Abstract
Cytokine stimulations of leukocytes many times result in transient activation of the p21 Rho family of small GTPases. The role of these molecules during cell migration and chemotaxis is well established. The traditional approach to study the activation dynamics of these proteins involves affinity pull-downs that are often cumbersome and prone to errors. Here, we describe a reagent and a method of simple “mix-and-measure” approach useful for determining the activation status of endogenous Cdc42 GTPase from cell lysates.
Keywords: Cdc42 activity, Biosensor, MeroCBD, Fluorometry, Mix-and-measure
1 Introduction
Activation status of the Rho family GTPases is often cumbersome to determine using traditional approaches including affinity- reagent based pull-down experiments [1]. These are often technically involved and challenging approaches, and thus could lead to possible artifacts due to the procedural variability and difficulty. Here, we describe a facile, “mix and measure” assay for determining the activation status of Cdc42 GTPase from whole-cell lysates. The approach we discuss here is based on the system described previously [2], in which a fluorescent biosensor that could detect the activation status of endogenous Cdc42 in living cells was used also in a lysate-based assay for neutrophils [2]. This biosensor (meroCBD) is based on a GTPase binding domain derived from Wiskott Aldrich Syndrome protein (WASp) with an organic dye capable of fluorescence modulation dependent on the local solvent polarity changes. The small binding domain is derivatized with a merocyanine dye, and upon binding of this dye-labeled domain to the target (endogenous, active Cdc42), the local solvent polarity changes from aqueous to more hydrophobic increasing the fluorescence emission [3, 4]. This sensor system is useful for in vitro characterization of the GTP-loading state of Cdc42 such as for the guanine nucleotide exchange assays, as well as for live-cell imaging experiments where the activation dynamics of Cdc42 can be monitored in real time [2]. Here, we discuss step-by-step, how to produce this meroCBD biosensor, characterize it in vitro, and then to use it to measure the endogenous Cdc42 activity changes in macrophage lysates during the time course of a cytokine stimulation experiment.
2 Materials
2.1 MeroCBD Preparation
LB medium.
Carbenicillin 100 mg/mL stock.
IPTG 1 M stock.
BL21(DE3) competent bacteria (see Note 1).
Talon Buffer: 50 mM NaH2PO4 (monobasic), 5 mM MgCl2, 10 % glycerol, pH 7.4; must be at 4 °C.
Dialysis/labeling Buffer: 50 mM NaH2PO4 (monobasic), pH 7.4.
StorageBuffer:50mMTris–HCl,50mMNaH2PO4(monobasic), 5 mM MgCl2, pH 7.4.
Amicon Ultra-4 5000MWCO spin columns.
5 M NaCl solution.
PMSF stock: 200 mM PMSF in methanol.
β-Mercaptoethanol.
DMSO.
100 mM DTT stock.
EDTA 0.5 M, pH 8.0, stock.
Mero-87-IAA (www.hahnlab.com).
Talon resin (Clontech).
Sephadex G-15.
Imidazole.
Poly-prep chromatography column.
2.2 MeroCBD Characterization
GTPase lysis buffer: 30 mM Tris–HCl, pH 7.8 (room temperature), 250 mM NaCl, 5 mM MgCl2, 10 % glycerol. Just before lysis, add PMSF to final concentration of 1 mM, β-mercaptoethanol to 2 mM, and 1 μM GTPγS.
GTPase dialysis buffer: 50 mM Tris–HCl, pH 7.6, 50 mM NaCl, 0.5 mM EDTA, pH 7.0, 5 mM MgCl2, 1 mM DTT, 10 nM GDP, 10 % glycerol.
Imidazole.
Talon resin (Clontech).
Amicon Ultra-4 5000 MW CO columns.
GTPase measurement buffer: 50 mM NaCl, 50 mM Tris–HCl, 5 mM MgCl, pH 7.6.
2.3 Cell Lysate Preparation
1 M Tris–HCl, pH 7.4.
2 M NaCl.
1 M MgCl2.
10 % Triton X-100.
