Abstract
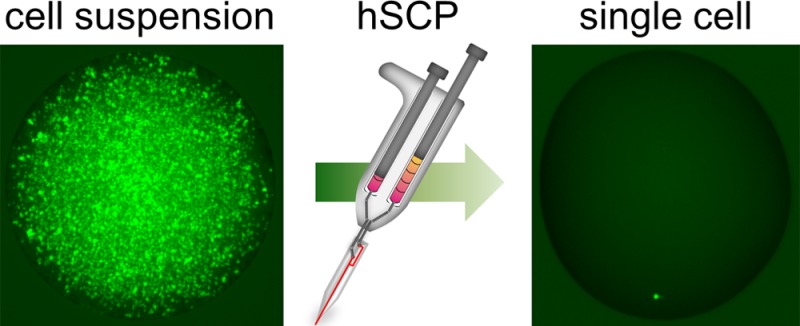
Successful single-cell isolation is a primary step for subsequent chemical and biological analyses of single cells. Conventional single-cell isolation methods often encounter operational complexity, limited efficiency, deterioration of cell viability, incompetence in the isolation of a single-cell into nanoliter liquid, and/or inability to select single adherent cells with specific phenotypes. Here, we develop a hand-held single-cell pipet (hSCP) that is rapid, operationally simple, highly efficient, and inexpensive for unbiased isolation of single viable suspended cells directly from submicroliter cell suspensions into nanoliter droplets without the assistance of any additional equipment. An integrated SCP (iSCP) has also been developed for selective isolation of single suspended and adherent cells according to the fluorescence imaging and morphological features. The isolated single cells can be conveniently transferred into standard 96-/384-well plates, Petri dishes, or vials for cloning, PCR, and other single-cell biochemical assays.
Due to cellular heterogeneity, information collected from cell populations represents averaged values, potentially masking important but rare events, such as dramatic variations in gene expression at the single-cell level.1 Molecular analyses of single cells are required to better understand the function and development of heterogeneous cell populations, including cancer cells, stem cells, immune cells, and other functional cells.2−7 The successful isolation of live single cells from these cell populations followed by maintenance of cell viability and transfer into a designated container with extremely low volume (<1 μL) is the first and key step for downstream cellular and molecular analyses including single-cell cloning,8 PCR,9 protein expression,10 and gene sequencing.11−13 Currently, four main approaches are available for single-cell isolation: serial dilution,14 micromanipulation (including mouth pipet),13,15 fluorescence-activated cell sorting,16 and laser-capture microdissection.17 These strategies are plagued by time spent, operational complexity, limited efficiency, deterioration of cell viability, incompetence in the isolation of single cells into nanoliter liquids, inability to select single adherent cells with distinct phenotypes, and/or the requirement of expensive instruments.
New methods for cell manipulation are necessary for advanced techniques in single-cell biochemical analysis.18 Recently, microfluidic technologies have been developed to handle single cells in integrated devices;4,8−10,19−24 however a direct and stand-alone approach for the isolation and transfer of single cells into standard 96-/384-well plates, Petri dishes, or vials remains unavailable. Here, we describe the development of a hand-held single-cell pipet (hSCP) that enables rapid, convenient, and highly efficient isolation of live single suspended cells directly from microliter or even nanoliter cell suspensions. After initial isolation, the hSCP conveniently transfers the single-cell droplets into designated containers (Figure 1, Movies S1, S2 (Supporting Information (SI))).
Figure 1.
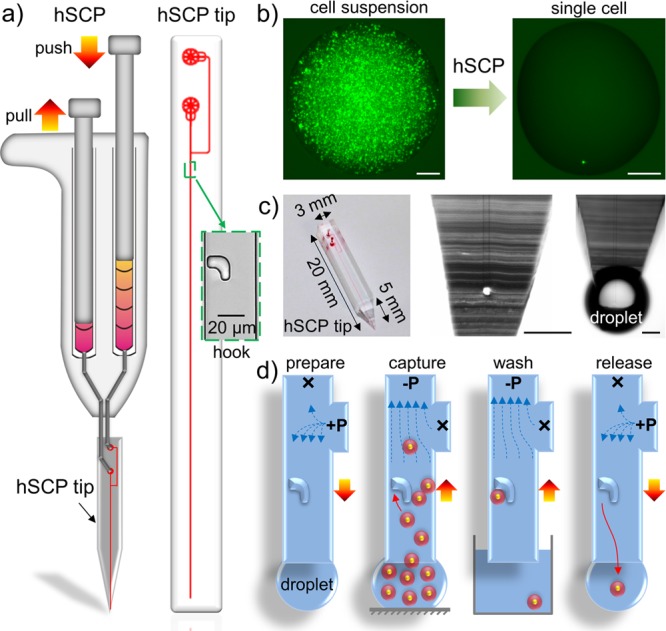
Design and mechanism of the hand-held single-cell pipet (hSCP). (a) The hSCP involves one dual-channel pipet and one hSCP tip. A magnified hook for single-cell capture is shown. (b) A single calcein-labeled SK-BR-3 cell is isolated directly from a dense cell suspension by hSCP. (c) The hSCP tip with a conical end and two magnified tip ends shown before and after extrusion of aqueous solution. (d) Work flow for single-cell isolation using hSCP. Scale bar, 200 μm.
