Abstract
Ionizing radiation exposure induces highly lethal DNA double-strand breaks (DSBs) in all phases of the cell cycle. After DSBs are detected by the cellular machinery, these breaks are repaired by either of two mechanisms: (1) nonhomologous end joining (NHEJ), which re-ligates the broken ends of the DNA and (2) homologous recombination (HR), that makes use of an undamaged identical DNA sequence as a template to maintain the fidelity of DNA repair. DNA DSB repair must occur within the context of the natural cellular DNA structure. Among the major factors influencing DNA organization are specific histone and nonhistone proteins that form chromatin. The overall chromatin structure regulates DNA damage responses since chromatin status can impede DNA damage site access by repair proteins. During the process of DNA DSB repair, several chromatin alterations are required to sense damage and facilitate accessibility of the repair machinery. The DNA DSB response is also facilitated by hierarchical signaling networks that orchestrate chromatin structural changes that may coordinate cell-cycle checkpoints involving multiple enzymatic activities to repair broken DNA ends. During DNA damage sensing and repair, histones undergo posttranslational modifications (PTMs) including phosphorylation, acetylation, methylation and ubiquitylation. Such histone modifications represent a histone code that directs the recruitment of proteins involved in DNA damage sensing and repair processes. In this review, we summarize histone modifications that occur during DNA DSB repair processes.
INTRODUCTION
Ionizing radiation (IR) induces many types of DNA damage but the most lethal lesions are DNA double-strand breaks (DSBs), the repair of which is critical for cell survival. One of the major challenges of radiotherapy related repair research is the lack of mechanistic and structural details about DNA damage repair as it occurs in the natural context of the cell, i.e., within chromatin. Chromatin, the physiological packaging structure of histones and DNA, is now gaining appreciation as a relevant regulator of multiple signaling pathways (1, 2). The spatial and temporal control of DSB repair may be critically dependent on histone modifiers and specific histone modifications (3-6) as many reports link chromatin structure to IR sensitivity (7-20). It has been suggested that the packing and accessibility of DNA in chromatin are major factors influencing IR sensitivity (14). Consistent with this proposition is that cells derived from individuals with ataxia-telangiectasia are radiation sensitive and have a higher rate of conversion of DNA DSBs into chromosome breaks postirradiation (17, 18, 20). Cells deficient for ATM have an increased frequency of chromosome aberrations that has been attributed to an altered chromatin status is observed at both the global chromatin level as well as specifically in telomeric chromatin (16, 21-24). It has also been reported that DNA damage renders chromatin more sensitive to micrococal nuclease digestion (25) and leads to chromatin decondensation at the local as well as global level (26, 27). Such chromatin structural alterations are an essential requirement for activation of the DNA damage response (DDR) and subsequent DSB repair. Recent studies have begun to reveal that eukaryotic cells orchestrate a complex array of responses to sense DNA damage (28) and these responses during DNA repair specifically involve chromatin structure alterations. This review will summarize the status of histone modifications in relationship to modulation of the DDR and DSB repair.
Chromatin Structure
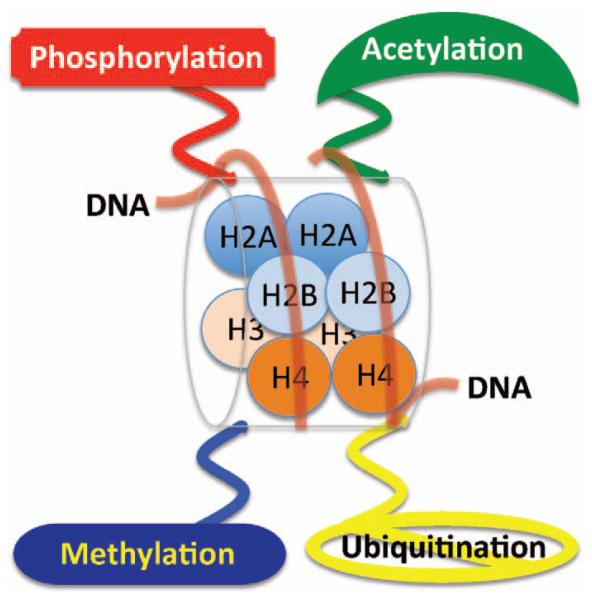
The basic repeating unit of chromatin, the nucleosome, consists of approximately 146 bp of DNA wound around an octamer of histone proteins that includes two molecules each of histones H3, H4, H2A and H2B (Fig. 1) (29). Histone-DNA interactions within the nucleosomes can be transiently broken by ATP-dependent nucleosome remodelers (30) and such histone-DNA interactions are regulated by enzymes through acetylation, methylation, phosphorylation and ubiquitination of specific amino acid residues (Fig. 2). Histone post-translational modifications usually function to recruit specific proteins to chromatin, the identity of which is determined by the type of post-translational modifications and the specific histone residue that is modified (31). Damaged DNA can be fully exposed by total removal of histones from DNA, a process mediated by histone chaperones (32). Thus histone modifications can influence DNA damage responses during signaling as well as during the opening and restoration of chromatin to its original native state. In addition to the post-translational modifications occurring after DNA damage, the initial state of histone modifications prior to DNA DSBs induction can also influence the DDR.
FIG. 1.
Structure of eukaryotic nucleosome consisting of DNA wound in sequence around four histone protein cores with covalent modifications of phosphorylation, acetylation, methylation and ubiquitination.