10× Tris-buffered saline (TBS): 1.37 M NaCl, 250 mM Tris- Base, adjusted to pH 7.4.
RAW/LR5 cells derived from RAW 264.7 [5].
RPMI 1640 medium with glutamine.
10 cm cell culture dishes.
Fractalkine/CX3CL1 (FKN) (R&D Systems); reconstituted at 50 μg/mL with PBS and stored at −80 °C.
Cell lysis buffer: 50 mM Tris–HCl, pH 7.4, 500 mM NaCl, 50 mM MgCl2, 1 % Triton X-100. Store at 4 °C. Prepare on the day before use to cool overnight. Cool on ice on the day of stimulation.
1× TBS: 137 mM NaCl, 25 mM Tris-Base; pH 7.4; prepare on the day before use to cool overnight. Cool on ice on the day of stimulation.
-
Protease inhibitor stock: prepare in double-distilled water and store at −20 °C.
1 M Benzamidine.
1 mg/mL Aprotinin.
1 mg/mL Leupeptin.
Cell scraper.
2.4 Fluorometry of Lysates
Bradford reagent.
Spetrofluorometer capable of measuring the fluorescence response of the sensor (i.e., Horiba-Jobin-Yvon Fluorolog system).
3 Methods
3.1 MeroCBD Preparation
Transform 100 μL of BL21(DE3) bacteria with 1 ;L of plasmid DNA following the manufacturer’s protocols. For the plasmid DNA, pET21-CBD-EGFP, or pET21-CBD-MBP can be used (see Note 2). These constructs are available from Addgene.com. The bacteria should be plated out onto 4× LB agar plates overnight.
Following day, prepare 2× 520 mL of LB broth in 2 L baffled flasks, with 100 ;g/mL final concentration of carbenicillin (1:1,000 dilution from the carbenicillin stock solution). Taking the colonies grown from the day before, rinse using this LB medium and place the bacterial suspension into the flask using 2× agar plates worth of colonies per one flask (see Note 3). Culture the bacteria in the shaker incubator at 37 °C, 225 rpm.
When the bacterial OD600 reading reaches 0.9–1.0, protein synthesis is induced. IPTG is added to the cultures to make the final concentration of 0.2 mM from the 1 M stock IPTG solution. The bacterial cultures are then transferred to a room temperature shaker and incubated for additional 6 h to overnight.
At the conclusion of the induction period, the bacterial suspension is centrifuged at 6,000 × g for 10 min at 4 °C. The pellet can be frozen at this point at -80 °C for processing at a later date.
On the day of the protein purification, prepare 200 mL of the chilled (4 °C) Talon buffer with 300 mM NaCl (see Note 4). This will be used from step 6 on.
Taking 50 mL of the Talon buffer, add PMSF to 1 mM final concentration, and β-mercaptoethanol to 2 mM final concentration just prior to resuspending the bacterial pellet (see Note 5). Here, the bacterial pellet should be weighed. We typically process up to 2 g of bacterial pellet into 30 mL of this buffer. Scale accordingly.
When the pellet is fully resuspended, proceed to ultrasonication. Use 4 × 30 s ultrasonic pulses with 30 s to 1 min rest on ice in between pulses. Users must optimize the ultrasonic amplitudes for their individual equipment and the probe specification. We typically use a ½ in. tip probe at 60 % amplitude setting (see Note 6).
When the ultrasonication is complete, collect 100 μL as a sample to run on a PAGE-gel for analysis of the protein expression, label “Crude lysate.” The crude lysate is spun at 22,000 × g for 25 min at 4 °C.
While the centrifugation is in progress, prepare the Talon resin. The resin is at 1:1 (v:v) mixture with the storage buffer containing methanol. For 2–3 g bacterial pellet, 2 g of resin is required, thus 4 mL of the resin suspension is pipetted into a 50 mL conical tube. This is then spun at 750 × g for 3 min at room temperature. The suspension buffer is carefully decanted and the resin is washed twice using 20 mL of the cold Talon buffer without PMSF or β-mercaptoethanol.
