Abstract
Transcripts of pak4, the zebrafish ortholog of p21-activated kinase 4 (PAK4), are most abundant in the egg and fall to low levels by the end of gastrulation, after which expression is essentially ubiquitous. Translation of maternal mRNA into pak4 protein is first detectable at high stage (3.3 hpf). Splice-blocking morpholino oligonucleotides (MOs) were used to prevent zygotic pak4 expression. This had no discernable effect on development through larval stages. In contrast, a translation-blocking MO, alone or in combination with the splice MOs, resulted in a complex lethal phenotype. In addition to disrupted somite development and other morphogenetic abnormalities, the knockdown of maternal pak4 expression led to alterations in regulatory gene expression in the primitive hematopoietic domains, leading to deficiencies in granulocyte and leukocyte lineages. At least some of the effects of pak4 knockdown on gene expression could be mimicked by treatment with actin depolymerization agents, suggesting a mechanistic link between regulation of microfilament dynamics by pak4 and regulation of gene expression in primitive myeloid cell differentiation.
Keywords: p21-Activated kinase 4, Maternal, Myelopoiesis, Granulocyte, Leukocyte, Actin
1. Introduction
The Rho GTPase subgroup of small GTPases (Rho, Rac and Cdc42) play central roles in the regulation of cell locomotion and cytoarchitectural reorganization (Bokoch, 2003; Edwards et al., 1999). Key effectors of the Rho GTPases are the p21-activated kinase (PAK) factors (Abo et al., 1998; Wells and Jones, 2010). There are six PAKs in mammals categorized into two groups according to their cellular responses to GTPase activation. Both group I (PAK1–3) and group II (PAK4–6) PAKs contain a GTPase-binding domain (CRIB, also named PBD), guanine nucleotide exchange factor (GEF-H1) interaction domain (GID), proline-rich regions and a C-terminal serine/threonine protein kinase domain (Callow et al., 2005; Paliouras et al., 2009; Zenke et al., 2004). So far the defined autoinhibitory domain (AID) has only been identified in group I PAKs (Ching et al., 2003; Jaffer and Chernoff, 2002; Zenke et al., 1999). Group I PAKs are activated via autophosphorylation upon binding to activated Rho GTPases while the catalytic activity of group II PAKs is not altered by this interaction (Bokoch, 2003; Eswaran et al., 2008). One possibility is that group II PAKs may be regulated via subcellular localization in response to signaling. For example, PAK4 has been reported to translocate to the Golgi apparatus in the presence of active Cdc42 (Abo et al., 1998). However, this may not be the case for other group II PAKs: PAK5 (also called PAK7) and PAK6 are localized to mitochondria in CHO cells and nucleus in CV-1 cells, respectively, but this is independent of kinase activity or binding with Rho GTPases (Cotteret et al., 2003; Yang et al., 2001).
PAK proteins have been shown to mediate cytoskeletal restructuring and cell morphology. PAK1 controls actin organization and associates with the microtubule organizing center (MTOC) while PAK4 is implicated in actin polymerization and formation of stress fibers (Dan et al., 2001; Eswaran et al., 2008; Sells et al., 1997; Zenke et al., 2004). PAK4 phosphorylates LIM kinase 1 and induces its activity to phosphorylate cofilin, leading to cell shape changes (Dan et al., 2001). During cell migration, PAK4 mediates formation of filopodia at the leading edge upon stimulation by activated Cdc42 (Abo et al., 1998). In association with Rho-family guanine nucleotide exchange factor (GEF-H1), PAK4 promotes formation of lamellipodia as well as destabilization of microtubules in cultured cells (Callow et al., 2005). PAK4 also enhances epithelial cell motility and invasion mediated by Gab1, a Met receptor tyrosine kinase, in response to hepatocyte growth factor (Paliouras et al., 2009). The roles of PAK proteins in cytoskeletal organization have also been implicated in vascular development. PAK1 mediates lamellipodia formation in endothelial cell migration and adhesion (Kiosses et al., 1999). In zebrafish, loss of pak2a and pak2b expression resulted in defects in cranial vascular integrity leading to cerebral hemorrhage (Buchner et al., 2007; Liu et al., 2007).
In mouse, loss of PAK4 resulted in an embryonic lethal phenotype (Qu et al., 2003). A major abnormality in PAK4 null embryos was defects in both embryonic and extraembryonic vasculature, where the abnormally vascularized epiblast was possibly the primary cause of death (Tian et al., 2009). Formation of early vessels was not affected by PAK4 deletion but the further vascular invasion in the labyrinthine layer of placenta was missing. PAK4-null embryos died with heart failure and thinning of the myocardial wall before embryonic day 11.5. The PAK4 knockouts also had multiple neuronal defects including failure in differentiation and migration of spinal cord motor neurons and interneurons. The caudal neural tube underwent improper folding, resulting in parted neural lumens in the PAK4 mutants. These data suggest that PAK4 has multiple functions in normal development as well as a complex role in epiblast/trophectoderm interaction (Tian et al., 2009).
While loss of function in the mouse provides valuable insight on the developmental or physiological function of a particular gene product, there are important questions in evolutionary, developmental and cell biology that can be addressed by comparing the functions of orthologous proteins in different species. For example, gene redundancy might mask interesting functions in one species that could be revealed by loss of function analysis in another. For this reason we have characterized the expression and function of pak4 in zebrafish, a highly tractable model for vertebrate development and genetics. We find pak4 to be essential for fish embryogenesis, as it is for mouse, but with important distinctions. As in mammals, zebrafish pak4 is required for normal development, but the effected tissues are different. Furthermore, zygotic expression of pak4 appears to be dispensable in zebrafish, whereas translation of maternal pak4 mRNA is essential for differentiation events in early development, including primitive myelopoiesis, which is the focus of this paper.
2. Materials and methods
2.1. Animals
Zebrafish of background Ekkwill (EK) were used in the current study. Vasculature was visualized in transgenic zebrafish (fli1a:eGFP, a gift from B. Weinstein; Lawson and Weinstein, 2002). Embryos were cultured according to standard procedures (Westerfield, 2000) and staged as previously described by Kimmel et al. (1995). All experimental procedures with zebrafish were approved by the NICHD IACUC (ASP # 09-039).
2.2. Plasmid constructs
A cDNA clone containing the full open reading frame of zebrafish pak4 (Genbank Accession NM_001002222) was obtained from Open Biosystems (Thermofisher Scientific). To construct wild type pak4 expression plasmids, PCR primers PAK4-F-EcoRI (GAG AAT TCT CTG CTG CAG CCA TGT TCA GCA) and PAK4-R-XhoI (CTC TCG AGT CAT CTC ATG CGG TTT TGT CTC) were designed at the 5′- and 3′-ends of the predicted open reading frame of pak4, respectively. Expand High Fidelity Enzyme Mix (Roche) was used in the PCR reaction. The amplification product was digested and cloned into the EcoRI-XhoI sites of pCS2+-myc tag vector to generate pCS2+-myc-PAK4, which was used for synthesis of myc-tag pak4 RNA. A 1.0-kb fragment of pak4 cDNA was subcloned into pCS2+ which was used to synthesize RNA probes for pak4. Plasmid clones for syntheses of the other riboprobes were gifts from Drs. Igor Dawid, Brant Weinstein (NICHD, NIH) and Raman Sood (NHGRI, NIH).
2.3. RNAs and antisense morpholinos (MOs)
Plasmid constructs for in vitro transcription were first linearized by restriction digestion, purified and used as templates for RNA synthesis reactions. Capped mRNAs were synthesized by using mMessage mMachine kits (Ambion) with SP6, T7 or T3 RNA polymerase according to the manufacturer’s instructions. With reference to the genomic sequence of zebrafish pak4 deposited in the EnsEMBL genome database (Zv9, ENSDARG00000018110), two splice-blocking morpholino oligonucleotides (MO-1 and MO-2) and one translation-blocking MO (MO-3) were designed. MO sequences: MO-1, GCC AGT CCA TTA GTC TTA TAT TTC G; MO-2, GAG ACT TTC ACT GAG ACC CTC TTG; MO-3, TGA AGA GTG ATG TCC AGA CTA CGG G. Initially, we titrated the dosage of splice MOs to determine the minimum concentration that would yield complete inhibition of pak4 RNA splicing. This was 1.5 ng of MO-1 and 1.5 ng of MO-2, and this dosage was used in all experiments. Target sites of MOs on the pak4 gene are shown in Fig. 2. For controls, either uninjected embryos were used, or a MO with a scrambled nucleotide sequence was injected, as indicated in the figure legends. A translation-blocking MO for p53 (GCG CCA TTG CTT TGC AAG AAT TG) was used to test for apoptosis-dependent effects of MO injection. MOs were designed and synthesized by Gene Tools LLC and prepared for injection according to the manufacturer’s instructions.