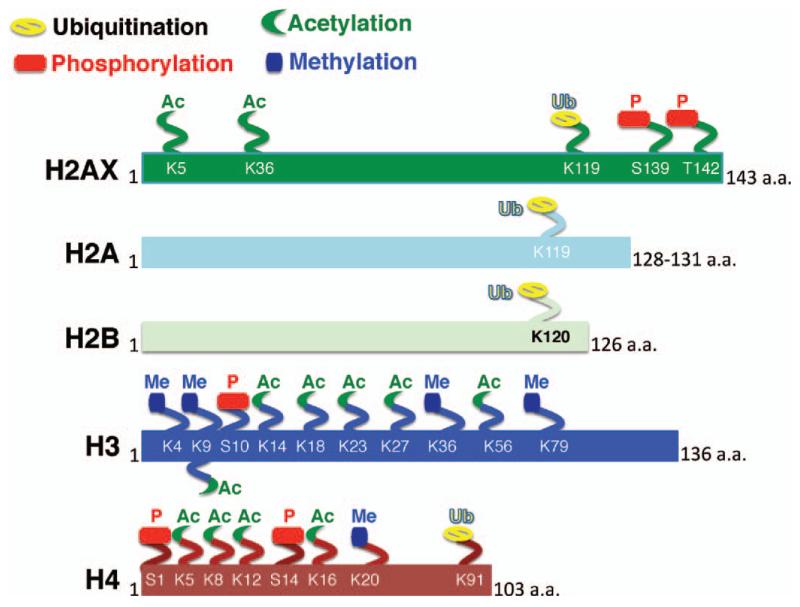
FIG. 2.
Major types of histone modification of specific histone residues linked with the DNA damage response (DDR) and DSB repair.
Role of Pre-Existing Histone Modifications in DDR
Histone modifications have been shown to play a primary role in DDR by facilitating repair protein access to DNA breaks (20, 33-35). Several studies indicate that pre-existing histone modifications play an important role in DDR. For example, repair proteins (53BP1, Schizosaccharomyces pombe Crb2 [SpCrb2] and Saccharomyces cerevisiae Rad9 [ScRad9]) require methylated histone H3 Lys79 (H3K79) (36) or methylated histone H4 Lys20 (H4K20) and/or CBP/p300-mediated acetylation of histone H3 lysine 56 (H3K56) (36-40) for focus formation at DNA-damage sites. All three of these histone modifications are normally present on chromatin and do not change in response to IR-induced DNA damage. Most histone acetylation modifications do not change appreciably after genotoxic stress, however, some studies have indicated that histone H3 acetylated at K9 (H3K9ac) and H3K56ac are rapidly deacetylated in response to DNA damage in human cells (41). Whether preexisting modifications can influence DNA damage responses at the level of signaling, chromatin relaxation or opening and recruitment of repair proteins, and the importance of restoring chromatin to the original native state has just begun to be recognized. In human cells, histone modifications such as H2AX S139 (phosphorylation), H4K16 (acetylation), H3K79 and H4K20 (methylation) have been linked with damage signaling; H3K9, H4K16 and H2A(X) are linked with chromatin opening and modifications like H4S1 and H2B S14 (phosphorylation), H2AX S139 (dephosphorylation), H3 K14, K23, K56 and H4 K5, K8, K12, K16, K91 (acetylation), H3/H4 Ks (deactylation), and H2A K119 (mono-ubiquitination) have been associated with chromatin restoration (42). The following is an example of how a pre-existing histone modification impacts DNA damage responses and DSB repair after irradiation. The amino-terminal tail of histone H4 is a well-described target for posttranslational modification, including acetylation (4, 19, 82). Transcriptionally active gene regions have histones that are post-transcriptionally modified and these regions are thought to have preferential DNA DSB repair. In support of this, studies in yeast of HO endonuclease induced DSBs generated at the ADH1 and MAT-loci have shown that DNA repair is coupled to transcription. Faster DSB repair at the highly active ADH1 locus compared to the nearly silent MAT locus suggests preferential DSB repair at active genes in vivo (43).
Direct evidence linking histone modification and transcription comes from the examination of males absent on the first (MOF) gene function, which is a highly conserved histone acetyl-transferase that specifically acetylates histone H4 at K16 (44). Acetylation of H4K16 is prevalent in Drosophila on the hyperactive male Drosophila polytene X chromosomes (45), where it contributes to transcriptional upregulation (46) although in yeast, H4K16ac does not correlate with active genes (47). However, all K5, K8 and K12 acetylation marks on histone H4 are linked with enhanced transcription (48). Among the 4 histone H4 acetylation sites, the H4K16ac modification poses a structural constraint on the formation of higher-order chromatin (49). Characterization of histone H4K16 acetylation function in mammalian cells by generating mutations has been difficult but both chemical and genetic approaches have validated its role in the DNA damage response where it potentially serves as a platform structure to generate proper signaling for DDR (50). MOF, as well as H4K16 acetylation, localizes on the X chromosome of male Drosophila, which has almost a twofold greater transcription level when compared to the X chromosomes of female Drosophila. Depletion of MOF in human cells reduced H4K16ac levels and such cells have reduced levels of ATM activation (51) as well as defective appearance of postirradiation γ-H2AX foci (50). Additional studies using either the deacetylase inhibitor trichostatin A or deletion of the SirT2 gene have also confirmed that H4K16ac levels are critical for a timely DDR. Furthermore, Sharma et al. (50) observed a relationship between H4K16ac levels and DNA damage responses in differentiated HL60 cells where reduced H4K16ac levels and a decreased frequency of γ-H2AX foci per cell following irradiation are observed, supporting the concept that the initial levels of H4K16ac are critical for sensing the DNA damage.