When the bacterial centrifugation is complete, transfer the supernatant into the tube containing the prepared and washed Talon resin. Be sure to collect 100 μL sample of the supernatant prior to mixing with the Talon resin for later analysis, labeled “Cleared lysate.” The pellet should also be resuspended in Talon buffer to the same volume used to resuspend the original bacterial pellet. 100 μL sample of this resuspended pellet should then be kept for later analysis, labeled “Pellet.” The cleared lysate–Talon resin suspension should be covered in aluminum foil and placed on a rotating wheel (or a Nutator) for 1 h at room temperature.
When the binding reaction is complete, spin at 750 × g for 3 min at room temperature. Keep 100 μL sample of the unbound fraction (supernatant) for later analysis, labeled “Unbound.” Discard the unbound supernatant. Wash with 20 mL of the Talon buffer twice. At every wash, spin down and keep taking the 100 μL samples for analysis, labeled “Wash1” and “Wash2.”
The final wash should be done in Talon buffer containing 5 mM Imidazole, for 5 min at room temperature on a rotating wheel (or a Nutator). The imidazole stock solution (1 M) should be made fresh, just prior to use, in the Talon buffer being used here). The third wash is then spun and 100 μL sample of the supernatant is kept, labeled “Wash3.”
Using 5 mL of 100 mM imidazole in the Talon buffer, the protein is eluted from the Talon resin by incubating for 5 min at room temperature on a rotating wheel (or on a Nutator). The mixture is then spun, and 100 μL of the eluted protein (supernatant) is kept for analysis, labeled “Elution” (see Note 7). The supernatant is carefully removed from the Talon resin by filtration through a Poly-prep chromatography column (Bio-Rad) to remove any trace of the Talon resin.
The eluted protein is dialyzed overnight in 2 L of the cold dialysis buffer at 4 °C for proceeding directly to dye labeling the following day. Otherwise, it can be dialyzed against 2 L of the storage buffer for freezing and storage at −80 °C for up to 1 year.
-
Following the overnight dialysis, the protein solution is collected and concentration is determined (see Note 8). The concentration is determined by measuring the absorbance at 280 nm. The equation is:
The molar extinction coefficient at 280 nm for CBD-EGFP is 28,260 M−1 cm−1, and for CBD-MBP is 72,970 M−1 cm−1. The target concentration ideal for the dye labeling reaction is 100 μM. If the protein solution is too dilute, use the Amicon Ultra-4 5000MWCO spin columns to concentrate the protein.
Prepare the reactive dye mixture. The crystals of mero-87-IAA should be brought up to room temperature, wrapped in foil, prior to opening the glass vial. Use 30 μL of fresh, anhydrous DMSO to dissolve the dye crystals, then transfer the dye mixture into a 0.2 mL Eppendorf tube, spin in a benchtop microcentrifuge at 18,000 rcf at room temperature for 2 min to pellet any undissolved material. Transfer the supernatant into a fresh 0.2 mL tube. The concentration of this dye mixture must be determined. The dye solution should be diluted in anhydrous DMSO at 1:5,000, and absorbance should be measured at 596 nm. The molar extinction coefficient for mero-87 in DMSO at 596 nm is 135,301 M−1 cm−1. The same equation as in step 15 can be used to determine the concentration. Ideally, the reactive dye solution should be at 10–20 mM concentration.
In labeling the protein, the protein to dye ratio and the reaction time must be optimized. As a good starting point, we recommend 1:5 protein to dye ratio, and 2 h reaction at room temperature. The goal here is to approach 100 % reaction efficiency at the single, target cysteine site on the CBD molecule (see Note 9). Place 300 μL of the purified protein solution (in Dialysis/labeling buffer) into a 2 mL round-bottom Eppendorf tube, calculate correct amount of the reactive dye solution to add directly to this solution in order to obtain the desired protein to dye ratio. The dye solution is added in one quick shot and immediately the mixture is vortexed. This is then wrapped in aluminum foil and placed on a rotating wheel or a Nutator for 2 h at room temperature. At the conclusion of the reaction, the reaction must be quenched by adding 1 μL of β-mercaptoethanol, allowing the mixture to be quenched for 5 min, and then column-purified over Sephadex G-15 column.