The hSCP is a dual-channel pipet with dimensions similar to those of a hand-held air-displacement pipet (hADP; Figure S1 (SI)). The hSCP contains a negative-pressure channel (−P) operated by pulling the left knob and a positive-pressure channel (+P) operated by pushing the right knob (Figure 1a), and these channels transfer one cell during each operation (Figure 1b). The hSCP tip was designed using AutoCAD and was fabricated by photolithography and polydimethylsiloxane (PDMS) molding techniques. The tip contains two ports (−P and +P) connected to the two hSCP channels, one Y-shaped microchannel, and one microscale hook with the open end pointing to the tip end (Figure S2 (SI)). The hSCP tip end is a conical shape with a 5 mm length and an approximately 300 μm minimal width, allowing direct sampling from 500 nL cell suspensions (Figures 1c, S3 (SI)).
The hSCP-mediated cell transfer mechanism employs a procedure similar to that of hADP-mediated liquid transfer (Figure S4 (SI)) in order to minimize operational complexity and cost (<$1). Single-cell transfer is achieved in four steps (Figure 1d): preparation, capture, washing, and release and transfer. (1) Preparation. Both hSCP channels are prefilled with 200–500 μL of cell-free medium. The hSCP tip is first assembled on the hSCP by connecting the two loading ports to the −P and +P channels. A gentle push on the +P knob drives the medium into the Y-shaped microchannel and replaces the air trapped in the hSCP tip. An approximately 300 nL droplet forms on the hSCP tip and is reserved as the contact point for aspirating the cell suspension. The droplet at the hSCP tip always forms a tiny sphere shape due to the PDMS hydrophobicity. (2) Capture. A 0.5–2 μL cell suspension (106–107 cells/mL) is aspirated into the upright microchannel by pulling the −P knob on the hSCP. Because of the flowing network design, no cells flow into the side microchannel (Figure S5a (SI)), and only one cell is captured by the single hook. The size and shape of the hooks have been optimized to effectively capture single cells in the size range of 8–20 μm.19 (3) Washing. The hSCP tip is quickly dipped into cell-free medium while pulling the −P knob, which washes out the uncaptured cells to the −P port (Figure S5b (SI)). (4) Release and transfer. As PDMS is not adhesive to cells, the captured single cells are easily released into nanoliter droplets by applying a gentle pushing force to the +P port (Figure S5c (SI)). Subsequently, single-cell droplets are conveniently transferred into designated containers, such as standard 96-/384-well plates, Petri dishes, and vials. Only one pressure source is applied in each step. The entire procedure can be completed within 10 s.
Two flow paths around the hook are possible. The capture path has a narrow gap, whereas the bypass path has a wide gap (Figure S2 (SI)). Numerical simulations of the flow profile show that the flow rate along the bypass path is much greater than that along the capture path (Figure 2a). This indicates that single cells preferentially flow along the bypass path and are not captured (Figure S6 (SI)), although a single cell is occasionally captured due to the laminar-flow effect (Figure S7, Movie S3 (SI)). In most cases, single-cell capture occurs only when the bypass path is temporarily blocked by other cells (Figure S8 (SI)). A model of single-cell capture was proposed based on the principle of temporary blocking (Figure 2b). Then, various parameters were investigated to determine the optimum conditions for single-cell capture, including fluid-resistance ratios, aspiration time, and cell concentration (Figures 2c–d, S9 (SI)).
Figure 2.