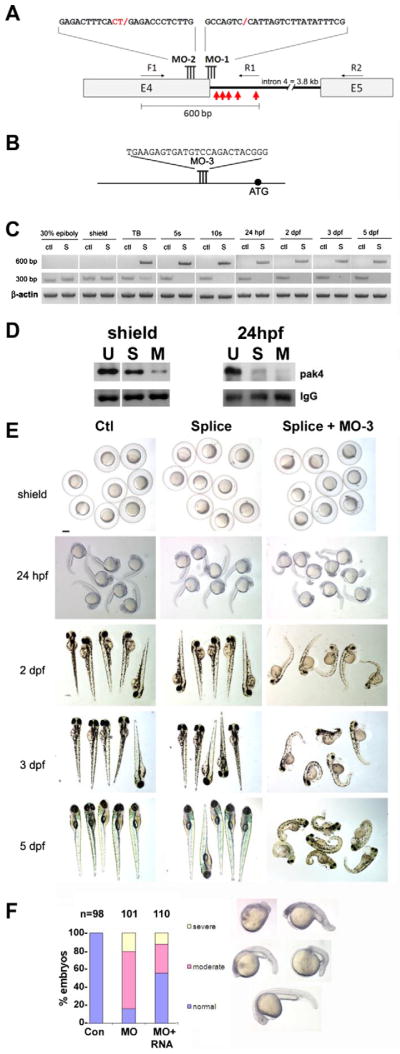
Fig. 2.
Loss of function analysis. (A) Splice-blocking MO design. The sequences of MO-1 and MO-2 and their target sites at the 3′-end of pak4 exon 4 are shown. Two MOs were necessary for complete knockdown due to the unmasking of a cryptic splice donor. F1, R1 and R2 refer to PCR primers used to monitor splicing (see Section 2). The red arrows indicate the positions of in-frame stop codons in the abortively spliced mRNA. (B) Translation-blocking MO of the indicated sequence targeting a site upstream from the start codon (black circle). (C) RT-PCR analysis of splice-blocking. Embryonic RNA from the indicated stages was isolated after injection of the control MO (ctl) or combined splice-blocking MOs (S) and used as template for PCR. The 300-bp PCR product was amplified by the primer pair F1 + R2 and represents the correctly spliced pak4 mRNA, while the 600-bp product (primers F1 + R1) represents the abortively spliced mRNA. The 600-bp product appeared at tailbud stage (TB), indicating pak4 transcription from the zygotic genome. At this stage the 300-bp product had declined in splice-inhibited embryos and had disappeared by the 5-somite stage (5s). This pattern persisted through the final stage tested, 5 dpf. As a positive control a fragment of β-actin was amplified in the corresponding samples. (D) MO effectiveness. Pak4 protein was detected by immunoprecipitation/western blotting from equal numbers of embryos injected with the splice MOs (S; MO-1, MO-2) or a mixture of splice- and translation-blocking MOs (M; MO-3), or from uninjected embryos (U). Equal protein input for immunoprecipitations was confirmed by BCA assays. At shield stage the splice MOs reduced pak4 partially, while the splice + translation-blocking mixture had a much stronger effect. Both treatments gave essentially complete inhibition at 24 hpf, although some residual pak4 protein was detected in the splice morphants. Ponceau S staining of the IgG heavy chain band (IgG) is shown below as a control for immunoprecipitation recovery. (E) Representative embryos at various stages injected with control (Ctl), splice-blocking MOs (Splice; 1.5 + 1.5 ng), or a combination of the translation- (3 ng) and splice-blocking (1.5 + 1.5 ng) MOs (Splice + MO-3). Only the latter showed disrupted morphology, as observed at 24 hpf and steadily worsening through 5 dpf. Scale bar, 250 μM. (F) Summary of pak4 mRNA rescue data at 24 hpf. Morphant embryos were classified as normal, moderate (bent axis, small head and eyes), or severe (kinked and shortened axis, small head and eyes, unidentified forebrain and hindbrain, or missing head and tail – “monster”). The proportion and severity of affected embryos were both decreased substantially by co-injection of 900 pg pak4 mRNA.
2.4. RT-PCR and quantitative PCR
Total RNAs were isolated from embryos at different stages using guanidinium thiocyanate and phenol/chloroform extraction as previously described (Sargent et al., 1986). For dissection, embryos were fixed in 1% trichloroacetic acid on ice for 10 min, followed by cutting through the yolk and body of the embryo at the level of the anterior somites (see Fig. 3J) with iridectomy scissors. Reverse transcription was carried out with 800 ng total RNA by using Superscript III Reverse Transcriptase (Invitrogen) and oligo (dT)20 primer. Quantitative real-time PCR was performed with gene-specific primers using 1 μl single-stranded cDNA synthesized from above and the LightCycler 480 SYBR Green I Master reagent (Roche) in the LightCycler 480 System (Roche) according to the manufacturer’s recommendations. Primer sequences were: fli1b-F (CAG TGG ACT GTA GTG TGA CT) and fli1b-R (TGA TGC GCA TCA TGT CCT CT); flk1-F (CTG GTG GAG AGG CTA GGA GA) and flk1-R (TGA TCG GGA TGT AGT GCT TTC); l-plastin-F (TCA CAT CAG CAC AGA TGA GC) and l-plastin-R (TTC TCC AGA GCC TTG TTG AC); PAK4-F3 (ACG ATA AGA GGC CCA AAT CC) and PAK4-R3 (GAA GTT TGA CCA CCA CCG CT). Primers for scl and mpo detection were described previously (Hsu et al., 2004; Veldman and Lin, 2012). Relative gene expression was normalized to the β-actin level using primer pair β-actin-F (TGA GCG CAA ATA CTC CGT CTG GAT) and β-actin-R (ACT CCT GCT TGC TGA TCC ACA TCT) for pak4 mRNA, and to 18S rRNA using primers (18S-F, CCT GCG GCT TAA TTT GAC TC; and 18S-R, GAC AAA TCG CTC CAC CAA CT) for other genes in the corresponding cDNA samples. Primers used for monitoring the inhibition of splicing by MOs, were F1 (GGG AAT TCA CGA TAA GAG GCC CAA ATC C), R1 (TTC TCG AGG TGC ATC ACC GTT GTG CAT AG) and R2 (TTT CTC GAG GAA GTT TGA CCA CCA CCG CT).
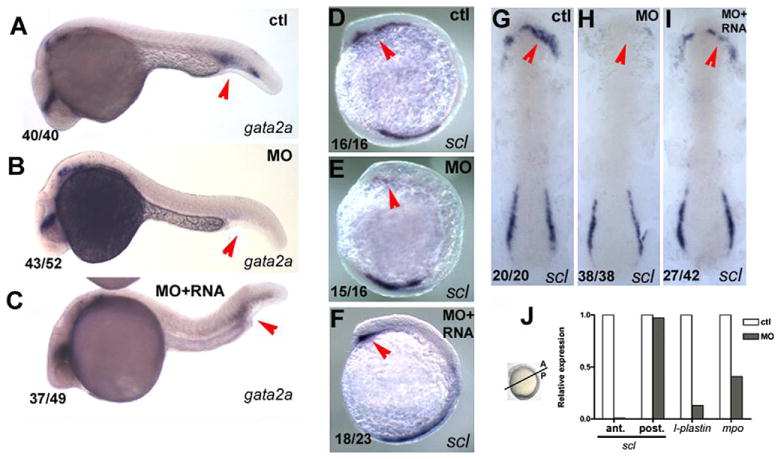
Fig. 3.
pak4 is required for primitive myelopoiesis. (A–C) Inhibition of gata2a expression by MZpak4 knockdown in the posterior blood island (red arrows) in 24-hpf embryos. (D–I) Inhibited expression of the hematopoietic regulatory factor scl in the rostral blood island (RBI; red arrows) at 5-somite stage, lateral view (D–F) and 7-somite stage flatmounts (G–I). (A, D and G) Control morphants, (B, E and H) MZpak4 knockdown embryos. (C, F and I) MZpak4 knockdown embryos rescued by pak4 mRNA injection. (J) Relative expression levels of scl in anterior and posterior regions at 5-somite stage, l-plastin at 15-somite and mpo at 20-somite stages. mRNA levels were measured by QPCR, normalized to 18S rRNA levels and represented as a ratio to the corresponding uninjected control embryos. Samples for anterior and posterior measurement of scl were dissected across the axis as shown while those for l-plastin and mpo were from whole embryos. Results shown are representatives from two individual experiments with similar results.
2.5. Whole-mount in situ hybridization
In situ hybridization of zebrafish embryos from various stages was performed according to published procedures (Thisse and Thisse, 2008). Antisense digoxigenin (DIG)-labeled riboprobes were synthesized using linearized plasmid clones as templates, SP6, T7 or T3 RNA polymerase and DIG RNA labeling Mix (Roche). DIG-labeled riboprobes were purified by using NucAway Spin Columns (Ambion) as described by the manufacturer.