The Sephadex G-15 column should be made during the dye labeling reaction. Sephadex slurry is equilibrated with fresh Dialysis/labeling buffer, and the slurry should be packed into a 1 cm diameter glass column, approximately 10 cm tall. The quenched reaction mixture should be loaded very carefully so as to maintain as straight and flat a meniscus as possible, which will enable cleaner separation of the labeled protein from the unreacted dyes. The labeled protein will travel faster than the unreacted dyes, so collect the labeled protein fraction when it reaches the bottom of the column. Once the meroCBD is collected, the concentration must be determined and the dye labeling efficiency must be determined.
The absorbance should be measured in the Dialysis/labeling buffer (aqueous) to determine the protein concentration, and in DMSO to determine the dye concentration. The mero-87 dye has small amount of absorbance at 280 nm, and this must be subtracted from the measured 280 nm absorbance in the aqueous condition. The correction factor for this is 0.07605. The peak absorbance at 596 nm for the dye is multiplied by the correction factor and then subtracted from the measured 280 nm absorbance to calculate the protein concentration. The dye concentration is in turn calculated from the measurement in DMSO, similar to step 16. Once the concentrations are determined, protein to labeled dye ratio can be calculated and the reaction condition can be further optimized if required.
The meroCBD is then aliquoted into 0.2 mL tubes (30 μL per tube) and flash-frozen in liquid nitrogen, and then stored at −80 °C until use.
3.2 MeroCBD Characterization
Transform 100 μL of BL21(DE3) bacteria with 1 μL of plasmid DNA following the manufacturer’s protocols. For the plasmid DNA, pET21-Cdc42wt is used. The bacterial propagation, media conditions, and induction conditions are identical to Subheading 3.1, steps 1 through 4.
The bacterial lysis and protein purifications steps are similar to Subheading 3.1, steps 5 through 15, exception being the GTPase lysis buffer is used, and the overnight dialysis buffer compositions are different. Furthermore, the buffers must contain guanosine phosphates to achieve stability of the purified GTPase. Here, we add 1 μM GTPγS to the lysis buffer, but during the 2× washes of the Talon resin following the protein binding, we exchange to 1 μM GDP, the final wash solution should contain 10 mM imidazole and 1 μM GDP. Following elution, concentration is determined by absorption at 280 nm and the extinction coefficient of 14,650 M−1 cm−1 is used. The stock protein can then be frozen at −80 °C.
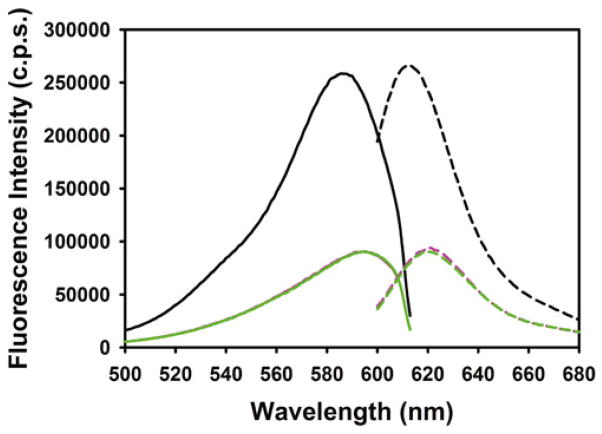
To test meroCBD for proper function, we perform an in vitro fluorometry using the purified Cdc42, loaded with GTPγS, GDP or no guanosine phosphate. We resuspend the purified Cdc42 into the GTPase measurement buffer at a final concentration of 1.4 μM, 200 μL per sample (GTPγS, GDP, no guanosine phosphates), together with 1 mM DTT and 10 mM EDTA. To 190 μL of this mixture, add 10 μL of the guanosine phosphate solutions (stock at 200 μM) so that the final concentration in the Cdc42 mixtures is 10 μM. These samples are then incubated at 30 °C for 30 min. The nucleotide loading reaction is quenched by addition of 3 μL of 1 M MgCl2 (final concentration 15 mM); vortex, wait for 5 min at 30 °C, and then spin down briefly, and put on ice. The meroCBD is prepared into a separate set of tubes containing 200 μL volume each of the