In addition to histone acetylation, chromatin contains other histone modifications that could impact DDR. The best studied histone modifications that occur post IR exposure or after induction of DNA DSBs are listed in Fig. 2 and Table 1. Histone modifications have been detected on serine/threonine residues as phosphorylation, on lysine as acetylation, methylation or ubiquitination modifications, and on arginine as methylation. Except for ubiquitination, these modifications alter histone/DNA electrostatic interactions and ultimately change chromatin dynamics and function by altering access of cellular factors involved in DDR. How histone modifications (Fig. 3) occur and influence repair of DSBs are summarized in the next section.
TABLE 1.
| Histone | Residue | Modifier | Role in DDR | References |
|---|---|---|---|---|
| Acetylation | ||||
| H2AX | K5 | TIP60 | Helps in K119ub of H2AX and removal of γ-H2AX |
(104,105) |
| H2AX | K36 | CBP/p300 | Recruits Ku during NHEJ | (88) |
| H3 | K9, K14, K23, K27 | GCN5, CBP/p300 | Recruits SW1/SNF, promotes spreading of γ- H2AX domain |
(76) |
| H3 | K18 | GCN5, CBP/p300 | Regulates Ku protein recruitment during DSB repair |
(75) |
| H3 | K56 | GCN5, CBP/p300 RTT109 | Depletes after IR to promote NHEJ, enriches K56ac after HR repair |
(106) |
| H4 | K5, K8, K12, K16 | MOF, TIP60, TRRAP, CBP/p300 |
Recruits DDR/repair proteins and SW1/SNF nucleosome remodeling complex |
(50, 51, 67) |
| Phosphorylation | ||||
| H2AX | S139 | ATM, ATR, DNA-PKcs | Recruits and accumulates DDR proteins (MDC1) to the repair lesion and promotes histone acetylation. |
(52, 64, 99, 107, 108) |
| H2AX | T142 | WSTF | Recruits activated ATM and MDC1. | (109,110) |
| H4 | S1 | CK2 | Role in DDR | (60, 61) |
| H4 | S14 | CK2 | Promotes NHEJ by 53BP1 recruitment and methylation of V(D) J recombination via RAG complex |
(111) |
| Methylation | ||||
| H3 | K4me3 | SET1 | Stimulates V(D) J recombination via RAG complex |
(112,113) |
| H3 | K9me3 | Suv3-9H1/Suv3-9H2 | Interacts with HP1β, phosphorylates damage induced HP1β and activates TIP60 |
(114, 115) |
| H3 | K36me2 | Metnase/SETMAR | Accumulates NBS1 and Ku to stimulate NHEJ |
(116,117) |
| H3 | K79me3 | DOT1 | Recruits RAD9 in budding yeast | (118) |
| H4 | K20me2 | Suv420H1/Suv420H2, MMSET |
Recruits DDR and repair proteins | (39, 101, 119) |
| Ubiquitination | ||||
| H2AX | K119ub/ K119ub2, K119poly-ub |
RNF8, RNF168, TIP60-UBC13 |
Recruits DDR proteins to the repair lesion | (84, 120, 121) |
| H2A | K119ub/ K119ub2, K119poly-ub |
RNF8, RNF168 | Accumulates BRAC1 and 53BP1 for DNA repair |
(105) |
| H2B | K120ub | RNF20-RNF40 | Recruits XRCC4 and Ku for NHEJ and BRCA1 and RAD51 FOR HR |
(111) |
| H4 | K91ub | BBAP | Induces H4K20me modification and recruits 53BP1 to promote NHEJ |
(93) |
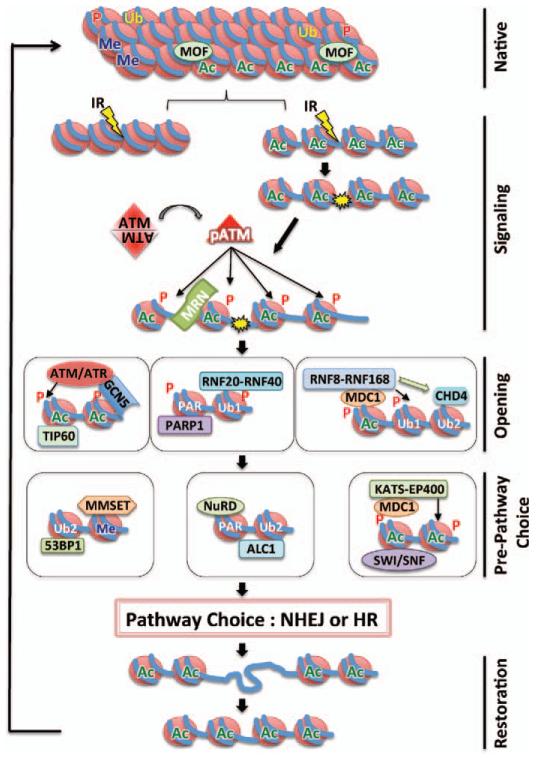
FIG. 3.
Role of histone modifications in response to ionizing radiation induced DSB sites. As described in the text, in response to IR DNA DSB induction, the histone modifications facilitate the relaxation of chromatin structure that triggers ATM kinase activation that phosphorylates H2AX enabling recruitment of MDC1 and other proteins. Histones (red) are surrounded by DNA (blue). Subsequent histone modifications, and the associated modifiers involved in sensing the damage and restoration of the native structure are also shown.