2.6. Immunoprecipitation and western blots
To prepare antibody to zebrafish pak4 protein, two peptides corresponding to amino acid residues 129–143 (TDRYGDSKEREKPRC) and 150–164 (PRQSQRPGRSSREDC) from a highly variable region of pak4 were synthesized, coupled to keyhole limpet hemocyanin, used to raise high-titer antibody in rabbits and affinity-purified by Genscript Inc. For immunoprecipitation and western analysis, chorionated embryos were homogenized in 10 μl per embryo Complysis Buffer (SignaGen) containing protease inhibitors (Roche) followed by centrifugation at 21,000g for 10 min. The supernatant from 50 embryos was combined with 2.5 μg purified anti-pak4 protein and incubated at 4 °C for 1 h. Then 15 μl of washed protein G Dynabeads (Novex) was added to each sample and mixed by rotation for one hour at 4 °C. Beads were removed by centrifugation and immobilized with amagnetic base, and washed five times with TBST (10 mM tris pH 7.4, 150 mM NaCl, 0.05% Tween 20). Protein was eluted by heating at 95° for 5 min in 1× gel loading buffer (Invitrogen) electrophoresed on gradient acrylamide gels (NuPAGE, Invitrogen) transferred to nitrocellulose and probed with 1:5000 dilution of the pak4 antibody. Detection was via HRP conjugated anti-rabbit IgG (ECL; GE Healthcare) and Immobilon chemiluminescent substrate (Millipore).
2.7. Imaging
Live and fixed embryos were mounted in 3% methyl-cellulose and 100% glycerol, respectively, for microscopy documentation. Embryos were examined and photographed by using the Leica MZ APO dissecting microscope with RETI-GA 1300 digital camera (Quantitative Imaging Corporation) and QCapture software. Fluorescence was observed in Leica MZ16F and documented with a Leica DFC500 camera. Flat mounts of fixed embryos were photographed using an Axio-plan2 Inverted microscope (Zeiss) and DFC490 camera (Leica).
2.8. Sequence analyses
For sequence comparison, multiple alignment and dendrogram construction, the following online programs were used: http://searchlauncher.bcm.tmc.edu/, http://blast.ncbi.nlm.nih.gov/Blast.cgi, and http://clustalw.genome.jp/.
2.9. Cytoskeletal inhibitor treatments
Dechorionated embryos were treated with inhibitors targeting microtubules (taxol; 39 nM, Sigma–Aldrich or nacodazole; 5 μg/ml; Sigma–Aldrich) or microfilament depolymerization drugs (cytochalasin B; 4 μg/ml; Sigma–Aldrich or latrunculin B; 0.25 μg/ml; Calbiochem). Drug dosages were determined from published reports (Cheng et al., 2004; Solnica-Krezel and Driever, 1994; Zalik et al., 1999) or by titration, in which case a dose was chosen that gave maximal effects but did not exceed 40% mortality. All reagents were dissolved in DMSO, which was used as a control. Embryos were treated continuously from tailbud through 5–6 somite stage, then fixed for in situ hybridization.
3. Results
3.1. Zebrafish pak4 identification and expression
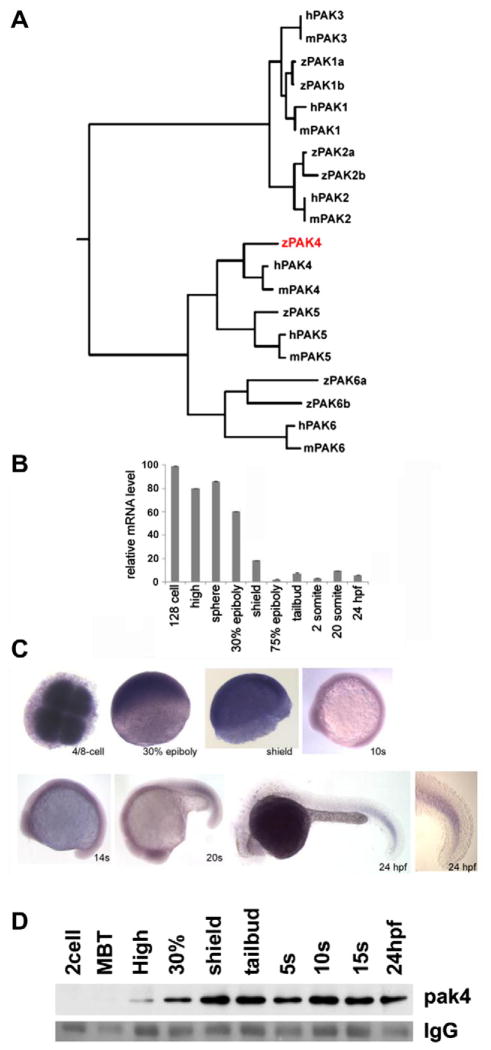
By searching the latest version of the zebrafish genome (Zv9 – http://www.ensembl.org), we were able to identify orthologs of mammalian PAK1, 2, 4, 5 and 6 that distinctly clustered with the index genes (Fig. 1A). Zebrafish pak1, 2 and 6 appeared duplicated, a common finding with genes in teleost species (Meyer and Van de Peer, 2005). We could find no zebrafish gene corresponding to mammalian PAK3. This was also the case for the medaka and stickleback genomes, suggesting this gene may be absent in teleosts. The single copy of PAK4 in zebrafish (pak4; ZFIN, zgc: 92014; Genbank Accession NM_001002222), mapped to chromosome 15 as annotated in the EnsEMBL zebrafish genome database (Gene ID ENSDARG00000018110). The predicted zebrafish pak4 protein has 65% amino acid sequence identity with the human ortholog, and exhibits the characteristic features of group II PAKs: a GTPase-binding domain (CRIB or PBD), a serine/threonine kinase domain, a guanine nucleotide exchange factor (GEF-H1) interaction domain (GID) and proline-rich regions. Zebrafish pak4 and other group II members were found to be more divergent from their human orthologs (52–65% sequence identity) compared to the group I PAKs (80–86% sequence identity).
Fig. 1.
Identification and expression of pak4 in zebrafish embryos. (A) Phylogeny of the pak family. Putative protein sequences of the kinase domains of human (h), mouse (m) and zebrafish (z) PAKs were analyzed using ClustalW (http://clustalw.genome.jp/). Results are shown as a dendrogram with branch length indicating divergence. PAK5 proteins were also named as PAK7 in Genbank and EnsEMBL databases. Genbank Accessions or EnsEMBL Gene IDs (Zv9): humanPAK1, NM_001128620; humanPAK2, NM_002577; humanPAK3, NM_001128166; humanPAK4, NM_005884; humanPAK5, NM_020341; humanPAK6, NM_020168; mousePAK1, NM_011035; mousePAK2, NM_177326; mousePAK3, NM_008778; mousePAK4, NM_027470; mousePAK5, NM_172858; mousePAK6, NM_001033254; zfPAK1a, ENSDARG0000004 2624; zPAK1b, NM_201328; zPAK2a, NM_001002717; zPAK2b, NM_001025456; zPAK4, NM_001002222; zPAK5, NM_212962; zfPAK6a, ENSDARG00000027564; zPAK6b, NM_001162487. Zebrafish PAK4 is indicated in red. (B) Quantitative analysis of zebrafish pak4 expression in development. Quantitative RT-PCR was carried out with total RNA (800 ng) extracted from specific embryonic stages and analyzed for pak4 expression. Maternal pak4 mRNA levels rapidly declined between 30% epiboly and shield stages, then remained approximately constant through 24 hpf. Results shown were collected from three individual experiments. (C) Whole-mount in situ hybridization for pak4. Stages are as indicated. 4/8-cell stage – animal pole view, others are lateral view. Signals at and after 30% epiboly were relatively weak and hence staining of these samples for alkaline phosphatase activity was prolonged. A higher magnification of the tail of the 24 hpf embryo is shown to highlight elevated expression in newly formed somites. (D) Immunoprecipitation/western analysis. Total soluble protein was isolated from embryos at multiple stages, immunoprecipitated and prepared for western blotting as described in Section 2. Equal numbers of embryos were used for each sample, which was confirmed by BCA protein assays. Ponceau S staining of the IgG heavy chain band (IgG) is shown below as a control for immunoprecipitation recovery.
Mammalian PAK4 expression has been reported to be ubiquitous, with highest abundance in prostate, testis and colon, and has been detected in mouse embryos as early as embryonic day 7 (Abo et al., 1998; Qu et al., 2003). We used quantitative RT-PCR (Fig. 1B) and traditional RT-PCR assays (not shown) to measure pak4 transcripts at various stages of development. Both methods revealed high levels of pak4 mRNA at cleavage through early epiboly stages, followed by a rapid decline during gastrulation to a much lower and relatively constant abundance throughout subsequent development. Whole-mount in situ hybridization (Fig. 1C) revealed that the maternal pak4 mRNA was uniformly distributed at early stages. This continued through 10–14-somite stages. At late somite stage (Fig. 1C, 20s) elevated pak4 mRNA was observed in the developing tail somites, with a gradient fading towards the anterior. This pattern persisted through 24 hpf. Thus, in the zebrafish embryo pak4 expression is predominantly maternal, and, as in mammalian species, zygotic expression is ubiquitous, although not uniform. To detect zebrafish pak4 protein, we generated a specific anti-peptide antibody (Section 2). To avoid interference from abundant co-migrating yolk proteins, pak4 protein was first immunoprecipitated from detergent lysates, then detected by western blot. pak4 Protein was first detectable at high stage, reaching a maximum at shield stage that was maintained through 24 hpf (Fig. 1D). Thus maternal pak4 mRNA appears not to be translated until after cleavage stages.
