurably slower in the absence of the C termini (Figs. 1 and 6). It is possible that the reduced rate of Rubisco folding inside the SRΔ526-GroES cavity is simply the result of reduced initial unfolding, so that the rate of all subsequent steps are impacted by the starting conformation of the protein at the moment of encapsulation. Indeed, it is remarkable that each of the kinetically distinct steps identified with the intramolecular FRET assay all slow to a similar extent in the absence of the C termini (Fig. 6 and Table 2). However, both cavity volume and character have also been suggested to impact intra-cavity folding, and both properties should be altered by deletion of the C-terminal tails (37, 39–41). Yet, the shift in the Rubisco binding position upon removal of the C-terminal tails is inconsistent with a volume effect alone. If the C termini primarily act via spatial constriction of the cavity, their removal would be expected to either have no effect on the Rubisco binding position or would allow it to occupy more of the cavity volume and thus bind more deeply into the cavity. However, we observe the opposite effect, i.e. removal of the C-terminal tails results in a more elevated average binding position (Fig. 3). This observation, in combination with our earlier cryo-EM observations (46), suggests a direct binding interaction with the Rubisco folding intermediate, whereby the C termini help pull and stretch the monomer toward the bottom of the GroEL cavity.
In confinement-based mechanisms, the spectrum of partially folded intermediates in the GroEL-GroES cavity should differ from the ensemble of states populated in free solution. Precisely how interactions between the C-terminal tails and a protein folding intermediate could impact this distribution is not understood. Transient interactions between the weakly hydrophobic interior of the GroEL cavity and a folding intermediate have been proposed to assist productive folding through an annealing process that prevents or destabilizes kinetically trapped states (9, 10). Consistent with a role for the C termini in such a process, we observe ongoing, although weakened, interactions between the Rubisco folding intermediate and the C-terminal tails during early stages of intra-cavity folding (46). Additionally, the amphipathic character of the C-terminal tails appears to be a key property of these unstructured segments as follows: modifications that make them either too polar or too hydrophobic inhibit folding (39, 45). Notably, Rubisco displays a striking ability to fold inside the GroEL-GroES cavity under conditions where the monomer in free solution folds slowly or not at all (15, 36, 37). More recently, a double mutant of the maltose-binding protein has been shown to display similar behavior (38). For stringent substrate proteins like Rubisco, it is tempting to speculate that a combination of unfolding and confinement might provide the most efficient method of stimulating productive folding. In a combined mechanism, unfolding and disruption of kinetically trapped, misfolded states would increase the chance of opening a productive folding pathway. Subsequent confinement of the partially unfolded intermediate within the GroEL-GroES cavity could then provide a maximally conducive environment, in which the probability of populating inhibitory conformational states is minimized, at least for the short duration of the GroEL-GroES complex.
Given the clear impact of the GroEL C termini on both protein encapsulation and folding, it is striking that these segments are not essential in vivo (44, 57). At the same time, the C-terminal tails are well conserved in the majority of chaperonins (58). These observations, combined with the near universal essentiality of chaperonins across phyla, suggest that the efficiency of protein folding by chaperonins is a tight evolutionary constraint, a conclusion supported by recent studies on the linkage between both fitness and protein evolutionary rates and chaperonin activity (59–63). In this view, even a modest loss of folding capacity, like that caused by the absence of the C-terminal tails, would result in a steep reduction in overall fitness. Indeed, one of the first studies to examine the role of the GroEL C termini observed just such an effect (44). In competition experiments between otherwise identical E. coli strains expressing either a full-length or a C-terminally truncated GroEL, cells forced to rely on a truncated GroEL rapidly lost out. Modifications of the C termini may also be linked to shifts in substrate specificity or activity of different chaperonin subtypes. Many microbial species maintain multiple, distinct chaperonin variants in the same cytoplasm, with the actinobacteria of particular note (64). In most cases, the essential housekeeping chaperonin (Cpn60.2) has a C-terminal tail that retains the sequence character and Gly-Gly-Met motifs common to eubacterial chaperonins like GroEL. Strikingly, a secondary chaperonin (Cpn60.1), thought to be a specialized variant important for biofilm formation and pathogenesis, possesses a modified tail in which the sequence character has been dramatically altered, and the typically conserved Gly-Gly-Met motifs have been replaced with sequences enriched in His (64). Thus, although the results we report here shed additional light on the role of the GroEL C termini, our understanding of the role of these important and characteristic chaperonin domains remains incomplete.
