Abstract
OBJECTIVES
Recent experimental evidence suggests that environmental microbial factors early in life determine susceptibility to allergic diseases through inappropriate chemotaxis and local activation of CD1d-restricted, invariant chain natural killer T (iNKT) cells. In this study, we analyzed the involvement of these pathways in pediatric patients with eosinophilic esophagitis (EoE) before and after dietary allergen elimination.
METHODS
mRNA expression levels of components of the C-X-C motif chemokine ligand 16 (CXCL16)–iNKT–CD1d axis were compared in esophageal biopsies from EoE patients vs. normal or inflammatory controls and before and after treatment.
RESULTS
CXCL16, iNKT cell–associated cell marker Vα24, and CD1d were significantly upregulated in esophageal biopsies from EoE patients and correlated with the expression of inflammatory mediators associated with allergy. Upregulation of each of these factors was significantly more pronounced in patients aged < 6 years at diagnosis, and this early-onset EoE subpopulation was characterized by a more prominent food allergic disease phenotype in a cohort-wide analysis. Successful, but not unsuccessful, treatment of early-onset EoE patients with dietary elimination of instigating allergens led to reduction in infiltrating iNKT cells and complete normalization of mRNA expression levels of CXCL16 and CD1d.
CONCLUSIONS
Our observations place iNKT cells at the center of allergic inflammation associated with EoE, which could have profound implications for our understanding, treatment and prevention of this and other human allergic diseases.
INTRODUCTION
A wide variety of allergic and immune-mediated inflammatory disorders are rapidly and globally increasing (1,2), suggesting that environmental factors are critical mediators of these changes (3). Moreover, the increasing incidence of these diseases is especially apparent among children (4–6), which implicates a role for the environment in the processes of education and development of the immune system that follow the acquisition of a commensal microbiota along mucosal surfaces early in life (7,8).
Recently, direct evidence has emerged from animal models of asthma and inflammatory bowel disease (IBD) that microbially derived signals during a critical neonatal time frame are important regulators of later-life susceptibility to immune-mediated diseases (9). Furthermore, environmentally induced disruptions of these signals, such as those that result from the use of antibiotics, conferred increased susceptibility to experimental asthma and IBD (9,10). These rodent models have focused particular attention on the role played by microbial-induced expression of chemoattractant chemokines that promote infiltration of mucosal tissues with invariant natural killer T (iNKT) cells. iNKT cells respond to host and microbial lipid antigens when presented by CD1d and rapidly express a variety of mediators that regulate downstream immune events and effector cells (11,12). In the absence of microbiota during neonatal, but not adult life, as in germ-free or antibiotic-treated mice, such tissues express increased quantities of C-X-C motif chemokine ligand 16 (CXCL16), a chemokine involved in iNKT cell trafficking (9,13). When these early-life microbial signals are not provided, an excessive and persistent accumulation of iNKT cells occurs in the colon and lungs. Consequently, these mucosal tissues are rendered more susceptible to later-life environmental triggers of iNKT cells, which are potent and rapid producers of T helper type 2 (Th2)-type cytokines such as interleukin (IL)-4, IL-5, and IL-13, which mediate allergic sensitization and tissue inflammation (9,12,14). These model studies suggest that microbially regulated immune events during early life, or lack thereof as a consequence of, for example, antibiotic administration, are critical determinants of later-life susceptibility to allergic disease. In addition, given that components of CD1d-restricted T-cell pathways and their regulating factors such as CXCL16 serve as mediators of this susceptibility, it is reasonable to expect that they have a potential role as markers for disease risk in humans.
To test the relevance of the hypothesis that early-life events determine the tone of CD1d-restricted T-cell pathways and susceptibility to immune-mediated diseases in humans, we turned our attention to pediatric patients with eosinophilic esophagitis (EoE). EoE is characterized by chronic eosinophilic inflammation of the esophagus and is strongly associated with allergies to inhaled and food-derived antigens (15,16). Like other allergic diseases, the disorder often manifests during childhood (17) and is undergoing a rapid increase in incidence and prevalence worldwide (6,18,19), suggesting a pathogenic role of yet-to-be defined environmental factors (16).
In addition to their previously established role in asthma and IBD (20,21), iNKT cells have recently been implicated in the pathogenesis of experimental EoE (22). To study the involvement of this pathway in EoE, here we analyzed the mRNA expression levels of CXCL16, as well as iNKT cells and their markers, in serum and esophageal biopsies that were obtained before and after treatment with an allergen-elimination diet.
METHODS
Study population
The patients included in this study are part of an observational longitudinal cohort study at Boston Children’s Hospital that aims at understanding the pathophysiology of EoE and is approved by the institutional Investigational Review Board (Harvard Medical School, Boston, MA) (23–25). Parents of children aged 0–18 years whose clinical presentation indicated possible EoE (e.g., feeding intolerance, food aversion, dysphagia, failure to thrive) were invited to participate. Before routine diagnostic endoscopy, caregivers were asked to fill out a questionnaire regarding the child’s past medical history, current and past symptomatology, allergic comorbidity, and dietary habits. We only considered children to be food allergic when foods were avoided based on the results of RAST (radioallergosorbent test), skin prick, or patch testing. Upon written informed consent, serum and additional esophageal biopsies were obtained. EoE was diagnosed according to the current consensus guidelines (26). After diagnosis, patients were treated independently of this study but were again asked to participate when admitted for follow-up endoscopy. Since 2008, 451 children have been included in the cohort, 160 of whom received a diagnosis of EoE. The 291 non-EoE patients had either normal esophageal tissue biopsies (N = 225) or inflammation that was classified by the hospital pathologist as consistent with reflux esophagitis (N = 66). Within this patient cohort, we focused on 31 biopsies available from patients aged < 6 years (18 EoE, 5 reflux esophagitis, and 8 normal histology) as well as 16 biopsies from patients aged ≥6 years old (10 EoE, 6 normal histology). Twelve follow-up biopsies from EoE patients aged < 6 years were available for analysis.
Sample preparation and assay
Study biopsies from the distal esophagus (approximately 2 cm from the gastro-esophageal junction) were collected in RNA later (Qiagen, Valencia, CA) and stored at −80 °C before processing. Tissue was homogenized in 350μl RLT Buffer (Qiagen) with β-mercaptoethanol (Sigma, St Louis, MO) in GentleMACS M tubes (Miltenyi Biotec, Auburn, CA), and RNA was extracted using the RNeasy plus kit (Qiagen). RT-qPCR (quantitative reverse transcriptase–PCR) was performed using Taqman gene expression assays (Applied Biosystems, Foster City, CA) for CXCL16, CD1d, CD1a, C-X-C motif chemokine receptor 6 (CXCR6), TRAV10 (Vα24), CD3ε, leukotriene C4 synthase (LTC4S), eotaxin-3, and periostin in duplex RT-qPCR reactions with the housekeeping gene GAPDH (glyceraldehyde 3-phosphate dehydrogenase; C1000 Thermal Cycler, Bio-Rad, Herculus, CA). Gene expression relative to GAPDH (ΔΔ) was calculated. Pooled cDNA from five control patients was used as an inter-run calibrator. CXCL16 protein levels were measured in serum using a commercially available enzyme-linked immunosorbent assay according to the manufacturer’s instructions (R & D Systems, Minneapolis, MN).