3.2. pak4 Loss of function analysis
Synthesis of pak4 protein was inhibited in embryos by injection of antisense morpholino oligonucleotides (MOs). Splicing of zygotic transcripts was inhibited by using MOs (MO-1 and MO-2) (Fig. 2A), Combined maternal and zygotic pak4 expression was suppressed using a translation-blocking MO (MO-3) targeting a site upstream from the initiating methionine codon (Fig. 2B). Two splice MOs were necessary because MO-1, which targeted the splice donor site of exon 4, uncovered a cryptic splice donor site located 33 nucleotides upstream, which would have resulted in an in-frame deletion of 11 amino acids. This corresponded to a highly variable region and could have resulted in a functional protein. A similar phenomenon of uncovering a cryptic site by MO was also previously observed in fgf8 knockdown (Draper et al., 2001). A second splice MO (MO-2) was thus designed to target the cryptic site. Using both splice MOs in combination resulted in formation of an abortive mRNA processing product with stop codons in all three frames. There is no evidence that this abortive RNA is exported to the cytoplasm or translated, but even if it were, the resulting protein would be truncated at amino acid 282, which would eliminate the kinase domain and would thus be likely to represent a loss of pak4 function. The splice MOs were highly effective in preventing pak4 mRNA processing, which enabled visualization of maternal pak4 RNA decay via RT-PCR. As shown in Fig. 2C, maternal transcripts were reduced but could be detected at the tailbud stage (300-bp band; TB, S lane), and had disappeared by the 5-somite stage, leaving only the abortive splice product resulting from zygotic expression (600 bp). This pattern persisted until 5 days post fertilization (dpf), indicating that the block to zygotic pak4 expression extended through all embryonic stages. By 7 dpf, spliced zygotic transcripts became detectable and reached a significant fraction of control levels by day 10 (data not shown). Immunoprecipitation/western analysis confirmed the loss of pak4 protein in MO-injected embryos at 24 hpf, although the splice MOs alone were ineffective at shield stage, presumably due to the predominance of maternal mRNA at that time (Fig. 2D).
3.3. Maternal, but not zygotic, pak4 expression is required for normal development
Embryos injected with the combined splice MOs developed normally through larval stage, compared to embryos injected with an identical dose of control MO (5 dpf; 100%, n = 84; Fig. 2E, columns 1 and 2). We observed no morphological effects, nor was there any evidence of abnormal behavior or locomotion (data not shown). These results conflicted with findings by (Fiedler et al., 2011), who reported vascular defects in zebrafish embryos injected with a splice-blocking morpholino targeting pak4 exon 5. The reason for this discrepancy is unclear, although we did observe similar vascular defects using combined translation/splice-blocking MOs (Fig. S1). In contrast to the splice-blocking MO experiments, when the translation-blocking MO was injected, development was severely perturbed. This result was obtained with the translation-blocking MO injected alone, but the severity and frequency were slightly higher when injected along with the pair of splice MOs, presumably due to incomplete inhibition by individual MOs (Fig. 2D). In the experiments shown in this paper, the splice + translation-blocking MO combination was used, referred to as MZpak4 knockdown. As can be seen in Fig. 2E (column 3), morphant embryos appeared normal through shield stage, but showed obvious defects by 24 hpf, including reduced head and tail size and kinked body axis. Further disruptions in morphogenesis continued at later stages, including axis defects, small heads and eyes, loss of lateral line neuromasts, edema, misshapen somites, and craniofacial cartilage abnormalities. Morphant embryos died by 7 dpf. To test the specificity of these effects, mRNA rescue experiments were performed. The splice + translation-blocking MO mixture was injected alone or with variable amounts of synthetic pak4 mRNA which lacked the translation-blocking MO target sequence and was thus not susceptible to inhibition by the MO. As shown in Fig. 2F, the morphant phenotype was substantially rescued by co-injection of pak4 mRNA, and was therefore due specifically to the loss of MZpak4 expression. Nevertheless, developmental defects in MO-injected zebrafish embryos can in some cases be due in part to off-target or toxic effects that lead to apoptosis (Gerety and Wilkinson, 2011; Robu et al., 2007). One way to identify such potential MO artifacts is to co-inject a MO designed to inhibit translation of the p53 protein, which is required for one of the major pathways of apoptosis. We performed such p53 MO experiments and found that several aspects of the morphant phenotype were rescued partially or completely. In some cases, for example the loss of neuromasts, the p53 MO rescue was more efficient than the pak4 mRNA rescue. Other phenotypes including somite dysmorphogenesis and disruption of myeloid gene expression, were not altered by the p53 MO. Craniofacial cartilage defects were not rescued by p53 knockdown, but could not be rescued by pak4 mRNA to a convincing level (data not shown). Mammalian PAK4 has been shown to inhibit caspase activity and thus has an anti-apoptotic function (Gnesutta et al., 2001), raising the possibility that some aspects of the MO phenotype might be due to enhancement of embryonic cell death. In this report we have focused on defects in myelopoiesis, which were rescued by pak4 mRNA injection and also p53-independent. MZpak4 function in specification of somites and other tissues will be addressed in separate studies.
3.4. Requirement for maternal pak4 expression in hematopoiesis
In order to identify more specific molecular targets corresponding to the MZpak4 morphant phenotype, we carried out in situ hybridization with a collection of probes for tissue-specific gene expression at various stages from gastrulation to 24 hpf. Most of these markers appeared to be unaffected (data not shown), but we observed a striking reduction of the hematopoietic regulatory factor gata2a in the posterior blood island (PBI, Fig. 3A and B) at 24 hpf, leading us to investigate the possibility that aspects of hematopoiesis in zebrafish might depend on maternal pak4 expression. Blood and vascular development takes place in two phases: primitive hematopoiesis which provides the embryo with blood cells and vessels, and definitive hematopoiesis that serves this purpose later in development and in the adult animal (Bertrand and Traver, 2009; de Jong and Zon, 2005). In zebrafish primitive hematopoiesis begins at the early somite stage, and is restricted to two spatial domains: anterior and posterior hematopoietic lineages arising from the lateral plate mesoderm. The anterior domain gives rise to the rostral blood island (RBI) and is the primary site for formation of myeloid cells. The posterior domain converges at the midline to form the intermediate cell mass (ICM) as bilateral stripes in the trunk. At this site hemangioblasts arise, some of which differentiate into endothelial cells that form the vasculature, and others that give rise to erythrocytes and the other circulating blood cells, including myeloid derivatives granulocytes and macrophages (Bertrand and Traver, 2009; de Jong and Zon, 2005; Metcalf, 2007).
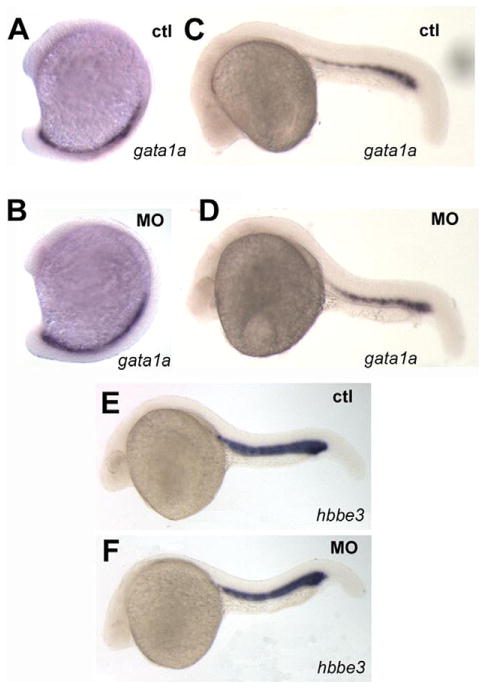
The basic HLH gene scl (stem cell leukemia; also named Tal-1; (Liao et al., 1998) is expressed in both hematopoietic domains, corresponding to the myeloid lineage in the anterior and erythroid and endothelial lineages in the posterior. Loss of MZpak4 expression also strongly inhibited scl expression in the anterior domain but had no effect on expression in the posterior at 5–7-somite stages (Fig. 3D–I). Expression of both scl in the anterior and gata2a in the posterior domain was restored by co-injection of pak4 mRNA (Fig. 3C, F, I; S2), suggesting a requirement for maternal pak4 in myelopoiesis. This was tested by measuring expression by QPCR of mpo (myeloperoxidase), and l-plastin (lymphocyte cytosolic plastin) as markers for granulocytes and leukocytes, respectively (Bennett et al., 2001; Lieschke et al., 2001; Meijer et al., 2008). As shown in Fig. 3J, both markers were strongly reduced in the MZpak4 morphants. This appeared to be due to a reduction in the number of expressing cells, as shown by in situ hybridization (Fig. S3), indicating a failure to form these major myeloid derivatives. To test for effects of MZpak4 depletion on other hematopoietic lineages, we monitored expression of gata1a and hbbe3 (hemoglobin β embryonic 3), two markers for erythropoiesis, and found no difference between control and MZpak4 morphants (Fig. 4). We conclude from these results that maternal pak4 expression is required specifically for primitive myelopoiesis but not erythropoiesis in the zebrafish.