Acknowledgment
We thank Dr. Chavela Carr for helpful discussions and editorial assistance.
This work was supported by National Institutes of Health Grant GM065421 (to H. S. R.).
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- EDANS or ED
- 5-(2-acetamidoethyl) aminonaphthalene 1-sulfonate
- fluorescein or F
- 5-iodoacetamidofluorescein.
REFERENCES
- 1. Dobson C. M. (2003) Protein folding and misfolding. Nature 426, 884–890 [DOI] [PubMed] [Google Scholar]
- 2. Anfinsen C. B. (1973) Principles that govern the folding of protein chains. Science 181, 223–230 [DOI] [PubMed] [Google Scholar]
- 3. Grantcharova V., Alm E. J., Baker D., Horwich A. (2001) Mechanisms of protein folding. Curr. Opin. Struct. Biol. 11, 70–82 [DOI] [PubMed] [Google Scholar]
- 4. Brockwell D. J., Radford S. E. (2007) Intermediates: ubiquitous species on folding energy landscapes? Curr. Opin. Struct. Biol. 17, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Powers E. T., Morimoto R. I., Dillin A., Kelly J. W., Balch W. E. (2009) Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78, 959–991 [DOI] [PubMed] [Google Scholar]
- 6. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 7. Powers E. T., Powers D. L., Gierasch L. M. (2012) FoldEco: a model for proteostasis in E. coli. Cell Rep. 1, 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horwich A. L., Fenton W. A. (2009) Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q. Rev. Biophys. 42, 83–116 [DOI] [PubMed] [Google Scholar]
- 9. Lin Z., Rye H. (2006) GroEL-mediated protein folding: making the impossible, possible. Crit. Rev. Biochem. Mol. Biol. 41, 211–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jewett A. I., Shea J.-E. (2010) Reconciling theories of chaperonin accelerated folding with experimental evidence. Cell. Mol. Life Sci. 67, 255–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Braig K., Otwinowski Z., Hegde R., Boisvert D. C., Joachimiak A., Horwich A. L., Sigler P. B. (1994) The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature 371, 578–586 [DOI] [PubMed] [Google Scholar]
- 12. Fenton W. A., Kashi Y., Furtak K., Horwich A. L. (1994) Residues in chaperonin GroEL required for polypeptide binding and release (see comments). Nature 371, 614–619 [DOI] [PubMed] [Google Scholar]
- 13. Kerner M. J., Naylor D. J., Ishihama Y., Maier T., Chang H. C., Stines A. P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Hartl F. U. (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122, 209–220 [DOI] [PubMed] [Google Scholar]
- 14. Fujiwara K., Ishihama Y., Nakahigashi K., Soga T., Taguchi H. (2010) A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 29, 1552–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rye H. S., Burston S. G., Fenton W. A., Beechem J. M., Xu Z., Sigler P. B., Horwich A. L. (1997) Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL (see comments). Nature 388, 792–798 [DOI] [PubMed] [Google Scholar]
- 16. Weissman J. S., Rye H. S., Fenton W. A., Beechem J. M., Horwich A. L. (1996) Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell 84, 481–490 [DOI] [PubMed] [Google Scholar]
- 17. Mayhew M., da Silva A. C., Martin J., Erdjument-Bromage H., Tempst P., Hartl F. U. (1996) Protein folding in the central cavity of the GroEL-GroES chaperonin complex. Nature 379, 420–426 [DOI] [PubMed] [Google Scholar]
- 18. Weissman J. S., Hohl C. M., Kovalenko O., Kashi Y., Chen S., Braig K., Saibil H. R., Fenton W. A., Horwich A. L. (1995) Mechanism of GroEL action: productive release of polypeptide from a sequestered position under GroES. Cell 83, 577–587 [DOI] [PubMed] [Google Scholar]