Statistical analysis
Distribution of serum and mRNA expression values was assessed with D’Agostino and Pearson omnibus normality test and patient groups were accordingly compared with either Student’s t-test for normally distributed or Mann–Whitney U test for non-parametric values unless otherwise indicated. Correlations were expressed as Pearson’s or Spearman’s ρ. Epidemiological data retrieved from questionnaires was analyzed with Fisher’s exact test. For logistic regression analysis, potential predictors of EoE that were estimated to relate directly to the report of early-life antibiotic use were analyzed with univariate logistic regression and subsequently included in a multivariate model, regardless of statistical significance. Analyses were performed with Stata 12.1 (StataCorp, College Station, TX) and Prism 5.0 (GraphPad Software, La Jolla, CA). Reported values represent mean ± s.e.m. All statistical testing was two-sided, with an α of 0.05.
RESULTS
The CXCL16–iNKT–CD1d axis is prominently involved in early-onset EoE patients
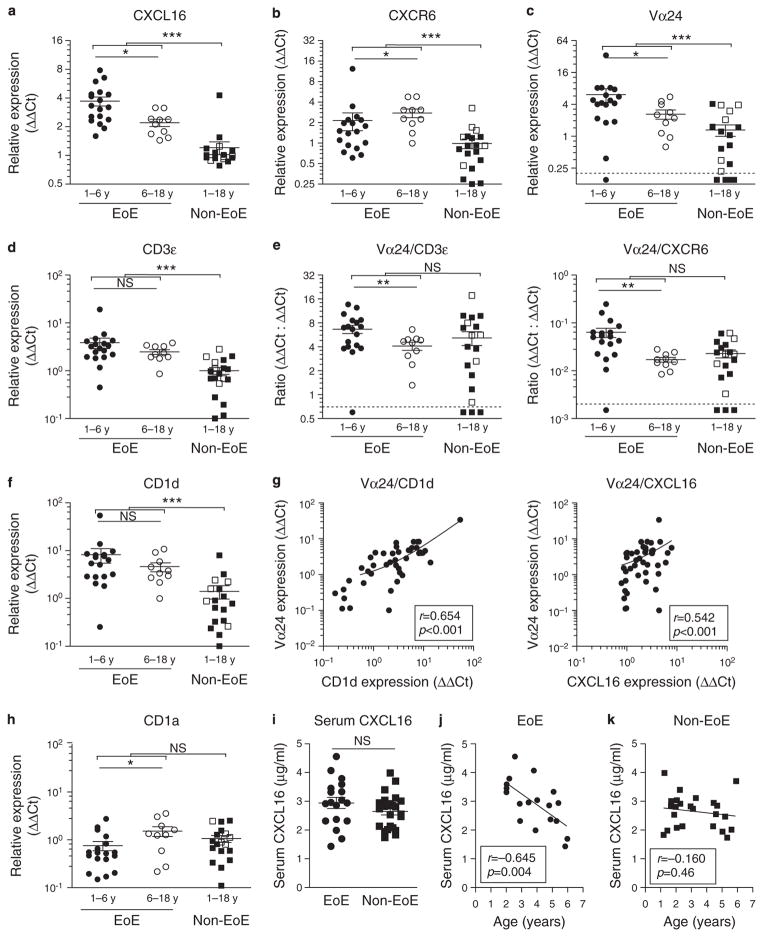
To investigate the contribution of CXCL16 to local disease in EoE, we analyzed mRNA expression levels of CXCL16 in esophageal biopsies at the time of diagnosis in 47 children. Patients were grouped according to age at diagnosis: (i) patients aged < 6 years (18 EoE vs. 13 non-EoE, of whom 5 with histological evidence of reflux esophagitis) and (ii) 6–18 years (10 EoE vs. 6 non-EoE with normal tissue histology). CXCL16 was expressed in all esophageal biopsies analyzed (cycle threshold range 26.6–32.7), but expression was significantly higher in EoE patients when compared with controls (3.2 ± 0.3 vs. 1.2 ± 0.2, P < 0.001, Figure 1a). Furthermore, expression levels were higher in early-onset EoE patients, which we defined as children diagnosed < 6 years of age, compared with their older EoE counterparts (3.7 ± 0.4 vs. 2.2 ± 0.2, P < 0.05). No age-dependent difference was seen in the non-EoE control patients (1.3 ± 0.3 vs. 1.0 ± 0.1, P = 0.52, Figure 1a).
Figure 1.
Components of the C-X-C motif chemokine ligand 16 (CXCL16)–iNKT cell (invariant chain natural killer T cell)–CD1d pathway are most prominently expressed in early-onset eosinophilic esophagitis (EoE) patients. Gene expression relative to housekeeping gene GAPDH (glyceraldehyde 3-phosphate dehydrogenase; expressed as ΔΔ Ct values) is shown in early-onset EoE patients (< 6 years of age, full circles), EoE patients 6–18 years (open circles), and non-EoE controls (patients aged < 6 years shown as full squares and patients aged 6–18 years as open squares) (a–f, h). mRNA expression levels were elevated in EoE patients compared with non-EoE controls for CXCL16 (a), C-X-C motif chemokine receptor 6 (CXCR6) (b), Vα24 T-cell antigen receptor (TCR) (c), CD3ε (d), and CD1d (f), but not for CD1a (h). Early-onset EoE patients express significantly higher levels of CXCL16 (a), CXCR6 (b), and Vα24 TCR (c) when compared with older EoE patients. Ratio of expression is plotted for Vα24/CD3ε and Vα24 / CXCR6 (e). (g) Vα24 TCR mRNA levels correlated significantly with CD1d and CXCL16. Expression values under the detection limit or the corresponding expression ratios are plotted under the dotted line when transcripts were not detected. (i) Serum CXCL16 levels of early-onset EoE patients were compared with age-matched non-EoE controls. (j, k) Serum CXCL16 was correlated to age at diagnosis. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant.
We analyzed the same biopsies for mRNA expression of CXCR6, the canonical receptor for CXCL16 that is expressed on a variety of T-cell subsets, including Th1-polarized CD4+ and CD8+ T cells (27), as well as iNKT cells (28). Although higher expression levels were seen in EoE patients compared with non-EoE controls (Figure 1b), the highest expression levels were observed in EoE patients aged 6–18 years. In light of the elevated CXCL16 levels observed in EoE patients < 6 years old, these results are consistent with observations that CXCR6 in T cells is downregulated upon activation (29) and suggest that CXCL16 engagement is highly active in early-onset EoE patients.