Fig. 4.
Primitive erythropoiesis is not affected by pak4 knockdown. Two specific markers for erythroid precursors, gata1a and hbbe3 in the posterior blood lineage and intermediate cell mass were unaffected by pak4 MO injection at 15-somite (A and B) or 24 hpf (C–F).
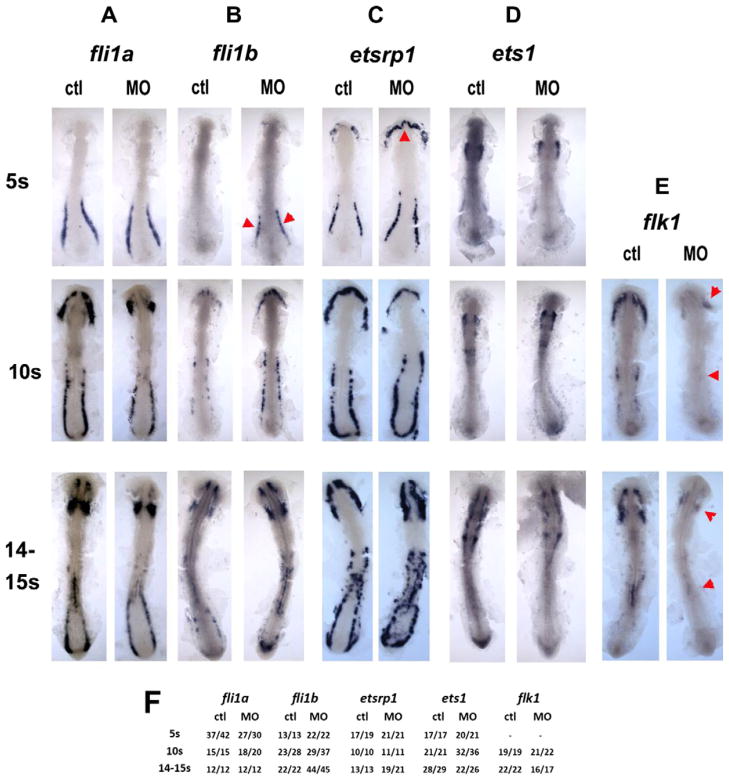
The developmental control of hematopoiesis has been studied extensively, and several components of a regulatory gene network have been identified (Berman et al., 2005; de Jong and Zon, 2005). The ets1, etsrp1 and fli1a/b genes encode ETS-class transcription factors that function at or near the top of this hierarchy, upstream from scl (Liu et al., 2008a; Pham et al., 2007; Ren et al., 2010). Expression patterns of fli1a, etsrp1 and ets1 were not reduced by MZpak4 knockdown at 5–15-somite stages (Figs. 5A, C, D). However, expression of fli1b in the posterior domain, and etsrp1 in the rostral domain were transiently enhanced at the 5-somite stage (Fig. 5B and C). We also examined expression of flk1, which encodes the receptor for VEGF (Habeck et al., 2002). This was markedly inhibited, in both the rostral and posterior domains (Fig. 5E). This could be a result of the inhibition of scl in the RBI (Patterson et al., 2005), but not in the ICM, since as discussed above, MZpak4 knockdown does not inhibit scl in the latter region. Thus we observe both increased and decreased expression of hematopoietic regulatory factors, in both domains, resulting from the loss of MZpak4.
Fig. 5.
Regulatory level of pak4 function. Flat mounts of in situ hybridization for ETS class transcription factors fli1a (column A) and fli1b (column B), etsrp1 (column C) and ets1 (column D), and the VEGF receptor flk1 (column E) reveal different dependence on pak4. Expression of the ETS transcription factors was not inhibited at any stage tested, however, expression of fli1b was transiently elevated at in the posterior domain, and etsrp1 in the rostral domain at the 5-somite stage (B and C; red arrowheads). flk1 Expression was reduced in both the anterior and posterior hematopoietic domains (E; red arrowheads). Frequency data are tabulated in (F).
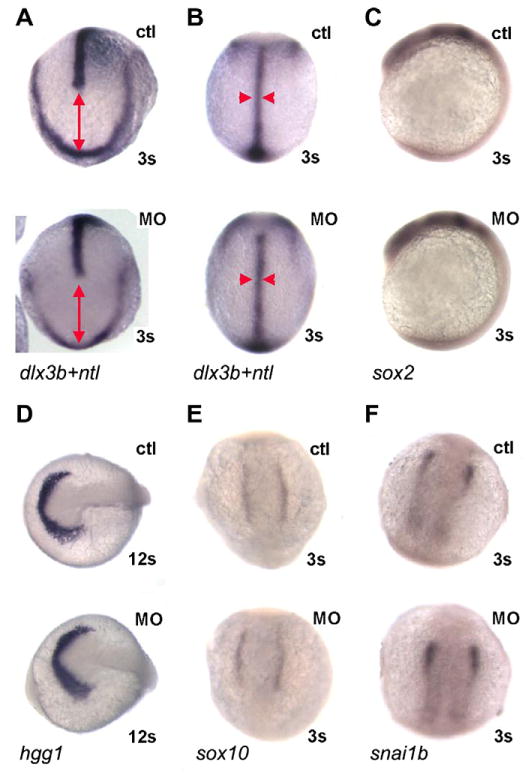
What could be the basis for these changes? Although we did not observe wholesale alteration in the pattern of gene expression at later stages, it seemed possible that loss of maternal pak4 might lead to disruption of gastrulation, as has been reported in Xenopus (Faure et al., 2005), or in early cell–cell signal transductions mechanisms such as MAPK signaling (Cammarano et al., 2005), which could lead indirectly to disruption of the hematopoietic regulatory hierarchy. To test these possibilities, we used in situ hybridization with appropriate probes to monitor convergent extension movements and the induction and patterning of neural plate, neural crest, and anterior development, of which depend on multiple signaling pathways during and shortly after gastrulation in all vertebrate embryos. As shown in Fig. 6, early patterning in MZpak4 knockdown embryos was normal. Extension and convergence of axial mesoderm, judged by the distance from the rostral end of the notochord and the anterior neural plate and the width of the notochord (detected by expression of ntl and dlx3b; Fig. 6A and B) was unaffected, as was the level and pattern of expression of sox2 (neural plate; Fig. 6C), hgg1 (hatching gland, an anterior marker; Fig. 6D), and sox10 and snai1b (neural crest; Fig 6E and F). We also examined expression of mpo in embryos homozygous for the ppt mutation (pipetail, wnt5a), which have convergent extension defects due to disruption of the non-canonical Wnt pathway (Westfall et al., 2003), and scl expression in casta56 (casanova; sox32) mutant embryos which fail to differentiate endoderm (Fig. S4). No difference was observed compared to control embryos in any of these comparisons. We conclude that in zebrafish MZpak4 is not required for the signaling pathways leading to embryonic axis formation and patterning, and is therefore likely to be a specific factor in primitive myeloid cell differentiation.
Fig. 6.
MZpak4 knockdown did not disturb early signaling pathways. (A and B) Convergent extension: double in situ hybridization with dlx3b and ntl probes was performed to outline the neural plate border and notochord at 3-somite stage. Dorsal view of anterior (A) and trunk (B) regions. The distance between dlx3b and the leading edge of ntl expression indicates extension (red line in A) while the width of ntl (red arrowheads in B) indicates convergence. The patterns in MZpak4 knockdown (bottom) are comparable to control embryos (top), indicating normal convergent extension in gastrulation. (C–F) Early markers for neural plate (sox2; C), hatching gland (hgg1; D) and neural crest (sox10; E and snai1b; F) were unaffected in pattern or intensity by MZpak4 knockdown (bottom embryos) compared to controls (top). Embryos were 12 somite stage in panel D, 3 somite stage in the other panels.