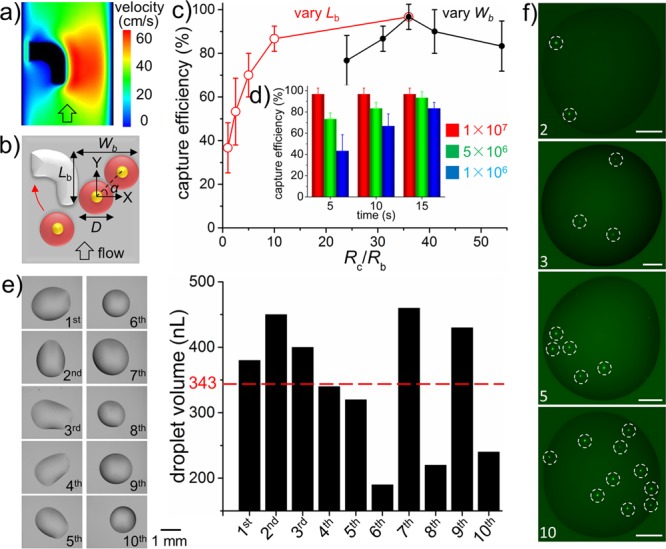
Optimization and characterization of hSCP for single-cell manipulation. (a) Numerical simulation of flow profile around the hook. (b) Model of single-cell capture by hook. Wb, width of bypass path; Lb, length of bypass path; D, diameter of cell. Single-cell capture efficiency under various fluid-resistance ratios (Rc/Rb) (c) as well as aspiration times and cell concentrations (d). Rc, fluid resistance of capture path; Rb, fluid resistance of bypass path. (e) Single-cell droplet volume evaluation. The average value (343 nL) is indicated by the red dotted line. (f) Quantitative single-cell isolation including 2, 3, 5, and 10 cells. The single cells are indicated by white dotted circles. Cells are SK-BR-3 (b–d) and MDA-MB-231/GFP (e–f). Scale bar, 200 μm.
The results of these investigations revealed that the highest capture efficiencies were achieved when the width of the bypass path (Wb) was 1.67 times the average cell diameter (D; Table S1 (SI)). Under these conditions, the included angle (α) between the x-axis and the line intersecting the nuclei of two cells was 45° (Figure 2b). With the optimum Wb, the ratio of fluid resistance along the capture path (Rc) to fluid resistance along the bypass path (Rb) was decreased by increasing the length of the bypass path (Lb) (Figure S9b (SI)). This alteration may increase the probability of cells flowing into the hook and becoming captured; however, the position of the captured single cell was not stable under the fluid force, and the cell was easily deformed and squeezed out from the hook, resulting in low capture efficiency (Table S2, Movie S4 (SI)). By contrast, captured single cells remained stable in hooks (Rc/Rb = 36) even with a flow velocity of 1 m/s. Thus, high efficiency (>95%) single-cell capture could be maintained by manual operation. Denser cell suspensions and greater aspiration times significantly increased the probability of single-cell capture (Figure 2d). Therefore, centrifugal enrichment was applied before aspiration for cell suspensions with concentrations below 106/mL. The presence of 1000 cells was considered acceptable because the hSCP can directly sample from less than 1 μL of a cell suspension. The single-cell capture efficiency was as high as 96.7% under conditions of Rc/Rb = 36, a 5 s aspiration time, and a cell suspension concentration of 107/mL. For rare cells, a pressure control system, such as a syringe pump, is recommended to replace manual operation to generate a weak and constant −P, in which case individual cells prefer to flow along the capture path due to Rc/Rb <1 while the captured single cell squeezing out of the hook is avoided. The hSCP can also combine with well-established cell enrichment methods, such as FACS and magnetic separation, for improved target single cell isolation.