To determine whether CXCL16 overexpression was indeed associated with infiltration of iNKT cells, we analyzed gene expression of Vα24, an essential component of the invariant T-cell antigen receptor (TCR) of iNKT cells (30). Vα24 TCR mRNA transcripts were detected in 17 of the 18 early-onset EoE patients, all 10 older EoE patients, and in 14 of the 19 control patients. In line with CXCL16 expression results, the highest levels of Vα24 TCR mRNA were detected in EoE patients aged < 6 years (Figure 1c). Increased Vα24 expression did not result from more pronounced overall T-cell infiltration, because expression levels of the ε chain of the TCR (CD3ε) were equivalent in both young and older EoE patients (Figure 1d), and normalization of Vα24 expression against CD3εor CXCR6 did not resolve the difference between old and young EoE patients (Figure 1e). These results suggest the existence of distinct differences in infiltrating T-cell subsets between early-onset and older EoE patients.
iNKT cells are restricted by recognition of lipid antigen presented through CD1d on antigen-presenting cells (12). We therefore examined CD1d mRNA and observed increased expression in EoE patients compared with non-EoE control patients, indicating that at the site of inflammation there is an enhanced capacity to activate infiltrating iNKT cells (Figure 1f). Indeed, we depicted a positive correlation between tissue mRNA expression of CD1d and Vα24 in esophageal biopsies (Spearman ρ = 0.654, P < 0.001, Figure 1g). Similarly, expression of CXCL16 and Vα24 correlated positively (Spearman ρ = 0.542, P < 0.001, Figure 1g). Conversely, expression levels of CD1a, a related glycoprotein on antigen-presenting cells, did not differ between EoE patients and controls (1.0 ± 0.2 vs. 1.2 ± 0.2, P = 0.38), arguing that increased CD1d expression in EoE is part of the disease-specific immune phenotype rather than the result of overall increase in tissue myeloid or inflammatory cells (Figure 1h). Combined, these data show local involvement of the CXCL16–iNKT–CD1d axis in EoE patients and that this involvement is significantly more prominent in early-onset EoE patients.
Serum CXCL16 levels in early-onset EoE patients are consistent with an age-dependent role in EoE pathogenesis
CXCL16 is present as a membrane-bound protein on epithelial and myeloid cells, as well as a soluble form, that can be retrieved from patient serum (31). We hypothesized that early-onset EoE patients might also be characterized by higher levels of CXCL16 in serum when compared with non-EoE control patients, as was observed in tissues (Figure 1a). If so, this might serve as a convenient biomarker for disease. Serum levels were similar between the patient groups (2.9 ± 0.2 μg / ml for EoE (N = 18) vs. 2.6 ± 0.1 μg/ml for non-EoE (N = 24), P = 0.19, Figure 1i). Interestingly, there was a significant negative correlation between serum CXCL16 levels and age of diagnosis in EoE (r = −0.65, P = 0.004, Figure 1j). Conversely, this relationship between age and serum CXCL16 levels was absent in the non-EoE patients (r = −0.16, P = 0.46, Figure 1j) and EoE patients aged 6–18 years (N = 8, r = 0.31, P = 0.45). These results further support an age-dependent role for CXCL16 in EoE pathophysiology, which is detectable in the serum.
Involvement of the CXCL16–iNKT–CD1d axis is associated with a more pronounced food allergic phenotype
As CXCL16 overexpression and iNKT cell infiltration were significantly more pronounced in the younger EoE patients, we asked whether early-onset EoE differs from EoE that becomes apparent during later childhood. Within our entire cohort of 451 patients, 44 of the 160 EoE patients (28 %) were < 6 years at the time of diagnosis. In line with data from multiple patient populations (6,19), we saw a strong male preponderance in EoE patients, as well as a higher occurrence of all investigated allergic diatheses compared with control patients (Table 1). Comparison between EoE patients aged < 6 and ≥6 years, however, revealed striking differences in the distribution of allergic comorbidity. Whereas older patients were more likely to be asthmatic (50 vs. 20 %, P = 0.003), early-onset EoE patients had significantly more often been diagnosed with food allergy against milk (32 vs. 14 %, P = 0.02) or any specific food allergen (48 vs. 25 %, P = 0.008) through skin prick, patch, or RAST testing. These epidemiological data are suggestive of a particularly dominant role of food allergic sensitization in early-onset EoE.
Table 1.
Symptomatology and allergic diatheses in early-onset vs. later onset EoE patients
| EoE
|
Non-EoE
|
EoE vs. non-EoE | EoE <6 vs. ≥6 years | |||
|---|---|---|---|---|---|---|
| <6 years | ≥6 years | <6 years | ≥6 years | |||
| N | 44 | 116 | 84 | 207 | ||
|
| ||||||
| Male sex | 32 (73%) | 79 (68%) | 45 (54%) | 89 (43%) | < 0.001 | 0.70 |
|
| ||||||
| Age (5th–50th–95th percentile) | 0.18–4.0–5.8 | 6.7–12.7–17.6 | 0.1–2.3–5.7 | 6.1–12.9–17.2 | 0.89 | |
|
| ||||||
| Preterm delivery (<36 weeks) | 10 (23%) | 33 (29%) | 18 (21%) | 55 (27%) | 0.74 | 0.55 |
|
| ||||||
| GI symptomatology | ||||||
|
| ||||||
| Pain with swallowing | 8 (19%) | 31 (27%) | 15 (18%) | 42 (21%) | 0.23 | 0.41 |
|
| ||||||
| Food getting stuck | 12 (28%) | 53 (46%) | 20 (24%) | 67 (34%) | 0.02 | 0.07 |
|
| ||||||
| Dysphagia | 10 (23%) | 43 (38%) | 19 (23%) | 66 (33%) | 0.39 | 0.09 |
|
| ||||||
| Abdominal pain | 14 (33%) | 58 (51%) | 36 (44%) | 141 (69%) | 0.002 | 0.049 |
|
| ||||||
| Constipation | 17 (40%) | 29 (26%) | 40 (48%) | 82 (41%) | 0.008 | 0.11 |
|
| ||||||
| Diarrhea | 16 (37%) | 34 (30%) | 38 (45%) | 44 (22%) | 0.45 | 0.44 |
|
| ||||||
| Weight loss | 11 (25%) | 24 (21%) | 20 (24%) | 36 (18%) | 0.54 | 0.67 |
|
| ||||||
| Regurgitation | 12 (29%) | 34 (30%) | 29 (35%) | 109 (55%) | <0.001 | 1.0 |
|
| ||||||
| Allergic diatheses | ||||||
|
| ||||||
| Milk allergy (skin or RAST tested) | 14 (32%) | 16 (14%) | 15 (18%) | 2 (1%) | <0.001 | 0.02 |
|
| ||||||
| Any food allergy (skin or RAST tested) | 21 (48%) | 29 (25%) | 17 (20%) | 5 (2%) | <0.001 | 0.008 |
|
| ||||||
| Wheezinga | 14 (39%) | 49 (50%) | 26 (36%) | 65 (35%) | 0.03 | 0.33 |
|
| ||||||
| Asthmaa | 7 (20%) | 49 (50%) | 7 (10%) | 53 (29%) | <0.001 | 0.003 |
|
| ||||||
| Hay fevera | 15 (44%) | 40 (45%) | 6 (9%) | 34 (20%) | <0.001 | 1.0 |
|
| ||||||
| Eczemaa | 20 (56%) | 46 (48%) | 33 (46%) | 45 (25%) | <0.001 | 0.56 |
EoE, eosinophilic esophagitis, GI, gastrointestinal; RAST, radioallergosorbent test.
Added later to the questionnaire, responses available for 383 patients (132 EoE).
P-values are based upon two-sided testing with Fisher’s exact test.
Bold italic values represent statistically significant results.