3.5. Cytoskeletal-transcriptional Linkage
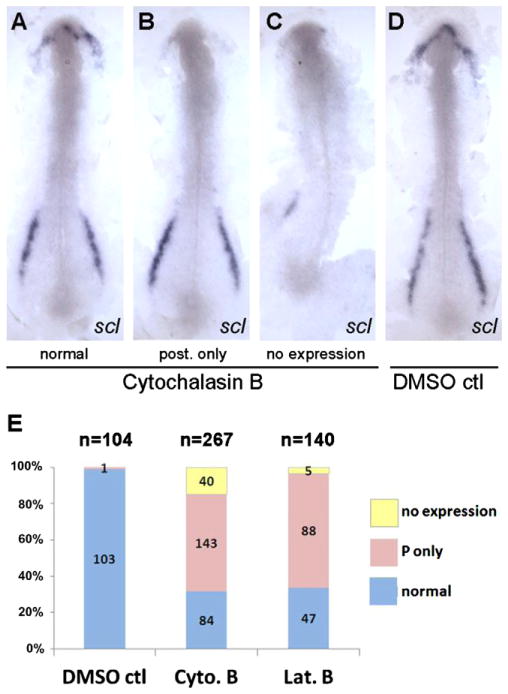
The major, although not exclusive, function of pak4 is a regulator of cytoskeletal dynamics, particularly of microfilaments, suggesting a possible link between cytoskeleton and transcriptional control of myelopoietic regulatory factors. To test this hypothesis we treated embryos at the tailbud stage with cytoskeletal disruption agents, then monitored scl expression by in situ hybridization at 5–6 somite stage. This time window was selected because treatment at earlier stages led to gastrulation failure (Solnica-Krezel and Driever, 1994; Zalik et al., 1999), and scl expression is first reliably detectable at 5 somite stage. Treatment with anti-microtubule drugs nacodazole and taxol caused some morphological defects but did not alter the scl expression pattern (data not shown). In contrast, the actin depolymerizing agents cytochalasin B and latrunculin B resulted in a selective inhibition of scl expression in the rostral hematopoietic domain, similar to what we observed in the MZpak4 knockdown embryos (Fig. 7). Therefore, we conclude that microfilament integrity between tailbud and 5-somite stage is necessary for the expression of specific regulatory genes required for primitive myeloid cell differentiation, and that this depends on maternal – but not zygotic – expression of pak4.
Fig. 7.
Disruption of actin microfilaments leads to loss of scl expression in the RBI. Dechorinated embryos at tailbud stage were exposed to cytochalasin B (4 μg/ml) or latrunculin B (0.25 μg/ml) and fixed at 5–6 somite stage for whole-mount in situ hybridization using the scl gene probe. 30–40% Mortality was observed in all drug exposures due to rupture of the enveloping layer at variable times. Dead embryos were not analyzed further. Flat mounts of representative embryos from cytochalasin B exposure (A– C; normal, posterior expression only, no expression) and DMSO control (D) were prepared to show the anterior and posterior expression of scl. (E) Data were summarized from four independent experiments which show that the majority of embryos specifically lost the anterior scl expression after drug treatments. Posterior-specific loss of expression was never observed.
4. Discussion
Since its initial identification in 1998 by Minden and colleagues (Abo et al., 1998) as an effector of Cdc42 signaling, PAK4 has been linked to a multitude of cellular and developmental functions, most involving, either directly or indirectly, the regulation of cytoarchitecture and cell movement, but also apoptosis and signal transduction (Barac et al., 2004; Gnesutta et al., 2001; Li and Minden, 2005; Liu et al., 2010, 2008b). It is thus not surprising to find that loss of pak4 expression during early development leads to disruption of multiple cells and tissues in the zebrafish embryo. Targeted disruption of PAK4 in mouse also resulted in a complex lethal phenotype (Qu et al., 2003). Interestingly, this included deficient vascularization of extraembryonic tissue, probably resulting from primary defects in the pak4−/− embryo (Tian et al., 2009). Fiedler et al. (2011) reported vascular defects in pak4 morphants, and we observed a similar phenotype in MZpak4 knockdown embryos (Fig. S1). However, we saw no such defects, or indeed any defects whatsoever, in splice-inhibited morphants. We cannot provide a definitive reason for this discrepancy, although it is worth noting that our splice-blocking MO virtually eliminated both pak4 mRNA and protein by 24 hpf. Additional experiments will be necessary to resolve this issue, preferably based on some methodology other than MO treatments, for example, gene targeting. Furthermore, the vascular defects in MZpak4 morphants could be attributable to the alterations in hematopoietic regulators we observed, but we cannot exclude the possibility that these vessel defects were secondary to morphological disruption of the somites, through which these vessels must navigate (Clements et al., 2011; Lawson et al., 2002). In this context it would be interesting to examine myelopoiesis in the PAK4 knockout mouse, and expression of the mouse homologs of the hematopoietic regulatory genes we found to be disturbed in the fish (i.e. scl, fli1b and flk1). Loss of function experiments with pak4 have also been carried out in Xenopus (originally named pak5; we use the designation pak4 for this gene, see Fig. 1A). In this earlier work, overexpression of a kinase-dead mutation of pak4 was employed as a dominant-negative inhibitor of endogenous pak4 (Faure et al., 2005). Gastrulation movements were abnormal, and cell adhesion was also affected, but there are few obvious parallels with zebrafish or mouse loss of function phenotypes. To some extent this could be due to the dominant negative strategy: since some pak4 functions are kinase-independent, over-expression of a kinase dead mutant might result in a combination of loss and gain of functions, complicating interpretation. Morpholino knockdown of Xenopus pak4 has not been reported, so this remains an open question.
In spite of the abundance of maternal pak4 mRNA in the zebrafish egg, we did not anticipate that zygotic pak4 expression would be dispensable. Maternal effect genes are defined by mutations (or other loss of function) in which the phenotype depends on the egg but not on the zygotic genotype. This phenomenon is most commonly found in species with mosaic development, such as many insects and invertebrates, in which there is differential inheritance of egg cytoplasmic determinants during cleavage that leads to specific developmental fates. Maternal effects are also known in vertebrates, however, where they tend to affect fertility and early cleavage [for a recent review see (Lindeman and Pelegri, 2010)]. Vertebrate maternal effect phenotypes involving later developmental processes are less common but have been identified. The pak4 splice-blocking MOs prevented processing of zygotic pak4 transcripts, leading to a single oversized band detectable by RT-PCR, which has stop codons in all three reading frames within a few hundred nucleotides of unprocessed intron sequence (Fig. 2A). Immunoprecipitation/western blotting also indicated essentially complete loss of pak4 protein by 24 hpf using the splice MOs and at all stages using the splice/translation-blocking cocktail (Fig. 2D). It is not known if the abortively spliced mRNA was translated, but even if it were, this would yield a truncated pak4 protein lacking the entire kinase domain. While it is conceivable that such a shortened protein could retain some biological activities, it is more likely that the splice MO treatment resulted in an effective loss of zygotic pak4 function. Since this treatment did not result in any detectable deficiencies or alterations in development through 10 dpf (Fig. 2E and data not shown), we conclude that zygotic pak4 expression is dispensable through larval stages. Testing for pak4 function at later stages and in adult fish will require other strategies, such as TILLING (Moens et al., 2008; Winkler et al., 2011) or gene targeting (Foley et al., 2009; Meng et al., 2008) to disrupt the pak4 gene. Taken with the predominantly maternal expression of pak4 in development, we conclude that pak4 is a maternal effect gene in zebrafish.
The loss of anterior scl expression would be expected to lead to reductions in macrophages and other myeloid lineages (Sumanas et al., 2008). Likewise, loss of granulocytes can be partially attributed to reduced scl in the rostral domain, but the granulocytic lineage also originates from the posterior domain where scl was not affected. In this region, inhibition of gata2a expression in MZpak4 morphants (Fig. 3A–C) could be responsible. The lack of an effect on posterior scl expression is consistent with the apparently normal primitive erythropoiesis of MZpak4 morphants, since this process is limited to the ICM, which derives from the posterior domain (Davidson and Zon, 2004). The lower expression of the endothelial marker flk1 also points to defects in both the rostral and posterior domains (Fig. 5), and as with the myeloid markers, loss of anterior flk1 could be attributed to loss of scl, but something else must be responsible for the posterior reduction. Thus maternal pak4 expression is likely to be required at multiple points in the primitive myeloid cell regulatory program.
The myeloid defects in MZpak4 were presumably due to pak4 functions occurring at or before the 5-somite stage when the maternal pak4 mRNA decay was complete. The major hematopoietic regulatory genes have been activated by this stage, consistent with a role for pak4. How could the loss of maternal pak4 yield the observed effects on blood development? Numerous functions have been assigned to PAK4 in mammals, involving control of cytoarchitecture, cell shape and motility, signal transduction and apoptosis (Abo et al., 1998; Barac et al., 2004; Callow et al., 2002, 2005; Cammarano et al., 2005; Dan et al., 2001; Liu et al., 2010; Paliouras et al., 2009; Wallace et al., 2010; Wells et al., 2010). However, aberrant cell migration is unlikely to be the basis for the observed myeloid deficiencies, since the spatial expression pattern of several hematopoietic regulatory genes was not affected in either rostral or posterior domain. Likewise, a direct effect on major signal transduction pathways in the early embryo would be difficult to reconcile with the normal pattern of gastrulation and associated gene expression observed in MZpak4 morphants at stages prior to when the myeloid deficiencies arise (Fig. 6).