After efficient capture by hooks, single cells could be easily released into nanoliter droplets by hSCP. In order to measure the precise volume, a single-cell droplet was first deposited onto a Petri dish by hSCP and then rapidly weighed by a precise electronic balance. The volume was calculated by assuming a droplet density of 1 mg/μL. A total of 10 single-cell droplets were optically imaged and weighed. All the volumes of single-cell droplets were calculated as <500 nL, and the average value was 343 nL (Figure 2e). Moreover, any defined cell numbers, such as 2, 3, 5 and 10, could easily be achieved with the hSCP attached to specially designed hSCP tips (Figures 2f, S10 (SI)). The capability to acquire a defined number of cells is useful for replacing the existing tedious and inaccurate serial dilution method for cell number-dependent research, such as validation of gene amplification linearity and testing of circulating tumor cell capture efficiency.25
The hSCP is compatible with commercial tissue culture plates, as demonstrated by the distribution of single cells into a 4 × 4 array region in a 384-well plate (Figure S11 (SI)). The viability of seeded single cells is high because the fluid shear stress around the captured cells is maintained in a low range (Figure S12 (SI)). The hSCP was first used for the distribution of single cells into 96-well plates for cloning analysis. After seeding, the single MDA-MB-231/GFP cells proliferated normally (Figure S13 (SI)). Subsequently, 50 single MDA-MB-231/GFP cells were plated in the wells of a 384-well plate filled with 80 μL of fresh medium in advance, and cell counts at various time points were determined (Figure 3a). Cell heterogeneity was an obvious factor in single-cell cloning. Interestingly, 18% of the single cells did not divide during this period or died and 10% of them divided only once while other cells had average doubling times8 of 28 h. The minimum doubling time was 20 h, and the maximum doubling time was 76 h (Figure 3b). Such a cell-doubling assay confirmed heterogeneity in cell proliferation. Note that clonal growth of a single cell can be promoted by using conditioned medium containing cytokines produced by the same cell population to replace fresh medium.
Figure 3.
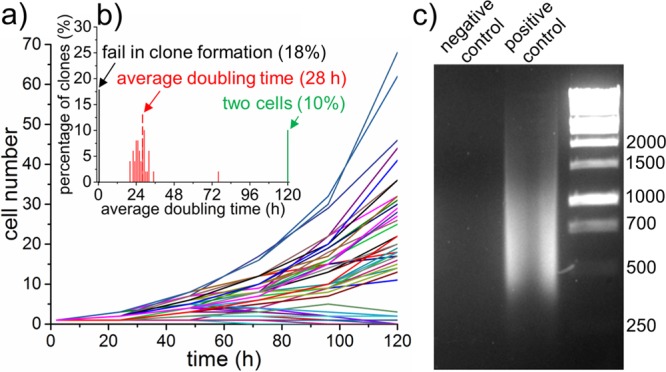
Single-cell cloning and PCR. (a) Cell numbers per clone versus time. (b) Distribution of average doubling time of clonal cells. Only cell numbers over 2 are calculated. (c) Analysis of 2% whole-genome DNA amplification from a single cell after 25 cycles. MDA-MB-231/GFP cells were used.
The hSCP was also successful for application in single-cell PCR analysis, in which single cells require isolation and lysis in the smallest possible volume. As a proof-of-concept experiment, one SUM 159 cell was first captured, then released into a nanoliter droplet, and finally transferred into a PCR vial by hSCP. After single-cell lysis, DNA fragmentation, and DNA amplification, the quality of the whole-genome amplification was determined by separation of 2% of the PCR product volume onto a 3% agarose gel (Figure 3c). The negative control (no cell) lacked visible bands while the positive control (single cell isolated by hSCP) migrated as a wide band of DNA primarily ranging from 300 to 1500 bp. The PCR products of the genomic DNA can be used for downstream applications such as sequencing and single nucleotide polymorphism analysis.
The hSCP provides unbiased isolation of single suspended cells and can be combined with statistical downstream single-cell analysis for cell heterogeneity studies. In other situations, selective isolation and analysis of single suspended and adherent cells10,26 via fluorescence imaging and morphological observation are required, and these two needs can be simultaneously met by integrated SCP (iSCP; Figure 4). In this setup, the 50-plex iSCP tips are evenly distributed into a round PDMS slab and share the same cell inlet/outlet (Figures 4a, S14 (SI)). The PDMS slab is placed onto a Petri dish or a glass slide without thermal or oxygen plasma treatment, allowing the captured single suspended cells to immediately image and spread for subsequent morphological phenotype identification. The work flow of selective iSCP is similar to the operation of the unbiased hSCP (Figures 4b, S15 (SI)). The main difference is that the captured single cells were allowed to adhere and spread normally within the iSCP tips. Cells can be morphologically heterogeneous, and morphological features of single cells can be easily identified under a microscope. After selection, the target single adherent cell is released into the cell outlet using trypsin loaded into the port connected to the bypass microchannel followed by transfer into a designated container via hADP (Figure 4c). The iSCP is easy to operate and can achieve close to 100% isolation of 50 single suspended cells within 1 min. After approximately 3 h in culture, the thin and long membrane protrusions of 50 single cells can be clearly identified under a fluorescence microscope (Figures 4d, S16 (SI)).
Figure 4.