Histone Phosphorylation
One of the first histone modification events linked with the DNA damage responses is H2A phosphorylation (52), specifically the H2A variant H2AX (phosphorylated H2AX is referred as γ-H2AX), which occurs within minutes after exposure to ionizing radiation. H2AX phosphorylation is carried out mostly by ATM or ATR and is thought to modify higher order chromatin structure at the DNA damage site. Phosphorylation of H2AX is critical for signaling/repair protein recruitment to DNA damage sites since IR induced formation of γ-H2AX foci is rapid, precedes repair factor assembly into repairosome foci and is required for subsequent foci formation by 53BP1, NBS1, BRCA1 and MDC1 [reviewed in ref. (53)] (Fig. 3). Loss of H2AX increases genomic instability, possibly because γ-H2AX physically interacts with NBS1, 53BP1 and MDC1, further supporting the critical role of γ-H2AX in DDR [reviewed in ref. (53)]. H2AX phosphorylation occurs in all phases of the cell cycle, consistent with IR-induced ATM activation occurring in all phases of the cell cycle (54). Thus H2AX phosphorylation is required for both DNA DSBrepair pathways e.g., NHEJ and HR (Fig. 3). For example, phosphorylation of H2AX facilitates damage site recruitment of the DDR component MDC1, during HR, which binds to γ-H2AX via its BRCT domain (55). γ-H2AX-recruited MDC1 is phosphorylated by ATM and also by casein kinase 2 (56, 57). Furthermore, binding of MDC1 to γ-H2AX is modulated by MOF dependent H4K16 acetylation (58). As shown in Fig. 3, H2AX phosphorylation facilitates the recruitment of SW1/SNF and RSC remodeling complexes and several other repair proteins including RAD16, CSB/RAD26, RAD5 and RAD54 that belong to the SWI2/SNF2 family of helicases involved in DNA repair (4). As summarized in Table 1, H2B phosphorylation at Ser14 and N-terminal phosphorylation of H4 also occur as part of the DDR (59). Both of these histones are abundant, present in close proximity to double-strand breaks (60, 61), and are modified by sterile 20 kinase (Mst1) (62) and casein kinase 2 (60, 61). Most of phosphorylation events occurring on histone H2AX and H2A have been linked with the sensing and opening of the DNA damage sites while H4 modification has been linked with chromatin condensation.
Histone Acetylation
The second most common histone modification linked with both transcription and DNA damage repair is histone acetylation. Acetylation neutralizes positively charged lysine residues, thus altering chromatin fiber intra- and internucleosomal interactions to facilitate chromatin decondensation and enhance access to nucleosomal DNA (63, 64). The major acetylated histone residues, the modifying enzymes and the function of the histone modification are summarized in Table 1 and Fig. 3. Acetylation is a dynamic histone marker regulated by the balance between histone acetyltransferases (HATs) and histone deacetylases (HDACs). HATs and HDACs transfer an acetyl moiety from acetyl-coenzyme A to the ε-amino group of lysine and remove the acetyl-group, respectively (65). Histone acetyl transferases are of two types, type-A acetylate histones in the nucleus whereas type-B enzymes acetylate cytoplasmic histones. HATs of type-A are responsible for chromatin dynamics in the nucleus and belong to GNAT superfamily and MYST family. Individual well-characterized HATs are MOF, TIP60, GCN5, KATS-EP400, SW1/SNF and p300/CBP. Histone H3 and H4 acetylation increases interactions with transcriptional machinery components containing bromodomains, H3 lysine 9 methylation allows heterochromatin protein 1 (HP1) binding via the chromodomain to chromatin, thereby blocking DNA binding of the transcription machinery. Acetylation at the H3 and H4 N termini and at H3K56 plays a critical role in DNA metabolism involving DNA replication, genomic stability and in the binding of the chromatin assembly factor (CAF1)-PCNA complex (66). The N-terminal tail of histone H4 is acetylated at K5, K8, K12 and K16, and H4K16ac is observed on the Drosophila male X chromosome (45) in relationship to control of gene dosage compensation and has also been implicated in the chromatin structure responsible for interaction of other proteins (49). The HAT responsible for histone H4 acetylation at K16 is MOF (50, 51, 67, 68) and acetylation at H4K16 has been implicated in the proper compaction of chromatin 30-nm fibers (49). Interestingly, MOF depletion also influences ATM functions (51) like delayed appearance of IR-induced γ-H2AX foci (50), thus providing more direct evidence for the role of histone modifications in the activation and function of DDR components. Since histone H4K16 acetylation levels correlate with ATM activation, HDAC inhibitor treatment results in global ATM activation even in the absence of DNA damage (69), supporting the argument that the histone modifications direct regulatory pathways sensing and processing DNA damage for repair. The current understanding of histone acetylation is still rudimentary and the field would be advanced if the acetylated residues could be mutated for analysis of their functional role in DDR.
Histones H3 and H4 are reportedly acetylated on the N-terminal lysines during repair by the NHEJ pathway [reviewed in ref. (3)]. At DSBs, histone H4 is acetylated by NuA4-TIP60 and this modification has been linked with improved DSB repair (70, 71). Similarly, pre-existing H4K16ac, prior to IR exposure, influences repair as cells with decreased levels of H4K16ac have a reduced efficiency of DNA DSB repair by NHEJ (50). For recruitment of Ku70/80 at DNA DSB sites, the necessary histone modifications are carried out by the INO80, SWR1 and RSC complexes (72, 73). At DSBs, the RSC complex recruits Mre11, which is followed by ATPase-dependent remodeling to facilitate access to the site by proteins required for NHEJ mediated repair (74). Histone H3 and H4 acetylation by CBP and p300 cooperate with the SW1/SNF complex to facilitate recruitment of NHEJ proteins such as Ku70/80 (75). Thus histone acetylation occurs during the initial stages of NHEJ to facilitate the chromatin opening and subsequent access of repair proteins to the DNA DSB sites. During HR mediated repair, a number of acetylation events occur on histones H3 and H4 with the proteins implicated in the modification being GCN5, NuA4 and HAT1 (71, 76). GCN5 also participates in pathway choice for DSB repair as DNA-PKcs phosphorylates GCN5 to inactivate its HAT domain. In addition, GCN5 also interacts with BRCA1 through a mechanism that is dependent upon its HAT activity implicating a role in HR repair of DNA DSBs (77).