In recent years it has become clear that the cytoskeleton and cell–cell adhesion molecules play regulatory as well as structural roles. For example, actin subunits can act as cofactors for transcriptional regulation via MRTF (myocardin-related transcription factor) and SRF (serum response factor), linking the transcription of a specific set of target genes to the polymerization state of the microfilament network, which is to a major extent regulated by Rho GTPase signaling mediated by PAK factors, including PAK4 (Olson and Nordheim, 2010). The observation that actin depolymerization drugs could partially phenocopy the effect of MZpak4 knockdown (i.e. scl expression in the RBI) suggests that a mechanism based on microfilaments could be involved in regulating myeloid gene expression. There are several candidates for how such a linkage could be accomplished. For example, the group II PAKs have been found to bind directly and phosphorylate p120-catenin (Wong et al., 2010), which functions as an adherens junction component in the cadherin complex and modulates Rho GTPase signaling and thus cytoskeletal organization (Stepniak et al., 2009). In Drosophila, a PAK4 ortholog, Mbt, phosphorylates the armadillo/β-catenin protein, affecting adherens junction stability in the eye (Menzel et al., 2007, 2008; Schneeberger and Raabe, 2003). p120-catenin also has a role in modulation of MAPK and NFκB signaling, as shown in p120-catenin deficient mice in which the ectopic activation of these two pathways was observed in epidermis (Perez-Moreno et al., 2006, 2008). Thus there are multiple mechanisms by which PAK4 might influence gene expression in myelopoiesis by virtue of its function as a cytoskeletal regulator.
It was surprising to find that the essential functions of pak4 in zebrafish appear to be exclusively maternal, i.e. zygotic expression of this gene seems dispensable. Gene redundancy conceivably could explain the lack of phenotype in splice morphants, but such potentially compensating factors must be absent at early stages, leading to the MZpak4 morphant phenotype. It is also remarkable that loss of pak4 expression had such specific effects on gene activity in myeloid development, as opposed, for example to the migratory or morphogenetic effects that have been previously associated with this gene (Faure et al., 2005; Fiedler et al., 2011; Qu et al., 2003; Tian et al., 2009). Understanding how actin dynamics, mediated by maternal pak4, participates in the control of myeloid gene expression will be an important problem for future investigation.
Supplementary Material
Acknowledgments
We thank the Burgess, Chitnis, Dawid and Weinstein laboratories in the Program on Genomics of Differentiation, NICHD, for discussion and materials, and Drs. T. Luo, H. Ro, Y. Ogawa and V. Virta for helpful discussions. This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.mod.2012.09.005.
References
- Abo A, Qu J, Cammarano MS, Dan C, Fritsch A, Baud V, Belisle B, Minden A. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 1998;17:6527–6540. doi: 10.1093/emboj/17.22.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barac A, Basile J, Vazquez-Prado J, Gao Y, Zheng Y, Gutkind JS. Direct interaction of p21-activated kinase 4 with PDZ-RhoGEF, a G protein-linked Rho guanine exchange factor. J Biol Chem. 2004;279:6182–6189. doi: 10.1074/jbc.M309579200. [DOI] [PubMed] [Google Scholar]
- Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, Langenau DM, Delahaye-Brown A, Zon LI, Fleming MD, Look AT. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98:643–651. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- Berman JN, Kanki JP, Look AT. Zebrafish as a model for myelopoiesis during embryogenesis. Exp Hematol. 2005;33:997– 1006. doi: 10.1016/j.exphem.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Traver D. Hematopoietic cell development in the zebrafish embryo. Curr Opin Hematol. 2009;16:243–248. doi: 10.1097/MOH.0b013e32832c05e4. [DOI] [PubMed] [Google Scholar]
- Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- Buchner DA, Su F, Yamaoka JS, Kamei M, Shavit JA, Barthel LK, McGee B, Amigo JD, Kim S, Hanosh AW, Jagadeeswaran P, Goldman D, Lawson ND, Raymond PA, Weinstein BM, Ginsburg D, Lyons SE. Pak2a mutations cause cerebral hemorrhage in redhead zebrafish. Proc Natl Acad Sci USA. 2007;104:13996–14001. doi: 10.1073/pnas.0700947104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow MG, Clairvoyant F, Zhu S, Schryver B, Whyte DB, Bischoff JR, Jallal B, Smeal T. Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J Biol Chem. 2002;277:550–558. doi: 10.1074/jbc.M105732200. [DOI] [PubMed] [Google Scholar]
- Callow MG, Zozulya S, Gishizky ML, Jallal B, Smeal T. PAK4 mediates morphological changes through the regulation of GEF-H1. J Cell Sci. 2005;118:1861–1872. doi: 10.1242/jcs.02313. [DOI] [PubMed] [Google Scholar]
- Cammarano MS, Nekrasova T, Noel B, Minden A. Pak4 induces premature senescence via a pathway requiring p16INK4/p19ARF and mitogen-activated protein kinase signaling. Mol Cell Biol. 2005;25:9532–9542. doi: 10.1128/MCB.25.21.9532-9542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JC, Miller AL, Webb SE. Organization and function of microfilaments during late epiboly in zebrafish embryos. Dev Dyn. 2004;231:313–323. doi: 10.1002/dvdy.20144. [DOI] [PubMed] [Google Scholar]
- Ching YP, Leong VY, Wong CM, Kung HF. Identification of an autoinhibitory domain of p21-activated protein kinase 5. J Biol Chem. 2003;278:33621–33624. doi: 10.1074/jbc.C300234200. [DOI] [PubMed] [Google Scholar]
- Clements WK, Kim AD, Ong KG, Moore JC, Lawson ND, Traver D. A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature. 2011;474:220–224. doi: 10.1038/nature10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotteret S, Jaffer ZM, Beeser A, Chernoff J. p21-activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol Cell Biol. 2003;23:5526–5539. doi: 10.1128/MCB.23.16.5526-5539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan C, Kelly A, Bernard O, Minden A. Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J Biol Chem. 2001;276:32115–32121. doi: 10.1074/jbc.M100871200. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Zon LI. The ‘definitive’ (and ’primitive’) guide to zebrafish hematopoiesis. Oncogene. 2004;23:7233–7246. doi: 10.1038/sj.onc.1207943. [DOI] [PubMed] [Google Scholar]
- de Jong JL, Zon LI. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet. 2005;39:481–501. doi: 10.1146/annurev.genet.39.073003.095931. [DOI] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- Eswaran J, Soundararajan M, Kumar R, Knapp S. UnPAKing the class differences among p21-activated kinases. Trends Biochem Sci. 2008;33:394–403. doi: 10.1016/j.tibs.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Faure S, Cau J, de Santa Barbara P, Bigou S, Ge Q, Delsert C, Morin N. Xenopus p21-activated kinase 5 regulates blastomeres’ adhesive properties during convergent extension movements. Dev Biol. 2005;277:472–492. doi: 10.1016/j.ydbio.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Fiedler J, Jazbutyte V, Kirchmaier BC, Gupta SK, Lorenzen J, Hartmann D, Galuppo P, Kneitz S, Pena JT, Sohn-Lee C, Loyer X, Soutschek J, Brand T, Tuschl T, Heineke J, Martin U, Schulte-Merker S, Ertl G, Engelhardt S, Bauersachs J, Thum T. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 2011;124:720–730. doi: 10.1161/CIRCULATIONAHA.111.039008. [DOI] [PubMed] [Google Scholar]
- Foley JE, Maeder ML, Pearlberg J, Joung JK, Peterson RT, Yeh JR. Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat Protoc. 2009;4:1855–1867. doi: 10.1038/nprot.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerety SS, Wilkinson DG. Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Dev Biol. 2011;350:279–289. doi: 10.1016/j.ydbio.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnesutta N, Qu J, Minden A. The serine/threonine kinase PAK4 prevents caspase activation and protects cells from apoptosis. J Biol Chem. 2001;276:14414–14419. doi: 10.1074/jbc.M011046200. [DOI] [PubMed] [Google Scholar]
- Habeck H, Odenthal J, Walderich B, Maischein H, Schulte-Merker S. Analysis of a zebrafish VEGF receptor mutant reveals specific disruption of angiogenesis. Curr Biol. 2002;12:1405–1412. doi: 10.1016/s0960-9822(02)01044-8. [DOI] [PubMed] [Google Scholar]
- Hsu K, Traver D, Kutok JL, Hagen A, Liu TX, Paw BH, Rhodes J, Berman JN, Zon LI, Kanki JP, Look AT. The pu.1 promoter drives myeloid gene expression in zebrafish. Blood. 2004;104:1291–1297. doi: 10.1182/blood-2003-09-3105. [DOI] [PubMed] [Google Scholar]