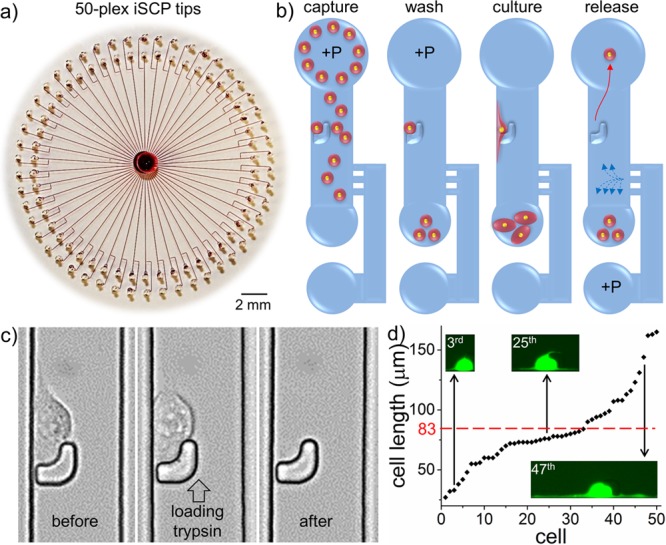
Design and mechanism of the integrated single-cell pipet (iSCP). (a) A rounded PDMS slab containing 50-plex iSCP tips was placed on a commercially available polystyrene Petri dish; red dye was injected to aid visualization. (b) Work flow of iSCP for selective isolation of single suspended and adherent cells. (c) Sequential images of single adherent cell release. After 3 h in culture, the single MDA-MB-231 cell in the 32nd iSCP tip displayed obvious adherent morphology and was then released by loading trypsin solution into the port connected to the bypass microchannel. (d) A plot of the lengths of 50 MDA-MB-231/GFP cells after 3 h in culture within the 50-plex iSCP-tips. The minimum cell length is 27 μm, and the maximum cell length is 165 μm. Average cell length (83 μm) is indicated by the red dotted line.
We recently found that breast cancer cells with the longest protrusions often exhibit the strongest metastatic and invasive capabilities.19 Therefore, the iSCP may be used to selectively isolate distinct single adherent cells according to their protrusion generation ability and used for downstream gene analyses, which may facilitate the study of genotype–phenotype relationships.10,27,28 In order to improve the quality of statistical data, integration of 100 hSCPs into one iSCP might be achieved by further optimizing the chip design. Moreover, the captured single cells can proliferate normally within the iSCP tips and can be utilized for long-term single-cell tracking assays (Figure S17 (SI)). Finally, a comprehensive comparison of h/iSCP with conventional single-cell isolation methods is presented (Table S3 (SI)).
In summary, we describe an hSCP for unbiased isolation of single suspended cells directly from cell suspensions, and this novel method has key features of speed, high efficiency, maintenance of cell viability, and low cost. This direct and stand-alone approach can be easily operated with one hand without the assistance of any additional equipment. The hSCP has the ability to sample directly from submicroliter cell suspensions and release the captured single cells into droplets smaller than 500 nL. The extremely small final single-cell suspension volumes are ideal microreactors for performing chemical and biological analyses of single cells. Furthermore, any defined cell number is attainable using an hSCP attached with specially designed hSCP tips, potentially providing an approach for quantitative cell analysis. In addition to the unbiased isolation of single suspended cells, selective isolation of single suspended or adherent cells can also be achieved via an iSCP. These methods provide novel tools for future biochemical and biological research, wherein simple and efficient transportation of single or defined numbers of suspended or adherent cells is required.
Acknowledgments
We are grateful for funding support from CPRIT-R1007, NIH-R01CA180083, NIH-U54CA143837, NIH-R01DA035868, and the Golfers against Cancer Foundation.