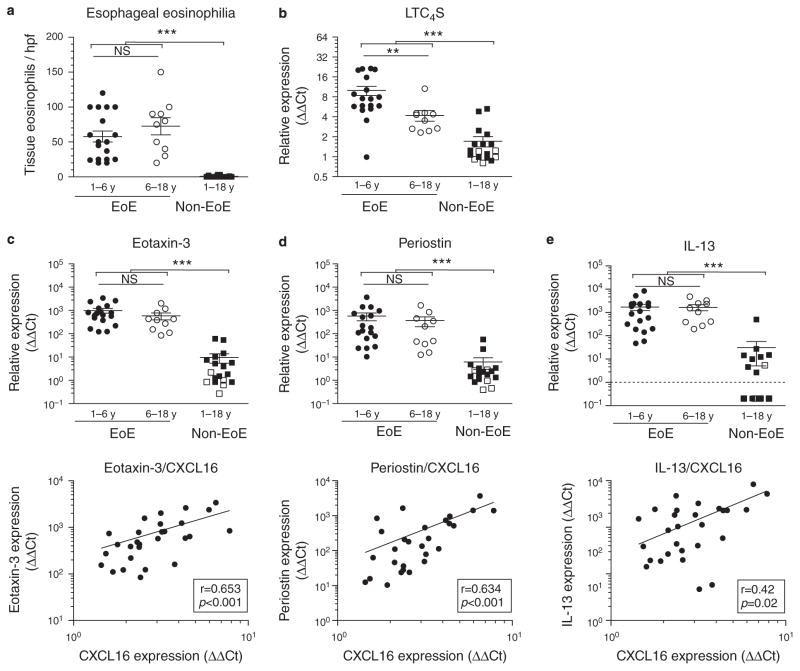
To further test this hypothesis at the tissue level, we again turned to tissue biopsies. We have recently demonstrated that elevated mRNA levels of LTC4S mark a subpopulation of EoE patients with a more pronounced local allergic phenotype (25). No difference was found in tissue eosinophil counts between early and later-onset EoE patients (Figure 2a). However, in line with our epidemiological data, we found that early-onset EoE patients had significantly higher levels of LTC4S mRNA expression (Figure 2b).
Figure 2.
C-X-C motif chemokine ligand 16 (CXCL16) correlates with allergic mediators in early-onset eosinophilic esophagitis (EoE). Early-onset EoE patients do not show (a) more severe tissue eosinophilia (counts / hpf (high power fields)) or increased expression of EoE-specific marker genes (c) eotaxin-3, (d) periostin, or (e) interleukin (IL)-13 but show a more pronounced allergic tissue phenotype as signified by increased expression of (b) leukotriene C4 synthase (LTC4S). In EoE patients, CXCL16 expression correlates with IL-13 and disease-specific, IL-13-dependent tissue markers eotaxin-3 (c) and periostin (d). **P < 0.01; ***P < 0.001; NS, not significant.
In addition, we analyzed tissue biopsies for EoE marker genes eotaxin-3, periostin, and IL-13, which were among the first genes to be identified as part of an EoE-specific esophageal transcriptome (32–34). We observed that all were increased in patients with EoE relative to the non-EoE subjects (Figures 3c–e). Although no age-dependent differences in expression were found, we observed a strongly positive correlation between tissue mRNA expression levels of CXCL16 and these marker genes (Figure 3c–e). Eotaxin-3 has been identified as one of the key eosinophil attractant cytokines in EoE (32), and periostin is described to facilitate eosinophil tissue infiltration (33). Expression of both effector molecules has been described to occur downstream of IL-13 signaling in EoE (33,34). Although IL-13 is produced by iNKT cells (12), we did not observe a significant correlation between expression of IL-13 and Vα24 (r = 0.13, P = 0.50), indicating that iNKT cells are unlikely to be the dominant cellular source of IL-13. These data suggest a link between the local expression of CXCL16 and effector pathways of EoE at the site of tissue inflammation.
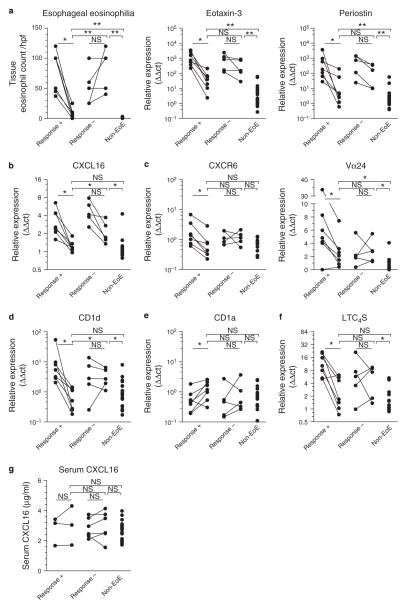
Figure 3.
The C-X-C motif chemokine ligand 16 (CXCL16)–iNKT cell (invariant chain natural killer T cell)–CD1d pathway responds to treatment with dietary elimination of allergens in early-onset eosinophilic esophagitis (EoE). (a)Tissue eosinophil counts decreased upon successful treatment (Response +, N = 7) but remained unaltered or increased in non-responders (Response−, N = 5). (b) CXCL16 mRNA expression levels decreased in all patients upon treatment but remained significantly elevated in non-responders over responders and controls. Comparable results were obtained for expression levels of (c) CXCR6 and Vα24 and (d) CD1d. (e) No decrease in CD1a expression was seen upon successful treatment. (f) Elimination of dietary allergens led to normalization of tissue leukotriene C4 synthase (LTC4S) mRNA levels in responding but not in non-responding patients. (g) Allergen avoidance does not affect serum CXCL16 levels in EoE patients. Asterisks represent the result of statistical testing with Wilcoxon signed rank test for before–after expression levels and Mann–Whitney U test for post-treatment expression levels as follows: * P < 0.05; * * P < 0.01; NS, not significant.
Treatment with elimination diet leads to normalization of the CXCL16–iNKT–CD1d axis in early-onset EoE patients
To analyze the response of the CXCL16–iNKT–CD1d axis to EoE treatment, we repeated mRNA measurements in biopsies that were obtained during follow-up endoscopies performed to monitor disease progression and therapeutic response. Follow-up biopsies were available from 12 of the 18 early-onset EoE patients after a median follow-up time of 10.1 months (range 1.9–24.8 months). Based on the degree of tissue eosinophilia obtained from routine histopathological analysis, 7 of the 12 EoE patients had significantly improved with treatment, with median eosinophil counts of 8 / high power field (hpf) (range 0–25) compared with the initial biopsies that showed a median of 100 eosinophils / hpf (range 37–120, P = 0.02 by Wilcoxon signed rank test, Figure 3a). Conversely, in the five non-responding patients, tissue eosinophilia at follow-up remained high, ranging from 40 to 120 eosinophils / hpf (median 100 / hpf, P = 0.25). In line with tissue eosinophil counts and previously published data (34), mRNA expression levels of eotaxin-3 and periostin at follow-up had decreased significantly in responding patients, whereas this reduction was smaller and not significant in the non-responders (Figure 3a). Patient and treatment details are summarized in Table 2.
Table 2.