- 19. Xu Z., Horwich A. L., Sigler P. B. (1997) The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex (see comments). Nature 388, 741–750 [DOI] [PubMed] [Google Scholar]
- 20. Rye H. S., Roseman A. M., Chen S., Furtak K., Fenton W. A., Saibil H. R., Horwich A. L. (1999) GroEL-GroES cycling: ATP and nonnative polypeptide direct alternation of folding-active rings. Cell 97, 325–338 [DOI] [PubMed] [Google Scholar]
- 21. Lin Z., Puchalla J., Shoup D., Rye H. S. (2013) Repetitive protein unfolding by the trans ring of the GroEL-GroES chaperonin complex stimulates folding. J. Biol. Chem. 288, 30944–30955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Madan D., Lin Z., Rye H. S. (2008) Triggering protein folding within the GroEL-GroES complex. J. Biol. Chem. 283, 32003–32013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin Z., Madan D., Rye H. S. (2008) GroEL stimulates protein folding through forced unfolding. Nat. Struct. Mol. Biol. 15, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burston S. G., Ranson N. A., Clarke A. R. (1995) The origins and consequences of asymmetry in the chaperonin reaction cycle. J. Mol. Biol. 249, 138–152 [DOI] [PubMed] [Google Scholar]
- 25. Todd M. J., Viitanen P. V., Lorimer G. H. (1994) Dynamics of the chaperonin ATPase cycle: implications for facilitated protein folding. Science 265, 659–666 [DOI] [PubMed] [Google Scholar]
- 26. Weissman J. S., Kashi Y., Fenton W. A., Horwich A. L. (1994) GroEL-mediated protein folding proceeds by multiple rounds of binding and release of nonnative forms. Cell 78, 693–702 [DOI] [PubMed] [Google Scholar]
- 27. Ranson N. A., Dunster N. J., Burston S. G., Clarke A. R. (1995) Chaperonins can catalyse the reversal of early aggregation steps when a protein misfolds. J. Mol. Biol. 250, 581–586 [DOI] [PubMed] [Google Scholar]
- 28. Ye X., Lorimer G. H. (2013) Substrate protein switches GroE chaperonins from asymmetric to symmetric cycling by catalyzing nucleotide exchange. Proc. Natl. Acad. Sci. U.S.A. 110, E4289–E4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Apetri A. C., Horwich A. L. (2008) Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc. Natl. Acad. Sci. U.S.A. 105, 17351–17355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Todd M. J., Lorimer G. H., Thirumalai D. (1996) Chaperonin-facilitated protein folding: optimization of rate and yield by an iterative annealing mechanism. Proc. Natl. Acad. Sci. U.S.A. 93, 4030–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Betancourt M. R., Thirumalai D. (1999) Exploring the kinetic requirements for enhancement of protein folding rates in the GroEL cavity. J. Mol. Biol. 287, 627–644 [DOI] [PubMed] [Google Scholar]
- 32. Horst R., Fenton W. A., Englander S. W., Wüthrich K., Horwich A. L. (2007) Folding trajectories of human dihydrofolate reductase inside the GroEL GroES chaperonin cavity and free in solution. Proc. Natl. Acad. Sci. U.S.A. 104, 20788–20792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Coyle J. E., Texter F. L., Ashcroft A. E., Masselos D., Robinson C. V., Radford S. E. (1999) GroEL accelerates the refolding of hen lysozyme without changing its folding mechanism. Nat. Struct. Biol. 6, 683–690 [DOI] [PubMed] [Google Scholar]
- 34. Chen J., Walter S., Horwich A. L., Smith D. L. (2001) Folding of malate dehydrogenase inside the GroEL-GroES cavity. Nat. Struct. Biol. 8, 721–728 [DOI] [PubMed] [Google Scholar]