Supporting Information Available
Experimental details and additional information. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Almendro V.; Marusyk A.; Polyak K. Annu. Rev. Pathol-Mech. 2013, 8, 277. [DOI] [PubMed] [Google Scholar]
- Meacham C. E.; Morrison S. J. Nature 2013, 501, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P.; Daley G. Q. Nat. Rev. Mol. Cell Biol. 2013, 14, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.; Fan R.; Ahmad H.; Shi Q.; Comin-Anduix B.; Chodon T.; Koya R. C.; Liu C. C.; Kwong G. A.; Radu C. G.; Ribas A.; Heath J. R. Nat. Med. 2011, 17, 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov A. K.; Radhakrishnan A.; Tsao D. S.; Bookwalter C. S.; Trybus K. M.; Diehl M. R. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, E334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K.; Zhong G. S.; Zhuang X. W. Science 2013, 339, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P. W.; Hwang K. V.; Lan T.; Lu Y. J. Am. Chem. Soc. 2013, 135, 5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecault V.; Vaninsberghe M.; Sekulovic S.; Knapp D. J.; Wohrer S.; Bowden W.; Viel F.; McLaughlin T.; Jarandehei A.; Miller M.; Falconnet D.; White A. K.; Kent D. G.; Copley M. R.; Taghipour F.; Eaves C. J.; Humphries R. K.; Piret J. M.; Hansen C. L. Nat. Methods 2011, 8, 581. [DOI] [PubMed] [Google Scholar]
- Sanchez-Freire V.; Ebert A. D.; Kalisky T.; Quake S. R.; Wu J. C. Nat. Protoc. 2012, 7, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A.; Kolitz S.; Lauffenburger D. A.; Han J. Nat. Commun. 2014, 5, 3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey P. C.; Quake S. R. Nat. Methods 2014, 11, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gole J.; Gore A.; Richards A.; Chiu Y. J.; Fung H. L.; Bushman D.; Chiang H. I.; Chun J.; Lo Y. H.; Zhang K. Nat. Biotechnol. 2013, 31, 1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong C.; Lu S.; Chapman A. R.; Xie X. S. Science 2012, 338, 1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma J. G.; Akhavan M.; Fernandes K. J.; Barnabe-Heider F.; Sadikot A.; Kaplan D. R.; Miller F. D. Nat. Cell Biol. 2001, 3, 778. [DOI] [PubMed] [Google Scholar]
- Kurimoto K.; Yabuta Y.; Ohinata Y.; Saitou M. Nat. Protoc. 2007, 2, 739. [DOI] [PubMed] [Google Scholar]
- Quintana E.; Shackleton M.; Sabel M. S.; Fullen D. R.; Johnson T. M.; Morrison S. J. Nature 2008, 456, 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espina V.; Milia J.; Wu G.; Cowherd S.; Liotta L. A. Methods Mol. Biol. 2006, 319, 213. [DOI] [PubMed] [Google Scholar]
- Trouillon R.; Passarelli M. K.; Wang J.; Kurczy M. E.; Ewing A. G. Anal. Chem. 2013, 85, 522. [DOI] [PubMed] [Google Scholar]
- Zhang K.; Chou C. K.; Xia X.; Hung M. C.; Qin L. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainla A.; Jansson E. T.; Stepanyants N.; Orwar O.; Jesorka A. Anal. Chem. 2010, 82, 4529. [DOI] [PubMed] [Google Scholar]
- Rowat A. C.; Bird J. C.; Agresti J. J.; Rando O. J.; Weitz D. A. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 18149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. U.; Olguin L. F.; Whyte G.; Scott D.; Babtie A.; Abell C.; Huck W. T. S.; Hollfelder F. J. Am. Chem. Soc. 2009, 131, 15251. [DOI] [PubMed] [Google Scholar]
- Borland L. M.; Kottegoda S.; Phillips K. S.; Allbritton N. L. Annu. Rev. Anal. Chem. 2008, 1, 191. [DOI] [PubMed] [Google Scholar]
- Lu Y.; Chen J. J.; Mu L. Y.; Xue Q.; Wu Y.; Wu P. H.; Li J.; Vortmeyer A. O.; Miller-Jensen K.; Wirtz D.; Fan R. Anal. Chem. 2013, 85, 2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagrath S.; Sequist L. V.; Maheswaran S.; Bell D. W.; Irimia D.; Ulkus L.; Smith M. R.; Kwak E. L.; Digumarthy S.; Muzikansky A.; Ryan P.; Balis U. J.; Tompkins R. G.; Haber D. A.; Toner M. Nature 2007, 450, 1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G. T.; Wang Y.; Young G.; Bachman M.; Sims C. E.; Li G. P.; Allbritton N. L. Anal. Chem. 2007, 79, 682. [DOI] [PubMed] [Google Scholar]
- Fu J.; Wang Y. K.; Yang M. T.; Desai R. A.; Yu X.; Liu Z.; Chen C. S. Nat. Methods 2010, 7, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giam L. R.; Massich M. D.; Hao L. L.; Wong L. S.; Mader C. C.; Mirkin C. A. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