Histone Ubiquitination
Similar to highly conserved DNA damage repair process, ubiquitylation is also a tightly regulated process involving the enzymatic activity of E1, E2 and E3 (78, 79). Ubiquitylation is among the more unique forms of post-translational modification in that a single ubiquitin monomer can be polyubiquitylated through one of seven lysines or through the amino terminus to create polyubiquitin chains. Ubiquitination conjugates a 76 amino acid protein, ubiquitin, to the lysine ε amino group of specific proteins as a result of which ubiquitin molecule acts to regulate protein function and stability by altering the activity of its target in a variety of ways, e.g., changing its localization or enzymatic activity to targeting it for degradation. For example, ubiquitination of H2A at K119 by the E3 ubiquitin-protein ligase RNF2 or RING2 is associated with transcriptional repression via subsequent binding of the polycomb repressive complex (80, 81) (Fig. 3) and is stimulated by RING finger domain containing proteins such as BMI-1 and RINGIA (82). In addition, BMI-1, RINGIA and RINGIB are also involved in DSB-associated H2A ubiquitination (83). Ubiquitination of nuclear histones occurs after irradiation by RNF8 and RNF168, which catalyze formation of lysine 63 linked polyubiquitination chains on histones H2A and H2AX (84, 85). As shown in Fig. 3, RNF8 is rapidly recruited to the sites of DNA damage in an MDC1-dependent manner through its functional FHA domain and RNF8 is required to recruit subsequent repair factors (86, 87). Although polyubiquitination generally results in protein degradation, RNF8 catalyzed ubiquitin modification does not because RNF8/UBC 13-mediated polyubiquitin synthesis produces a lysine-63 linkage, rather than the lysine-48 canonical signal for protein degradation. While RNF8 mediated ubiquitination has a role in maintaining genomic integrity, the role of postdamage monoubiquitylation in chromatin reassembly remains unclear (88). Another important ubiquitination event during DDR is performed by BRCA1 E3 ligase activity, which promotes BRCA2 recruitment that in turn promotes RAD51 recruitment during DNA strand resection in HR repair (89). BRCA1 is localized to DSBs through an interaction with a repair protein complex containing RAP80, a protein that contains tandem ubiquitin interaction motifs (90-93). The domains of ubiquitin interaction motifs recognize K63-linked ubiquitin over K48-linked structures, suggesting that BRCA1 modification would be functioning in nondegradative ubiquitin roles (92). Recent studies have begun to reveal that specific ubiquitination markers are linked with specific DSB repair pathways. The UBC13 E3 ligase seems to be required for homologous recombination because cells lacking UBC13 are defective in break resection as determined by RPA recruitment to DNA DSBs (94). These observations suggest a DSB-associated ubiquitin requirement during the initial stages of repair. The major histone sites of ubiquitination (95), the enzymes required and the role of the modification in the DNA damage response are summarized in Table 1 and Fig. 3.
Histone Methylation
More than forty years ago, histone methylation on lysine and arginine (96-99) was discovered (100), but, in contrast to acetylation, histone methylation does not alter the charge of arginine and lysine residues. Thus methylation does not directly modulate the nucleosomal interactions required for chromatin restructuring. Histone methylation is crucial for proper programming of the genome during development but the recent identification of histone demethylases such as LSD1/AOF2, JMJD1, JMJD2 and JHDM1 has also indicated methylation is reversible and also provides a rationale for a role in DNA DSB repair. Specific methylation DNA sites linked to the DNA damage response are summarized in Table 1. Methylations at multiple sites on H3 and H4 have been reported (mono- di- and trimethyl groups per residue) including K4, K9, K27, K36, K79 and R2, R8, R17, R26 for H3 and K20 and R3 for H4. The source of the methyl group required by histone methyl transferases is S-adenosyl-methionine. SET domain containing proteins (Dm Su(var)3-9), Enhancer of zeste (E(z)) and trithorax (trx) methylate lysine, while methylation of arginine is performed by coactivator arginine methyltransferase (CARM1) and arginine methyl transferase (PRMT1). In contrast to histone acetylated site detection by bromo-domain containing proteins, methylated sites are detected by proteins containing a chromodomain motif. In mammals, dimethylation of histone H4 lysine 20 (H4K20me2), is mediated by the histone methyltransferase MMSET (also known as NSD2 or WHSC1) (101), which does not seem to increase globally after DNA damage, however, it is critical for recruitment of 53BP1 at DSBs (101) and increases in vicinity to DSBs. Interestingly, MMSET depletion significantly decreases H4K20 methylation at DSBs as well as 53BP1 accumulation at DSBs. It has been shown that MMSET recruitment to DSBs requires γ-H2AX-MDC1 and involves the interaction between the MDC1 BRCT domain and phosphorylated Ser102 of MMSET (101). These observations suggest a pathway involving γ-H2AX-MDC1-MMSET regulates the induction of H4K20 methylation on histones around DSBs, which, then facilitates 53BP1 recruitment. Pre-existing chromatin modifications at the histone level can directly affect the initial DNA damage response and subsequent pathway choice for NHEJ and HR. Modifications such as H4K16ac or H4K20me2, regulated by MOF and MMSET respectively, are epigenetic markers that could act as a histone code since the absence of appropriate markers results in the loss of a DDR during DSB repair. Metnase, a methylase which dimethylates histone H3 residue K36 (H3K36me2) in the DNA DSB region, and the levels of H3K36me2 have been found to correspond positively to DSB repair efficiency [reviewed in ref. (3)]. Another modification, H3K4me3, carried by Set1p methyltransferase has been found at DSBs and the absence of this modification has been linked to defective DNA DSB repair (102).