- 35. Goldberg M. S., Zhang J., Sondek S., Matthews C. R., Fox R. O., Horwich A. L. (1997) Native-like structure of a protein-folding intermediate bound to the chaperonin GroEL. Proc. Natl. Acad. Sci. U.S.A. 94, 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin Z., Rye H. S. (2004) Expansion and compression of a protein folding intermediate by GroEL. Mol. Cell 16, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brinker A., Pfeifer G., Kerner M. J., Naylor D. J., Hartl F. U., Hayer-Hartl M. (2001) Dual function of protein confinement in chaperonin-assisted protein folding. Cell 107, 223–233 [DOI] [PubMed] [Google Scholar]
- 38. Chakraborty K., Chatila M., Sinha J., Shi Q., Poschner B. C., Sikor M., Jiang G., Lamb D. C., Hartl F. U., Hayer-Hartl M. (2010) Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell 142, 112–122 [DOI] [PubMed] [Google Scholar]
- 39. Tang Y. C., Chang H. C., Roeben A., Wischnewski D., Wischnewski N., Kerner M. J., Hartl F. U., Hayer-Hartl M. (2006) Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125, 903–914 [DOI] [PubMed] [Google Scholar]
- 40. Tang Y. C., Chang H. C., Chakraborty K., Hartl F. U., Hayer-Hartl M. (2008) Essential role of the chaperonin folding compartment in vivo. EMBO J. 27, 1458–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sharma S., Chakraborty K., Müller B. K., Astola N., Tang Y.-C., Lamb D. C., Hayer-Hartl M., Hartl F. U. (2008) Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell 133, 142–153 [DOI] [PubMed] [Google Scholar]
- 42. Motojima F., Motojima-Miyazaki Y., Yoshida M. (2012) Revisiting the contribution of negative charges on the chaperonin cage wall to the acceleration of protein folding. Proc. Natl. Acad. Sci. U.S.A. 109, 15740–15745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Farr G. W., Fenton W. A., Horwich A. L. (2007) Perturbed ATPase activity and not “close confinement” of substrate in the cis cavity affects rates of folding by tail-multiplied GroEL. Proc. Natl. Acad. Sci. U.S.A. 104, 5342–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McLennan N. F., Girshovich A. S., Lissin N. M., Charters Y., Masters M. (1993) The strongly conserved carboxyl-terminus glycine-methionine motif of the Escherichia coli GroEL chaperonin is dispensable. Mol. Microbiol. 7, 49–58 [DOI] [PubMed] [Google Scholar]
- 45. Machida K., Kono-Okada A., Hongo K., Mizobata T., Kawata Y. (2008) Hydrophilic residues 526KNDAAD531 in the flexible C-terminal region of the chaperonin GroEL are critical for substrate protein folding within the central cavity. J. Biol. Chem. 283, 6886–6896 [DOI] [PubMed] [Google Scholar]
- 46. Chen D.-H., Madan D., Weaver J., Lin Z., Schröder G. F., Chiu W., Rye H. S. (2013) Visualizing GroEL/ES in the act of encapsulating a folding protein. Cell 153, 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rye H. (2001) Application of fluorescence resonance energy transfer to the GroEL-GroES chaperonin reaction. Methods 24, 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. James D. R., Siemiarczuk A., Ware W. R. (1992) Stroboscopic optical boxcar technique for the determination of fluorescence lifetimes. Rev. Sci. Instrum. 63, 1710–1716 [Google Scholar]
- 49. Suzuki M., Ueno T., Iizuka R., Miura T., Zako T., Akahori R., Miyake T., Shimamoto N., Aoki M., Tanii T., Ohdomari I., Funatsu T. (2008) Effect of the C-terminal truncation on the functional cycle of chaperonin GroEL: implication that the C-terminal region facilitates the transition from the folding-arrested to the folding-competent state. J. Biol. Chem. 283, 23931–23939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gray T. E., Fersht A. R. (1991) Cooperativity in ATP hydrolysis by GroEL is increased by GroES. Correction (1992) FEBS Lett. 292, 254–258 [DOI] [PubMed] [Google Scholar]