Patient characteristics and treatment details of 12 early-onset EoE patients
| Patient | Sex/age | Dietary elimination | Pharmacological treatment | Time to followup (months) | Tissue eosinophil response | Reported symptomatology at follow-up | Comments |
|---|---|---|---|---|---|---|---|
| Responders | |||||||
| M, 3.5 | Milk, soy, eggs nuts, peas, beans, beef, pork | Swallowed budesonide | 17.5 | 25→5 | Improved appetite and symptoms | ||
| M, 2.9 | Milk | PPI | 5.9 | 37→0 | Improvement, normal growth velocity | Negative environmental allergen screen | |
| M, 2.0 | Milk, soy, oat, egg, sweet potatoes, chicken, carrots, grapes, strawberries, pears, string beans | None | 11.9 | 100→2 | No upper GI symptoms | Soy was reintroduced before followup endoscopy | |
| F, 4.9 | Corn, rye, pork, egg, wheat, dairy, nuts, soy, shellfish, beef | PPI | 10.6 | 120→10 | Asymptomatic | ||
| M, 2.2 | Elemental diet | PPI | 9.6 | 100→25 | Improved but not resolved on elemental diet | ||
| M, 4.0 | No restrictions | Swallowed budesonide, no PPI | 5.7 | 100→8 | Asymptomatic | ||
| M, 2.9 | Milk, eggs, wheat, turkey, nuts | PPI | 1.9 | 45→10 | Clear improvement | Inconsistent use of PPI | |
| Non-responders | |||||||
| F, 5.8 | Milk, soy | PPI | 24.8 | 25→125 | Persistent episodes of food getting stuck | Admitted poor compliance with diet | |
| F, 2.0 | Milk, soy, beef, oranges, eggs, wheat | None | 7.4 | 50→50 | Initially weight gain but recent flare of symptoms | Complete clinical resolution after elimination of corn | |
| M, 5.3 | Eggs, wheat, chicken, turkey, pear, apples, peas, corn, milk, onions, peanuts, tree nuts | PPI | 15.5 | 25→40 | Less dysphagia and reflux, though symptoms still occur | RAST positive to > 50 foods, only 12 of which were excluded | |
| M, 5.2 | No restrictions | PPI | 5.3 | 100→100 | Persistent symptoms | RAST positive for milk, though no specific dietary interventions | |
| M, 5.1 | None | PPI, 1 course of swallowed budesonide | 15.3 | 60→100 | Relatively asymptomatic | ||
EoE, eosinophilic esophagitis; F, female; GI, gastrointestinal; M, male; PPI, proton pump inhibitor; RAST, radioallergosorbent test.
We next turned our attention to the CXCL16–iNKT cell–CD1d axis in treated patients. We observed that, in the seven responders, CXCL16 mRNA expression completely normalized to the expression levels encountered in age-matched control patients (1.2 ± 0.1 vs. 1.3 ± 0.3, P = 0.88, Figure 3b). In contrast, CXCL16 expression remained significantly elevated in the five non-responders (2.3 ± 0.4) when compared with control patients (P = 0.01) as well as responding EoE patients (P = 0.03). CXCR6 and Vα24 TCR mRNA levels also decreased significantly in the seven responding patients (2.7-fold, P = 0.02 and 3.3-fold, P = 0.03, respectively) but not in the non-responders (P = 1.0 and P = 0.63, Figure 3c). Likewise, response to treatment was associated with a 17-fold reduction in CD1d expression (P = 0.01) compared with a 1.5-fold reduction in non-responders (P = 0.31, Figure 3d). No decrease, but rather an increase, in CD1a expression levels was observed upon successful treatment (2.0-fold increase, P = 0.03), in this case showing that reduction of CD1d mRNA levels did not result from decreased myeloid cell infiltration (Figure 3e). Finally, we found that LTC4S mRNA levels only decreased following successful treatment (4.0-fold, P = 0.03 compared with 1.3-fold, P = 0.81 in non-responders, Figure 3f). Allergen avoidance therapy did not affect the levels of serum CXCL16 (Figure 3g), confirming that upregulation of CXCL16 represents a local mucosal event that is not broadcasted systemically. Together, these data show that involvement of the CXCL16–iNKT cell–CD1d axis can be modified by dietary interventions that aim to eliminate common allergens from the diet.
EoE is potentially associated with use of antibiotics during the first year of life
Animal model experiments suggest that CXCL16 overexpression by epithelial cells can result from inadequate instruction from the commensal microbiota at an early age (9). We therefore hypothesized that antibiotic treatment during the first year of life may be associated with the development of EoE, as has been previously suggested for pediatric IBD (35) and asthma (36) and, very recently, also for EoE (37). Unfortunately, our questionnaires at the time of study enrollment did not directly address the extent of early-life antibiotic exposure or mode of delivery but instead inquired into medication use during the first year of life, in particular the use of antacids, corticosteroids, and prokinetics. Data on the prescription of antibiotics was obtained through an open-ended question asking parents to list additional medications during the first year of life. Questionnaires of 451 patients (160 EoE) were analyzed. Antibiotics in the first year of life were reported in 29 patients, of whom 20 were subsequently diagnosed with EoE (P < 0.001). Although it is very likely that under-reporting affected the total number of cases, the difference between EoE patients and controls is unlikely the result of recall bias. This is because report of non-antibiotic drug-use during the first year of life did not differ between cohorts (33 % of EoE patients vs. 41 % of non-EoE controls, P = 0.09), and questionnaires were filled out before diagnosis. Furthermore, a positive association (odds ratio 4.10, 95 % confidence interval 1.36–12.35) between EoE and reported antibiotics use during the first year of life remained after adjustment for sex, age, prematurity, hospitalization in the first year of life, allergic comorbidities, and non-antibiotic drug use in the first year of life (Table 3). Although statistical significance was reached for this association in our cohort, additional epidemiological studies designed to specifically address this question are needed to confirm a causative link between early-life antibiotic exposure and occurrence of EoE later in life.
Table 3.
Predictors in a cohort-wide logistic regression analysis for a diagnosis of EoE versus non-EoE before diagnostic endoscopy
| Predictor | Univariate analysis | Multivariate analysis (N =345) | ||
|---|---|---|---|---|
| Odds ratio (95 % CI) | P value | Odds ratio (95 % CI) | P value | |
| Male sex | 2.65 (1.77–3.99) | < 0.001 | 2.47 (1.42–4.28) | 0.001 |
| Age (per year increase) | 1.00 (0.97–1.04) | 0.89 | 1.00 (0.94–1.06) | 0.93 |
| Preterm delivery (< 36 weeks) | 1.08 (0.69–1.67) | 0.73 | 1.23 (0.67–2.34) | 0.51 |
| Hospitalization in the first year of life | 0.70 (0.45–1.10) | 0.13 | 0.59 (0.31–1.11) | 0.10 |
| Reported asthma | 2.35 (1.50–3.69) | < 0.001 | 1.45 (0.80–2.63) | 0.22 |
| Reported eczema | 2.22 (1.44–3.42) | < 0.001 | 1.60 (0.91–2.79) | 0.10 |
| Reported seasonal allergy | 4.08 (2.50–6.68) | < 0.001 | 2.90 (1.63–5.18) | < 0.001 |
| Any food allergy | 5.56 (3.21–9.62) | < 0.001 | 4.58 (2.19–9.57) | < 0.001 |
| > 1 annual ear infection requiring antibiotics | 1.41 (0.95–2.09) | 0.09 | 1.04 (0.61–1.80) | 0.87 |
| Reported antibiotic allergy | 1.54 (0.96–2.48) | 0.08 | 1.77 (0.91–3.48) | 0.09 |
| Non-antibiotic drugs in the first year of life | 0.70 (0.46–1.04) | 0.08 | 0.48 (0.27–0.87) | 0.02 |
| Reported antibiotics in the first year of life | 4.48 (1.98–10.1) | < 0.001 | 4.10 (1.36–12.35) | 0.01 |
CI, confidence interval; EoE, eosinophilic esophagitis.
Bold values represent statistically significant results.