Conclusion and Future Directions
The major protein components of chromatin are histones, which are subject to many types of posttranslational modifications especially on their flexible N-terminal tails. Based on the presence of specific interactions between histone modifications and proteins involved in cellular metabolism, these modifications may constitute a “histone code” and could be used to manage epigenetic information that determines the selection of DSB repair pathway choice. The pre-existing histone code for DNA damage responses may be critical for generating the damage identification signal and such a code could have implications for the recruitment of repairosome factors specific to particular pathways. In the future it will be important to establish the relative impact of epigenetic changes in the context of specific DNA sequences and their role in DNA DSB repair. The modulation of histone acetylation with HDAC inhibitors has already been shown to alter the response to IR exposure resulting in some instances to radioprotection while in other situations radiosensitization (103). This indicates that small molecule modulation of histone post-translational modifications is a potential route to enhance radiotherapy responses. As with other areas of basic research, a detailed mechanistic understanding of chromatin modifications (e.g., the histone code) that regulate gene expression and the DDR will help to develop effective patient specific radiotherapies based on their own tumor epigenetic phenotype.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health National Cancer Institute Grants R01CA129537, R01CA154320, U19A1091175 (TKP) and NASA Grants NNX11AC15G, NNJO5HD36G and NNX09AU95G (JWS). Thanks are due to Jessica Tyler for her valuable discussions and to Swati Pandita for graphics.
REFERENCES
- 1.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15(2):172–83. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 2.Legube G, Trouche D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003;4(10):944–7. doi: 10.1038/sj.embor.embor941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson EA, Wray JW, Bansal P, Hromas R. Overview for the histone codes for DNA repair. Prog Mol Biol Transl Sci. 2012;110:207–27. doi: 10.1016/B978-0-12-387665-2.00008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Widlak P, Pietrowska M, Lanuszewska J. The role of chromatin proteins in DNA damage recognition and repair. Histochem Cell Biol. 2006;125(1-2):119–26. doi: 10.1007/s00418-005-0053-5. [DOI] [PubMed] [Google Scholar]
- 5.Verger A, Crossley M. Chromatin modifiers in transcription and DNA repair. Cell Mol Life Sci. 2004;61(17):2154–62. doi: 10.1007/s00018-004-4176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vidanes GM, Bonilla CY, Toczyski DP. Complicated tails: histone modifications and the DNA damage response. Cell. 2005;121(7):973–6. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Arundel CM, Vines CM, Tofilon PJ. Chromatin modifications associated with N-methylformamide-induced radiosensitization of clone A cells. Cancer Res. 1988;48(20):5669–73. [PubMed] [Google Scholar]
- 8.Barone F, Belli M, Pazzaglia S, Sapora O, Tabocchini MA. Radiation damage and chromatin structure. Ann Ist Super Sanita. 1989;25(1):59–67. [PubMed] [Google Scholar]
- 9.Gordon DJ, Milner AE, Beaney RP, Grdina DJ, Vaughan AT. The increase in radioresistance of Chinese hamster cells cultured as spheroids is correlated to changes in nuclear morphology. Radiat Res. 1990;121(2):175–9. [PubMed] [Google Scholar]
- 10.Hittelman WN, Pandita TK. Possible role of chromatin alteration in the radiosensitivity of ataxia-telangiectasia. Int J Radiat Biol. 1994;66(6 Suppl):S109–13. [PubMed] [Google Scholar]
- 11.Johnston PJ, MacPhail SH, Stamato TD, Kirchgessner CU, Olive PL. Higher-order chromatin structure-dependent repair of DNA double-strand breaks: involvement of the V(D)J recombination double-strand break repair pathway. Radiat Res. 1998;149(5):455–62. [PubMed] [Google Scholar]
- 12.Misri S, Pandita S, Pandita TK. Detecting ATM-dependent chromatin modification in DNA damage and heat shock response. Methods Mol Biol. 2009;523:395–410. doi: 10.1007/978-1-59745-190-1_26. [DOI] [PubMed] [Google Scholar]
- 13.Mazurik VK, Mikhailov VF. Nekotorye biokhimicheskie determinanty i markery radiorezistentnosti organizma mlekopitaiushchikh. [Various biochemical determinants and radioresistance markers in the mammalian body] Radiat Biol Radioecol. 1997;37(4):512–21. [PubMed] [Google Scholar]
- 14.Nackerdien Z, Michie J, Bohm L. Chromatin decondensed by acetylation shows an elevated radiation response. Radiat. Res. 1989;117(2):234–44. [PubMed] [Google Scholar]
- 15.Oleinick NL, Chiu SM, Friedman LR. Gamma radiation as a probe of chromatin structure: damage to and repair of active chromatin in the metaphase chromosome. Radiat Res. 1984;98(3):629–41. [PubMed] [Google Scholar]
- 16.Pandita TK, Hittelman WN. Evidence of a chromatin basis for increased mutagen sensitivity associated with multiple primary malignancies of the head and neck. Int J Cancer. 1995;61(5):738–43. doi: 10.1002/ijc.2910610524. [DOI] [PubMed] [Google Scholar]
- 17.Pandita TK, Hittelman WN. The contribution of DNA and chromosome repair deficiencies to the radiosensitivity of ataxia-telangiectasia. Radiat Res. 1992;131(2):214–23. [PubMed] [Google Scholar]
- 18.Pandita TK, Hittelman WN. Initial chromosome damage but not DNA damage is greater in ataxia telangiectasia cells. Radiat Res. 1992;130(1):94–103. [PubMed] [Google Scholar]
- 19.Pandita TK, Hittelman WN. Increased initial levels of chromosome damage and heterogeneous chromosome repair in ataxia telangiectasia heterozygote cells. Mutat Res. 1994;310(1):1–13. doi: 10.1016/0027-5107(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 20.Pandita TK, Richardson C. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 2009;37(5):1363–77. doi: 10.1093/nar/gkn1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandita TK. The role of ATM in telomere structure and function. Radiat Res. 2001;156(5 Pt 2):642–7. doi: 10.1667/0033-7587(2001)156[0642:troait]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 22.Pandita TK. ATM function and telomere stability. Oncogene. 2002;21(4):611–8. doi: 10.1038/sj.onc.1205060. [DOI] [PubMed] [Google Scholar]
- 23.Pandita TK. A multifaceted role for ATM in genome maintenance. Expert Rev Mol Med. 2003;5(16):1–21. doi: 10.1017/S1462399403006318. [DOI] [PubMed] [Google Scholar]
- 24.Smilenov LB, Dhar S, Pandita TK. Altered telomere nuclear matrix interactions and nucleosomal periodicity in ataxia telangiectasia cells before and after ionizing radiation treatment. Mol Cell Biol. 1999;19(10):6963–71. doi: 10.1128/mcb.19.10.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telford DJ, Stewart BW. Micrococcal nuclease: its specificity and use for chromatin analysis. Int J Biochem. 1989;21(2):127–37. doi: 10.1016/0020-711x(89)90100-6. [DOI] [PubMed] [Google Scholar]