- Jaffer ZM, Chernoff J. p21-activated kinases: three more join the Pak. Int J Biochem Cell Biol. 2002;34:713–717. doi: 10.1016/s1357-2725(01)00158-3. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kiosses WB, Daniels RH, Otey C, Bokoch GM, Schwartz MA. A role for p21-activated kinase in endothelial cell migration. J Cell Biol. 1999;147:831–844. doi: 10.1083/jcb.147.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Vogel AM, Weinstein BM. Sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev Cell. 2002;3:127–136. doi: 10.1016/s1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- Li X, Minden A. PAK4 functions in tumor necrosis factor (TNF) alpha-induced survival pathways by facilitating TRADD binding to the TNF receptor. J Biol Chem. 2005;280:41192–41200. doi: 10.1074/jbc.M506884200. [DOI] [PubMed] [Google Scholar]
- Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 1998;12:621–626. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001;98:3087–3096. doi: 10.1182/blood.v98.10.3087. [DOI] [PubMed] [Google Scholar]
- Lindeman RE, Pelegri F. Vertebrate maternal-effect genes: insights into fertilization, early cleavage divisions, and germ cell determinant localization from studies in the zebrafish. Mol Reprod Dev. 2010;77:299–313. doi: 10.1002/mrd.21128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Fraser SD, Faloon PW, Rollins EL, Vom Berg J, Starovic-Subota O, Laliberte AL, Chen JN, Serluca FC, Childs SJ. A betaPix Pak2a signaling pathway regulates cerebral vascular stability in zebrafish. Proc Natl Acad Sci USA. 2007;104:13990–13995. doi: 10.1073/pnas.0700825104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Walmsley M, Rodaway A, Patient R. Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr Biol. 2008a;18:1234–1240. doi: 10.1016/j.cub.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Liu Y, Xiao H, Tian Y, Nekrasova T, Hao X, Lee HJ, Suh N, Yang CS, Minden A. The pak4 protein kinase plays a key role in cell survival and tumorigenesis in athymic mice. Mol Cancer Res. 2008b;6:1215–1224. doi: 10.1158/1541-7786.MCR-08-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen N, Cui X, Zheng X, Deng L, Price S, Karantza V, Minden A. The protein kinase Pak4 disrupts mammary acinar architecture and promotes mammary tumorigenesis. Oncogene. 2010;29:5883–5894. doi: 10.1038/onc.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AH, van der Sar AM, Cunha C, Lamers GE, Laplante MA, Kikuta H, Bitter W, Becker TS, Spaink HP. Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev Comp Immunol. 2008;32:36–49. doi: 10.1016/j.dci.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel N, Schneeberger D, Raabe T. The Drosophila p21 activated kinase Mbt regulates the actin cytoskeleton and adherens junctions to control photoreceptor cell morphogenesis. Mech Dev. 2007;124:78–90. doi: 10.1016/j.mod.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Menzel N, Melzer J, Waschke J, Lenz C, Wecklein H, Lochnit G, Drenckhahn D, Raabe T. The Drosophila p21-activated kinase Mbt modulates DE-cadherin-mediated cell adhesion by phosphorylation of Armadillo. Biochem J. 2008;416:231–241. doi: 10.1042/BJ20080465. [DOI] [PubMed] [Google Scholar]
- Metcalf D. On hematopoietic stem cell fate. Immunity. 2007;26:669–673. doi: 10.1016/j.immuni.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) BioEssays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Moens CB, Donn TM, Wolf-Saxon ER, Ma TP. Reverse genetics in zebrafish by TILLING. Brief Funct Genomic Proteomic. 2008;7:454–459. doi: 10.1093/bfgp/eln046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliouras GN, Naujokas MA, Park M. Pak4, a novel Gab1 binding partner, modulates cell migration and invasion by the Met receptor. Mol Cell Biol. 2009;29:3018–3032. doi: 10.1128/MCB.01286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson LJ, Gering M, Patient R. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood. 2005;105:3502–3511. doi: 10.1182/blood-2004-09-3547. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M, Davis MA, Wong E, Pasolli HA, Reynolds AB, Fuchs E. P120-catenin mediates inflammatory responses in the skin. Cell. 2006;124:631–644. doi: 10.1016/j.cell.2005.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno M, Song W, Pasolli HA, Williams SE, Fuchs E. Loss of p120 catenin and links to mitotic alterations, inflammation, and skin cancer. Proc Natl Acad Sci USA. 2008;105:15399–15404. doi: 10.1073/pnas.0807301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, Weinstein BM. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Li X, Novitch BG, Zheng Y, Kohn M, Xie JM, Kozinn S, Bronson R, Beg AA, Minden A. PAK4 kinase is essential for embryonic viability and for proper neuronal development. Mol Cell Biol. 2003;23:7122–7133. doi: 10.1128/MCB.23.20.7122-7133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Gomez GA, Zhang B, Lin S. Scl isoforms act downstream of etsrp to specify angioblasts and definitive hematopoietic stem cells. Blood. 2010;115:5338–5346. doi: 10.1182/blood-2009-09-244640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 Activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent TD, Jamrich M, Dawid IB. Cell interactions and the control of gene activity during early development of Xenopus laevis. Dev Biol. 1986;114:238–246. doi: 10.1016/0012-1606(86)90399-4. [DOI] [PubMed] [Google Scholar]
- Schneeberger D, Raabe T. Mbt, a Drosophila PAK protein, combines with Cdc42 to regulate photoreceptor cell morphogenesis. Development. 2003;130:427–437. doi: 10.1242/dev.00248. [DOI] [PubMed] [Google Scholar]
- Sells MA, Knaus UG, Bagrodia S, Ambrose DM, Bokoch GM, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L, Driever W. Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development. 1994;120:2443–2455. doi: 10.1242/dev.120.9.2443. [DOI] [PubMed] [Google Scholar]
- Stepniak E, Radice GL, Vasioukhin V. Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb Perspect Biol. 2009;1:a002949. doi: 10.1101/cshperspect.a002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanas S, Gomez G, Zhao Y, Park C, Choi K, Lin S. Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008;111:4500–4510. doi: 10.1182/blood-2007-09-110569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Tian Y, Lei L, Cammarano M, Nekrasova T, Minden A. Essential role for the Pak4 protein kinase in extraembryonic tissue development and vessel formation. Mech Dev. 2009;126:710– 720. doi: 10.1016/j.mod.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Veldman MB, Lin S. Etsrp/Etv2 is directly regulated by Foxc1a/b in the zebrafish angioblast. Circ Res. 2012;110:220–229. doi: 10.1161/CIRCRESAHA.111.251298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SW, Durgan J, Jin D, Hall A. Cdc42 regulates apical junction formation in human bronchial epithelial cells through PAK4 and Par6B. Mol Biol Cell. 2010;21:2996–3006. doi: 10.1091/mbc.E10-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CM, Jones GE. The emerging importance of group II PAKs. Biochem J. 2010;425:465–473. doi: 10.1042/BJ20091173. [DOI] [PubMed] [Google Scholar]
- Wells CM, Whale AD, Parsons M, Masters JR, Jones GE. PAK4: a pluripotent kinase that regulates prostate cancer cell adhesion. J Cell Sci. 2010;123:1663–1673. doi: 10.1242/jcs.055707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 4. University of Oregon Press; Eugene: 2000. The Zebrafish Book. [Google Scholar]
- Westfall TA, Brimeyer R, Twedt J, Gladon J, Olberding A, Furutani-Seiki M, Slusarski DC. Wnt-5/pipetail functions in vertebrate axis formation as a negative regulator of Wnt/beta-catenin activity. J Cell Biol. 2003;162:889–898. doi: 10.1083/jcb.200303107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler S, Gscheidel N, Brand M. Mutant generation in vertebrate model organisms by TILLING. Methods Mol Biol. 2011;770:475–504. doi: 10.1007/978-1-61779-210-6_19. [DOI] [PubMed] [Google Scholar]
- Wong LE, Reynolds AB, Dissanayaka NT, Minden A. P120-catenin is a binding partner and substrate for Group B Pak kinases. J Cell Biochem. 2010;110:1244–1254. doi: 10.1002/jcb.22639. [DOI] [PubMed] [Google Scholar]
- Yang F, Li X, Sharma M, Zarnegar M, Lim B, Sun Z. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J Biol Chem. 2001;276:15345–15353. doi: 10.1074/jbc.M010311200. [DOI] [PubMed] [Google Scholar]
- Zalik SE, Lewandowski E, Kam Z, Geiger B. Cell adhesion and the actin cytoskeleton of the enveloping layer in the zebrafish embryo during epiboly. Biochem Cell Biol. 1999;77:527–542. [PubMed] [Google Scholar]
- Zenke FT, King CC, Bohl BP, Bokoch GM. Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J Biol Chem. 1999;274:32565–32573. doi: 10.1074/jbc.274.46.32565. [DOI] [PubMed] [Google Scholar]
- Zenke FT, Krendel M, DerMardirossian C, King CC, Bohl BP, Bokoch GM. p21-Activated kinase 1 phosphorylates and regulates 14–3-3 binding to GEF-H1, a microtubule-localized Rho exchange factor. J Biol Chem. 2004;279:18392–18400. doi: 10.1074/jbc.M400084200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.