DISCUSSION
In the current study, we provide evidence that supports the involvement of CD1d-restricted iNKT cell pathways in the pathogenesis of pediatric EoE. There are several key elements associated with the findings contained here. First, our results provide a direct human corollary of previous observations in animal models that commensal microbes, in an age-dependent fashion, regulate iNKT cell accumulation in the colon and lung by altering expression of chemokine ligands that promote iNKT cell migration, such as CXCL16. Similar to this previous report, we found that all of the factors associated with iNKT cells such as the associated TCR chain (Vα24), the antigen-presenting molecule that regulates their activity (CD1d), and the chemokine ligand that determines their chemotaxis into tissues (CXCL16), are observed at significantly higher levels in the esophageal tissue of EoE patients compared with normal or inflammatory controls. Moreover, the highest levels were observed in children with early-onset EoE (< 6 years), and the elements of the CXCL16–iNKT cell–CD1d pathway correlated with the presence of inflammatory mediators associated with EoE-specific inflammation (eotaxin, periostin), pointing towards a direct role in disease pathogenesis. Indeed, successful dietary therapy was associated with the resolution of disease, together with a synchronous and proportionate decrease in the levels of CXCL16, CD1d, and Vα24, suggestive of a cause-and-effect relationship. Given the observations in animal models, these findings in humans with EoE support the hypothesis that early-life events that affect CD1d-restricted T-cell pathways at mucosal surfaces have an important role in the pathogenesis of EoE. Indeed, we detected an association between antibiotic use in the first year of life and the development of EoE, drawing additional and provocative similarities to the aforementioned animal studies. Together, these studies support the concept that a pathway involving environmentally induced changes in early life, in this case caused by antibiotic administration, cause perturbations in mucosal iNKT cell homeostasis in esophageal tissues and determines susceptibility to EoE.
Previous translational studies on the role of iNKT cells in human allergy have predominantly been performed in asthmatic patients, after it was shown that iNKT cells are required for the induction of experimental airway hyper-reactivity in mice (12,38,39). As efforts to demonstrate increased levels of iNKT cells in bronchoalveolar lavage fluid of asthmatic patients have not yielded clear differences with control populations (30,40,41), such a role has remained a matter of debate (21,42). These analyses of bronchoalveolar lavage fluid have, however, been performed in adult patients with a longstanding history of asthma and therefore do not rule out a potential role for iNKT cells in the early phase of disease pathogenesis. This concept is further supported by recent experimental work that suggests that rather than being the main effector cell in advanced asthma, iNKT cells are critically involved in the allergic sensitization phase of disease. Specifically, Scanlon et al. (14) showed that intratracheal exposure to bacterial lipid antigens resulted in rapid peribronchiolar infiltration of iNKT cells and formation of eosinophil- rich granulomas. Upon intratracheal co-administration of ovalbumin, a Th2 immune response was mounted against ovalbumin, including synthesis of anti-ovalbumin immunoglobulin E. Allergic sensitization persisted until months after the initial exposure and, once established, was independent of iNKT cells (14). These observations thus point towards iNKT cells as a crucial instigating cell type in allergic forms of inflammation and demonstrate that once allergic sensitization has occurred, cytokine production by iNKT cells is dispensable for disease pathogenesis. The latter finding likely explains the absence of a correlation between IL-13 and Vα24 expression in our cohort.
By studying pediatric EoE patients at the time of diagnosis, we were able to obtain affected mucosal tissues to directly correlate the involvement of iNKT cells and their mediators with disease phenotype. Like asthma, EoE is a chronic, immune/antigen-driven condition characterized by aberrant Th2 immune responses and allergic sensitization (26). Both food-derived and inhaled allergens have been implicated in its pathogenesis (26,43,44), which is reflected in high response rates upon initiation of an elemental or six-food elimination diet (45,46). As a corollary, increased esophageal eosinophilia is observed during the pollen season in pollen-sensitized patients (47,48). A likely role for iNKT cells in the process of allergic sensitization was suggested from studies that showed that food-allergen dependent, experimental murine EoE could not be induced in CD1d-deficient mice (22). Consistent with this, we have shown that individual components of the CXCL16–CD1d–iNKT axis are overexpressed in our entire pediatric EoE population, but increased expression was significantly more prominent in patients who developed the disease at a very young age. Epidemiological data from our cohort study further indicate that in patients diagnosed when < 6 years of age, EoE is characterized by a higher fraction of food-allergic sensitization. In addition, local mRNA levels of LTC4S, the enzyme that generates the allergic mediator leukotriene C4, were significantly higher in children with early-onset EoE than in their older counterparts. As such, these data suggest that this early-onset disease phenotype is likely to depend more heavily on local sensitization to food-derived allergens. Our results are therefore supportive of an association between local accumulation of iNKT cells and allergic sensitization to food antigens in human EoE. Additional support for our concept was provided recently by Jyonouchi et al., (49) who reported increased numbers of iNKT cells in esophageal biopsies of pediatric patients with active disease. Upon stimulation with milk-derived sphingolipids, iNKT cells from these EoE patients readily responded with increased IL-13 and IL-4, but not interferon-γ production, showing that iNKT cell–mediated responses to food-derived antigens can be responsible for Th2 skewing of immune responses (49).
Despite recent advances in our understanding of iNKT cell biology, the exact molecular mechanism that couples iNKT cell infiltration to allergic sensitization in human EoE patients remains to be further identified. iNKT cells are CD1d-restricted and can be activated in response to exogenous lipid antigens, including pollen and dust mite–associated lipids (50,51), as well as dietary antigens such as sphingomyelin derived from milk (52) or the allergen Ber e1 from Brazil nuts in the presence of a naturally occurring lipid fraction (53). The study by Olszak et al. (9) has furthermore implicated endogenous (self) antigens as a source of iNKT stimulation in a pathogenic environmental context and showed that microbes inhibit this auto-reactivity, at least in part, through dampening CXCL16 expression. It is therefore of interest that we identified a relationship between early-life antibiotic use and EoE, suggesting that microbial reduction during early life may also confer risk for the development of EoE as previously demonstrated for asthma (36,54) and IBD (35) in humans. Such antibiotic use may diminish the quantity and / or activity of microorganisms capable of regulating iNKT cells. Interestingly, in this regard is evidence that Bacteroides fragilis encodes a biosynthetic pathway for the generation of glycosphingolipids capable of inhibiting colonic iNKT cells, suggesting that antibiotics which diminish this or similar types of organisms may predispose to iNKT cell-mediated diseases (55). Our longitudinal cohort study was not designed to specifically address the potential association between antibiotics use and onset of EoE, which has resulted in potential under-reporting of early-life antibiotic use and the absence of data on the quantity or types of antibiotics prescribed. Although our data suggest that the association between early-life antibiotic exposure and development of EoE was not due to recall bias, we caution that additional epidemiological studies are needed to prove this important hypothesis. We propose that EoE is a suitable model disease for such studies, as an association with antibiotic exposure is less susceptible to reverse causation, which has hampered inference from pediatric asthmatic cohorts (56). Indeed, gastrointestinal symptoms are less likely to be treated with antibiotics than upper or lower airway infections, and symptomatology was not more pronounced in EoE patients compared with controls in our cohort. Supportive of our data is a recently published paper that has described an association of the same order of magnitude between EoE and antibiotic use during infancy as found in our study (37). Large European studies have further pointed out that exposure to a wide variety of environmental microorganisms confers protection against both atopy and asthma (7), making it reasonable to surmise that similar mechanisms are operative in EoE.