- 51. Jackson G. S., Staniforth R. A., Halsall D. J., Atkinson T., Holbrook J. J., Clarke A. R., Burston S. G. (1993) Binding and hydrolysis of nucleotides in the chaperonin catalytic cycle: implications for the mechanism of assisted protein folding. Biochemistry 32, 2554–2563 [DOI] [PubMed] [Google Scholar]
- 52. Yifrach O., Horovitz A. (1995) Nested cooperativity in the ATPase activity of the oligomeric chaperonin GroEL. Biochemistry 34, 5303–5308 [DOI] [PubMed] [Google Scholar]
- 53. Sameshima T., Iizuka R., Ueno T., Wada J., Aoki M., Shimamoto N., Ohdomari I., Tanii T., Funatsu T. (2010) Single-molecule study on the decay process of the football-shaped GroEL-GroES complex using zero-mode waveguides. J. Biol. Chem. 285, 23159–23164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sameshima T., Iizuka R., Ueno T., Funatsu T. (2010) Denatured proteins facilitate the formation of the football-shaped GroEL-(GroES)2 complex. Biochem. J. 427, 247–254 [DOI] [PubMed] [Google Scholar]
- 55. Yang D., Ye X., Lorimer G. H. (2013) Symmetric GroEL:GroES2 complexes are the protein-folding functional form of the chaperonin nanomachine. Proc. Natl. Acad. Sci. U.S.A. 110, E4298–E4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grason J. P., Gresham J. S., Lorimer G. H. (2008) Setting the chaperonin timer: A two-stroke, two-speed, protein machine. Proc. Natl. Acad. Sci. U.S.A. 105, 17339–17344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Burnett B. P., Horwich A. L., Low K. B. (1994) A carboxy-terminal deletion impairs the assembly of GroEL and confers a pleiotropic phenotype in Escherichia coli K-12. J. Bacteriol. 176, 6980–6985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brocchieri L., Karlin S. (2000) Conservation among HSP60 sequences in relation to structure, function, and evolution. Protein Sci. 9, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tokuriki N., Tawfik D. S. (2009) Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459, 668–673 [DOI] [PubMed] [Google Scholar]
- 60. Wyganowski K. T., Kaltenbach M., Tokuriki N. (2013) GroEL/ES buffering and compensatory mutations promote protein evolution by stabilizing folding intermediates. J. Mol. Biol. 425, 3403–3414 [DOI] [PubMed] [Google Scholar]
- 61. Williams T. A., Fares M. A. (2010) The effect of chaperonin buffering on protein evolution. Genome Biol. Evol. 2, 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cetinbaş M., Shakhnovich E. I. (2013) Catalysis of protein folding by chaperones accelerates evolutionary dynamics in adapting cell populations. PLoS Comput. Biol. 9, e1003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fares M. A., Ruiz-González M. X., Moya A., Elena S. F., Barrio E. (2002) Endosymbiotic bacteria: groEL buffers against deleterious mutations. Nature 417, 398. [DOI] [PubMed] [Google Scholar]
- 64. Lund P. A. (2009) Multiple chaperonins in bacteria–“ why so many? FEMS Microbiol. Rev. 33, 785–800 [DOI] [PubMed] [Google Scholar]
- 65. Murai N., Makino Y., Yoshida M. (1996) GroEL locked in a closed conformation by an interdomain cross-link can bind ATP and polypeptide but cannot process further reaction steps. J. Biol. Chem. 271, 28229–28234 [DOI] [PubMed] [Google Scholar]
- 66. Kad N. M., Ranson N. A., Cliff M. J., Clarke A. R. (1998) Asymmetry, commitment and inhibition in the GroE ATPase cycle impose alternating functions on the two GroEL rings. J. Mol. Biol. 278, 267–278 [DOI] [PubMed] [Google Scholar]