- 26.Dellaire G, Kepkay R, Bazett-Jones DP. High resolution imaging of changes in the structure and spatial organization of chromatin, gamma-H2A.X and the MRN complex within etoposide-induced DNA repair foci. Cell Cycle. 2009;8(22):3750–69. doi: 10.4161/cc.8.22.10065. [DOI] [PubMed] [Google Scholar]
- 27.Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172(6):823–34. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoeijmakers JH. DNA repair mechanisms. Maturitas. 2001;38(1):17–22. doi: 10.1016/s0378-5122(00)00188-2. [DOI] [PubMed] [Google Scholar]
- 29.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 30.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21(3):396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–99. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 32.Elsasser SJ, D’Arcy S. Towards a mechanism for histone chaperones. Biochim Biophys Acta. 2012;1819(3-4):211–21. doi: 10.1016/j.bbagrm.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Misri S, Pandita S, Kumar R, Pandita TK. Telomeres, histone code, and DNA damage response. Cytogenet Genome Res. 2008;122(3-4):297–307. doi: 10.1159/000167816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19(5):207–17. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 35.van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol. 2005;6(10):757–65. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 36.Huyen Y, Zgheib O, Ditullio RA, Jr., Gorgoulis VG, Zacharatos P, Petty TJ, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432(7015):406–11. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 37.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127(7):1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459(7243):113–7. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119(5):603–14. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 40.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121(6):859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 41.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28(13):1878–89. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossetto D, Truman AW, Kron SJ, Cote J. Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin Cancer Res. 2010;16(18):4543–52. doi: 10.1158/1078-0432.CCR-10-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaurasia P, Sen R, Pandita TK, Bhaumik SR. Preferential repair of DNA double-strand break at the active gene in vivo. J Biol Chem. 2012;287(43):36414–22. doi: 10.1074/jbc.M112.364661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhadra MP, Horikoshi N, Pushpavallipvalli SN, Sarkar A, Bag I, Krishnan A, et al. The role of MOF in the ionizing radiation response is conserved in Drosophila melanogaster. Chromosoma. 2012;121(1):79–90. doi: 10.1007/s00412-011-0344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner BM, Birley AJ, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69(2):375–84. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 46.Gelbart ME, Larschan E, Peng S, Park PJ, Kuroda MI. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol. 2009;16(8):825–32. doi: 10.1038/nsmb.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117(6):721–33. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 48.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A. 2005;102(15):5501–6. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 50.Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30(14):3582–95. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, et al. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25(12):5292–305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10(15):886–95. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 53.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004;3(8-9):959–67. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 54.Pandita TK, Lieberman HB, Lim DS, Dhar S, Zheng W, Taya Y, et al. Ionizing radiation activates the ATM kinase throughout the cell cycle. Oncogene. 2000;19(11):1386–91. doi: 10.1038/sj.onc.1203444. [DOI] [PubMed] [Google Scholar]
- 55.Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: to be or not to be. FEBS Lett. 2010;584(17):3717–24. doi: 10.1016/j.febslet.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Molecular cell. 2006;21(2):187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 57.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123(7):1213–26. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 58.Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, et al. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 2010;30(22):5335–47. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernandez-Capetillo O, Allis CD, Nussenzweig A. Phosphorylation of histone H2B at DNA double-strand breaks. J Exp Med. 2004;199(12):1671–7. doi: 10.1084/jem.20032247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheung WL, Turner FB, Krishnamoorthy T, Wolner B, Ahn SH, Foley M, et al. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr Biol. 2005;15(7):656–60. doi: 10.1016/j.cub.2005.02.049. [DOI] [PubMed] [Google Scholar]
- 61.Utley RT, Lacoste N, Jobin-Robitaille O, Allard S, Cote J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol Cell Biol. 2005;25(18):8179–90. doi: 10.1128/MCB.25.18.8179-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120(1):25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 63.Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol. 2001;21(5):1719–29. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19(6):1176–9. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. Epub 2007/03/17. [DOI] [PubMed] [Google Scholar]
- 66.Chen CC, Tyler J. Chromatin reassembly signals the end of DNA repair. Cell Cycle. 2008;7(24):3792–7. doi: 10.4161/cc.7.24.7188. Epub 2008/12/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol. 2008;28(1):397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25(21):9175–88. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 70.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419(6905):411–5. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 71.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8(1):91–9. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 72.van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO. 2007;26(18):4113–25. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shiloh Y. Ataxia-telangiectasia: closer to unraveling the mystery. Eur J Hum Genet. 1995;3(2):116–38. doi: 10.1159/000472285. [DOI] [PubMed] [Google Scholar]
- 74.Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cellular Biol. 2007;27(5):1602–13. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, et al. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 2011;30(18):2135–46. doi: 10.1038/onc.2010.592. [DOI] [PubMed] [Google Scholar]