In summary, we have assessed how recent findings from experimental studies in mouse models translate to human allergic disease in the context of EoE. Based on our results, we propose the following model of EoE pathophysiology: (1) Insufficient immune imprinting by environmental microorganisms results in esophageal upregulation of CXCL16 and chemotaxis of iNKT cells into the esophagus, making the esophagus susceptible to triggers of iNKT cell activation; (2) Lipid antigens derived from food, aeroallergens, microorganisms, and / or potentially self-antigens associated with the esophagus are inappropriately presented to iNKT cells by CD1d in a manner that stimulates their pathological activation; (3) Activated iNKT cells serve as an early source of IL-4, IL-5, and IL-13, facilitating Th2-mediated allergic sensitization and responses that promote and sustain inflammation. Our observations further suggest that in an esophageal iNKT cell–dominant disease phenotype, as occurs early in life, allergic inflammation is driven primarily by food antigens. Conversely, in EoE that manifests at a later age, esophageal inflammation is potentially less dependent on iNKT cells based upon our findings. This model of disease pathophysiology suggests that pharmacological interference with CXCL16 and CD1d, especially at a young age and at onset of disease, is a potent target in the prevention and treatment of allergic diseases such as EoE and may further serve as diagnostic markers in determining disease etiology and therapeutic response.
Study Highlights.
WHAT IS CURRENT KNOWLEDGE
The incidence of eosinophilic esophagitis is rapidly increasing.
Animal studies have demonstrated a role for invariant chain natural killer T (iNKT) cells in allergic sensitization and susceptibility to allergic immune disorders.
WHAT IS NEW HERE
iNKT cells, as well as the cellular components that regulate their chemotaxis and activation, are increased in esophageal tissue of eosinophilic esophagitis (EoE) patients.
Involvement of the iNKT cell pathway is particularly pronounced in patients with early-onset EoE who have high degrees of sensitization to food antigens.
Elimination of allergens from diet results in normalization of cellular markers of the iNKT cell pathway.
Acknowledgments
We are grateful to Ms Katharine Rooney and Mr Michael Pardo for assistance in patient recruitment and sample collection and thank Dr Jeremy Goettel for helpful discussions.
Financial support: This work was supported by the National Institutes of Health Grants R01AI075037 (to E.F.), DK044319, DK051362, DK053056 and DK088199 (to R.S.B.), and K24DK82792-1 (to S.N.). This work was also supported by the Harvard Digestive Diseases Center Grants P30 DK034854 and DK0034854 and the Research Council, Boston Children’s Hospital (pilot study, to E.F. and S.N.). W.L. is supported by grants from the Ter Meulen Fund of the Royal Netherlands Academy of Arts and Sciences and the Banning de Jong Fund in The Netherlands. None of the funding sources had a role in the design of the experiments.
Footnotes
CONFLICT OF INTEREST
Guarantor of the article: Edda Fiebiger, PhD.
Specific author contributions: Project conception and design, data acquisition / analysis/ interpretation, drafting of the article, critical revision: Willem S. Lexmond; data acquisition, critical revision: Joana F. Neves; patient identification, inclusion, and prospective cohort study design: Samuel Nurko; data interpretation and critical revision: Torsten Olszak and Mark Exley; project conception and design, data interpretation, drafting of the article, and critical revision: Richard S. Blumberg; project conception and design, data interpretation, prospective cohort study design, and critical revision: Edda Fiebiger. All authors approved the final submitted version of the manuscript.
Potential competing interests: Exley declares holding equity in NKT Therapeutics. All the other authors declare no conflict of interest.
References
- 1.Eder W, Ege MJ, Mutius von E. The asthma epidemic. N Engl J Med. 2006;355:2226–35. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 2.Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. e42. doi: 10.1053/j.gastro.2011.10.001. quiz e30. [DOI] [PubMed] [Google Scholar]
- 3.Garn H, Neves JF, Blumberg RS, et al. Effect of barrier microbes on organ-based inflammation. J Allergy Clin Immunol. 2013;131:1465–78. doi: 10.1016/j.jaci.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branum AM, Lukacs SL. Food allergy among children in the United States. Pediatrics. 2009;124:1549–55. doi: 10.1542/peds.2009-1210. [DOI] [PubMed] [Google Scholar]
- 5.Cherian S, Smith NM, Forbes DA. Rapidly increasing prevalence of eosinophilic oesophagitis in Western Australia. Arch Dis Child. 2006;91:1000–4. doi: 10.1136/adc.2006.100974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004;351:940–1. doi: 10.1056/NEJM200408263510924. [DOI] [PubMed] [Google Scholar]
- 7.Ege MJ, Mayer M, Normand A-C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–9. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 8.Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137rv7. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell SL, Gold MJ, Hartmann M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–7. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 12.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13:101–17. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 13.Germanov E, Veinotte L, Cullen R, et al. Critical role for the chemokine receptor CXCR6 in homeostasis and activation of CD1d-restricted NKT cells. J Immunol. 2008;181:81–91. doi: 10.4049/jimmunol.181.1.81. [DOI] [PubMed] [Google Scholar]
- 14.Scanlon ST, Thomas SY, Ferreira CM, et al. Airborne lipid antigens mobilize resident intravascular NKT cells to induce allergic airway inflammation. J Exp Med. 2011;208:2113–24. doi: 10.1084/jem.20110522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothenberg ME. Biology and treatment of eosinophilic esophagitis. Gastroenterology. 2009;137:1238–49. doi: 10.1053/j.gastro.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulder DJ, Justinich CJ. Understanding eosinophilic esophagitis: the cellular and molecular mechanisms of an emerging disease. Mucosal Immunol. 2011;4:139–47. doi: 10.1038/mi.2010.88. [DOI] [PubMed] [Google Scholar]
- 17.Assa’ad AH, Putnam PE, Collins MH, et al. Pediatric patients with eosinophilic esophagitis: an 8-year follow-up. J Allergy Clin Immunol. 2007;119:731–8. doi: 10.1016/j.jaci.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 18.Prasad GA, Alexander JA, Schleck CD, et al. Epidemiology of eosinophilic esophagitis over three decades in Olmsted County, Minnesota. Clin Gastroenterol Hepatol. 2009;7:1055–61. doi: 10.1016/j.cgh.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hruz P, Straumann A, Bussmann C, et al. Escalating incidence of eosinophilic esophagitis: a 20-year prospective, population-based study in Olten County, Switzerland. J Allergy Clin Immunol. 2011;128:1349–1350. e5. doi: 10.1016/j.jaci.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Fuss IJ, Heller F, Boirivant M, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest. 2004;113:1490–7. doi: 10.1172/JCI19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umetsu DT, DeKruyff RH. Natural killer T cells are important in the pathogenesis of asthma: the many pathways to asthma. J Allergy Clin Immunol. 2010;125:975–9. doi: 10.1016/j.jaci.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajavelu P, Rayapudi M, Moffitt M, et al. Significance of para-esophageal lymph nodes in food or aeroallergen-induced iNKT cell-mediated experimental eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;302:G645–54. doi: 10.1152/ajpgi.00223.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dehlink E, Platzer B, Baker AH, et al. A soluble form of the high affinity IgE receptor, Fc-epsilon-RI, circulates in human serum. PLoS One. 2011;6:e19098. doi: 10.1371/journal.pone.0019098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yen EH, Hornick JL, Dehlink E, et al. Comparative analysis of FcεRI expression patterns in patients with eosinophilic and reflux esophagitis. J Pediatr Gastroenterol Nutr. 2010;51:584–92. doi: 10.1097/MPG.0b013e3181de7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lexmond WS, Pardo M, Rooney K, et al. Elevated levels of leukotriene C4 synthase mRNA distinguish a subpopulation of eosinophilic oesophagitis patients. Clin Exp Allergy. 2013;43:902–13. doi: 10.1111/cea.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. e6. doi: 10.1016/j.jaci.2011.02.040. quiz 21–2. [DOI] [PubMed] [Google Scholar]
- 27.Kim CH, Kunkel EJ, Boisvert J, et al. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J Clin Invest. 2001;107:595–601. doi: 10.1172/JCI11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim CH, Johnston B, Butcher EC. Trafficking machinery of NKT cells: shared and differential chemokine receptor expression among V alpha 24(+)V beta 11(+) NKT cell subsets with distinct cytokine-producing capacity. Blood. 2002;100:11–6. doi: 10.1182/blood-2001-12-0196. [DOI] [PubMed] [Google Scholar]
- 29.Koprak S, Matheravidathu S, Springer M, et al. Down-regulation of cell surface CXCR6 expression during T cell activation is predominantly mediated by calcineurin. Cell Immunol. 2003;223:1–12. doi: 10.1016/s0008-8749(03)00130-8. [DOI] [PubMed] [Google Scholar]
- 30.Vijayanand P, Seumois G, Pickard C, et al. Invariant natural killer T cells in asthma and chronic obstructive pulmonary disease. N Engl J Med. 2007;356:1410–22. doi: 10.1056/NEJMoa064691. [DOI] [PubMed] [Google Scholar]
- 31.Diegelmann J, Seiderer J, Niess J-H, et al. Expression and regulation of the chemokine CXCL16 in Crohn’s disease and models of intestinal inflammation. Inflamm Bowel Dis. 2010;16:1871–81. doi: 10.1002/ibd.21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanchard C, Wang N, Stringer KF, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanchard C, Mingler MK, McBride M, et al. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal Immunol. 2008;1:289–96. doi: 10.1038/mi.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanchard C, Mingler MK, Vicario M, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 35.Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am J Gastroenterology. 2010;105:2687–92. doi: 10.1038/ajg.2010.398. [DOI] [PubMed] [Google Scholar]
- 36.Murk W, Risnes KR, Bracken MB. Prenatal or early-life exposure to antibiotics and risk of childhood asthma: a systematic review. Pediatrics. 2011;127:1125–38. doi: 10.1542/peds.2010-2092. [DOI] [PubMed] [Google Scholar]
- 37.Jensen ET, Kappelman MD, Kim H, et al. Early life exposures as risk factors for pediatric eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2013;57:67–71. doi: 10.1097/MPG.0b013e318290d15a. [DOI] [PubMed] [Google Scholar]
- 38.Akbari O, Stock P, Meyer E, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med. 2003;9:582–8. doi: 10.1038/nm851. [DOI] [PubMed] [Google Scholar]
- 39.Lisbonne M, Diem S, de Castro Keller A, et al. Cutting edge: invariant V alpha 14 NKT cells are required for allergen-induced airway inflammation and hyperreactivity in an experimental asthma model. J Immunol. 2003;171:1637–41. doi: 10.4049/jimmunol.171.4.1637. [DOI] [PubMed] [Google Scholar]
- 40.Thomas SY, Lilly CM, Luster AD. Invariant natural killer T cells in bronchial asthma. N Engl J Med. 2006;354:2613–6. doi: 10.1056/NEJMc066189. author reply 2613– 6. [DOI] [PubMed] [Google Scholar]
- 41.Mutalithas K, Croudace J, Guillen C, et al. Bronchoalveolar lavage invariant natural killer T cells are not increased in asthma. J Allergy Clin Immunol. 2007;119:1274–6. doi: 10.1016/j.jaci.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 42.Thomas SY, Chyung YH, Luster AD. Natural killer T cells are not the predominant T cell in asthma and likely modulate, not cause, asthma. J Allergy Clin Immunol. 2010;125:980–4. doi: 10.1016/j.jaci.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spergel JM, Brown-Whitehorn TF, Cianferoni A, et al. Identification of causative foods in children with eosinophilic esophagitis treated with an elimination diet. J Allergy Clin Immunol. 2012;130:461–467. e5. doi: 10.1016/j.jaci.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 44.Spergel JM. Eosinophilic oesophagitis and pollen. Clin Exp Allergy. 2005;35:1421–2. doi: 10.1111/j.1365-2222.2005.02372.x. [DOI] [PubMed] [Google Scholar]
- 45.Kagalwalla AF, Sentongo TA, Ritz S, et al. Effect of six-food elimination diet on clinical and histologic outcomes in eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2006;4:1097–102. doi: 10.1016/j.cgh.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 46.Gonsalves N, Yang G-Y, Doerfler B, et al. Elimination diet effectively treats eosinophilic esophagitis in adults; food reintroduction identifies causative factors. Gastroenterology. 2012;142:1451–1459.e1. doi: 10.1053/j.gastro.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Onbasi K, Sin AZ, Doganavsargil B, et al. Eosinophil infiltration of the oesophageal mucosa in patients with pollen allergy during the season. Clin Exp Allergy. 2005;35:1423–31. doi: 10.1111/j.1365-2222.2005.02351.x. [DOI] [PubMed] [Google Scholar]
- 48.Fogg MI, Ruchelli E, Spergel JM. Pollen and eosinophilic esophagitis. J Allergy Clin Immunol. 2003;112:796–7. doi: 10.1016/s0091-6749(03)01715-9. [DOI] [PubMed] [Google Scholar]
- 49.Jyonouchi S, Smith CL, Saretta F, et al. Invariant natural killer T cells in children with eosinophilic esophagitis. Clin Exp Allergy. 2014;44:58–68. doi: 10.1111/cea.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agea E, Russano A, Bistoni O, et al. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202:295–308. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wingender G, Rogers P, Batzer G, et al. Invariant NKT cells are required for airway inflammation induced by environmental antigens. J Exp Med. 2011;208:1151–62. doi: 10.1084/jem.20102229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jyonouchi S, Abraham V, Orange JS, et al. Invariant natural killer T cells from children with versus without food allergy exhibit differential responsiveness to milk-derived sphingomyelin. J Allergy Clin Immunol. 2011;128:102–109. e13. doi: 10.1016/j.jaci.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mirotti L, Florsheim E, Rundqvist L, et al. Lipids are required for the development of Brazil nut allergy: the role of mouse and human iNKT cells. Allergy. 2013;68:74–83. doi: 10.1111/all.12057. [DOI] [PubMed] [Google Scholar]
- 54.Kummeling I, Stelma FF, Dagnelie PC, et al. Early life exposure to antibiotics and the subsequent development of eczema, wheeze, and allergic sensitization in the first 2 years of life: the KOALA Birth Cohort Study. Pediatrics. 2007;119:e225–31. doi: 10.1542/peds.2006-0896. [DOI] [PubMed] [Google Scholar]
- 55.An D, Oh S, Olszak T, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–33. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kummeling I, Thijs C. Reverse causation and confounding-by-indication: do they or do they not explain the association between childhood antibiotic treatment and subsequent development of respiratory illness? Clin Exp Allergy. 2008;38:1249–51. doi: 10.1111/j.1365-2222.2008.03047.x. [DOI] [PubMed] [Google Scholar]