- 76.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25(12):4903–13. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oishi H, Kitagawa H, Wada O, Takezawa S, Tora L, Kouzu-Fujita M, et al. An hGCN5/TRRAP histone acetyltransferase complex co-activates BRCA1 transactivation function through histone modification. J Biol Chem. 2006;281(1):20–6. doi: 10.1074/jbc.M510157200. [DOI] [PubMed] [Google Scholar]
- 78.Hershko A, Ciechanover A, Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nat Med. 2000;6(10):1073–81. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 79.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 80.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7(5):663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 81.Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem. 2004;279(51):52812–5. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- 82.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Molecular Cell. 2005;20(6):845–54. doi: 10.1016/j.molcel.2005.12.002. Epub 2005/12/20. [DOI] [PubMed] [Google Scholar]
- 83.Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26. doi: 10.3389/fonc.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–46. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 85.Campbell SJ, Edwards RA, Leung CC, Neculai D, Hodge CD, Dhe-Paganon S, et al. Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation. J Biol Chem. 2012;287(28):23900–10. doi: 10.1074/jbc.M112.359653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mok MT, Henderson BR. Three-dimensional imaging reveals the spatial separation of gammaH2AX-MDC1-53BP1 and RNF8-RNF168-BRCA1-A complexes at ionizing radiation-induced foci. Radiother Oncol. 2012;103(3):415–20. doi: 10.1016/j.radonc.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 87.Shi W, Ma Z, Willers H, Akhtar K, Scott SP, Zhang J, et al. Disassembly of MDC1 foci is controlled by ubiquitin-protea-some-dependent degradation. J Biol Chem. 2008;283(46):31608–16. doi: 10.1074/jbc.M801082200. [DOI] [PubMed] [Google Scholar]
- 88.Deem AK, Li X, Tyler JK. Epigenetic regulation of genomic integrity. Chromosoma. 2012;121(2):131–51. doi: 10.1007/s00412-011-0358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qing Y, Yamazoe M, Hirota K, Dejsuphong D, Sakai W, Yamamoto KN, et al. The epistatic relationship between BRCA2 and the other RAD51 mediators in homologous recombination. PLoS Genetics. 2011;7(7):e1002148. doi: 10.1371/journal.pgen.1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316(5828):1202–5. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 91.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8(10):735–48. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 92.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316(5828):1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yan Q, Dutt S, Xu R, Graves K, Juszczynski P, Manis JP, et al. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Molecular Cell. 2009;36(1):110–20. doi: 10.1016/j.molcel.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Molecular Cell. 2007;25(5):663–75. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 95.Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187(3):319–26. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15(18):2343–60. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 97.Grant PA. A tale of histone modifications. Genome Biol. 2001;2(4) doi: 10.1186/gb-2001-2-4-reviews0003. reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438(7071):1116–22. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 99.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 100.Murray K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry. 1964;3:10–5. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 101.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470(7332):124–8. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Faucher D, Wellinger RJ. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genetics. 2010;6(8) doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Camphausen K, Tofilon PJ. Inhibition of histone deacetylation: a strategy for tumor radiosensitization. J Clin Oncol. 2007;25(26):4051–6. doi: 10.1200/JCO.2007.11.6202. [DOI] [PubMed] [Google Scholar]
- 104.Kusch T, Florens L, MacDonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306(5704):2084–7. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 105.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27(20):7028–40. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17(9):1144–51. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Foster ER, Downs JA. Histone H2A phosphorylation in DNA double-strand break repair. Febs J. 2005;272(13):3231–40. doi: 10.1111/j.1742-4658.2005.04741.x. [DOI] [PubMed] [Google Scholar]
- 108.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 109.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458(7238):591–6. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457(7225):57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell. 2003;4(4):497–508. doi: 10.1016/s1534-5807(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 112.Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459(7248):847–51. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stanlie A, Aida M, Muramatsu M, Honjo T, Begum NA. Histone3 lysine4 trimethylation regulated by the facilitates chromatin transcription complex is critical for DNA cleavage in class switch recombination. Proc Natl Acad Sci U S A. 2010;107(51):22190–5. doi: 10.1073/pnas.1016923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453(7195):682–6. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 115.Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11(11):1376–82. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Beck BD, Lee SS, Williamson E, Hromas RA, Lee SH. Biochemical characterization of metnase’s endonuclease activity and its role in NHEJ repair. Biochemistry. 2011;50(20):4360–70. doi: 10.1021/bi200333k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.De Haro LP, Wray J, Williamson EA, Durant ST, Corwin L, Gentry AC, et al. Metnase promotes restart and repair of stalled and collapsed replication forks. Nucleic Acids Res. 2010;38(17):5681–91. doi: 10.1093/nar/gkq339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bostelman LJ, Keller AM, Albrecht AM, Arat A, Thompson JS. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2007;6(3):383–95. doi: 10.1016/j.dnarep.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 119.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18(11):1251–62. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131(5):887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 121.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136(3):420–34. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 122.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Molecular Cell. 2011;41(5):529–42. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Molecular Cell. 2011;41(5):515–28. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]