GTPase measurement buffer so that the final concentration of meroCBD will be 300 nM, with 15 mM MgCl2, and placed on ice, shielded from ambient light. Just prior to the fluorometric measurements, the prepared meroCBD suspension and the Cdc42 samples are mixed, and the total volume of 400 μL is loaded into the cuvette and excitation/emission spectra are measured. For the excitation spectral measurement, the emission wavelength is set to 630 nm, and excitation wavelength is scanned from 400 to 623 nm. For the emission spectral scan, the excitation wavelength is set to 596 nm, and the emission wavelength is scanned from 603 to 800 nm (Fig. 1) The peak emission intensities are compared between various guanosine phosphate loaded conditions to determine the total fold-change in fluorescence emission as a function of binding to active versus inactive Cdc42.
Fig. 1.
Excitation and emission spectra of meroCBD in response to GTPgS-loaded (black ), GDP-loaded (magenta) and Apo (green) wild-type Cdc42. Excitation spectra are shown with solid lines and the emission spectra are shown with dashed lines. The protein to dye label ratio for the meroCBD used in this characterization was 69 %
3.3 Cell Lysate Preparation
Cool cell lysis buffer and 1× TBS on ice.
For serum starvation, replace media on RAW/LR5 cells, which were plated the night before in 10 cm dishes at approximately 70 % confluent, with RPMI 1640 for at least 4 h.
Pre-warm RPMI 1640 to 37 °C for stimulation. Add FKN at 2× concentration to pre-warmed RPMI 1640 right before use.
-
Add protease inhibitors to cell lysis buffer right before use; working concentrations:
1 mM Benzamidine.
1 μg/mL Aprotinin.
1 μg/mL Leupeptin.
Transfer dish to 37 °C water bath.
Add equal volume of 2× FKN solution to cells, bringing FKN solution to 1× (50 ng/mL) (e.g., add 5 mL 2× FKN solution to 5 mL serum-starving cells) and swirl gently to mix. Start timer as adding solution. If there are multiple time points, perform one time point at a time.
To stop reaction, quickly aspirate media, place dish on ice and add ice-cold 1× TBS right at the time when stimulation ends. It takes ~5 s to aspirate media; start aspirating media while still in water bath ~5 s before stimulation time ends, so 1× TBS can be added immediately as the dish is placed on ice.
Immediately aspirate 1× TBS and add ice-cold 0.5 mL of cell lysis buffer (containing protease inhibitors).
Scrape cells in dish and transfer lysate to 1.5 mL microcentrifuge tubes. Keep on ice until all time points have been performed.
Clear lysates of DNA by centrifuging at 22,000 rcf for 10 min at 4 °C in a microcentrifuge.
Transfer supernatant to clean 1.5 mL microcentrifuge. Avoid carrying over the DNA pellet. Keep on ice until performing fluorometry with MeroCBD. Alternatively, lysates can be stored at −80 °C until use.
3.4 Fluorometry of Lysates See (Note 10)
Measure the relative concentration differences among the lysates using the Bradford reagent. Here, protein concentration standards are not necessary as long as one operates within the linear range of the detector response of the UV-Visible spectrophotometer. Based on the relative absorbance at 595 nm, determine the correct dilutions necessary to achieve uniform concentrations across all the samples being measured (see Note 11).
Once the concentrations are adjusted to be uniform, aliquot 400 μL of the cell lysate each into Eppendorf tube on ice (see Note 12). Based on the concentration of the meroCBD solution, calculate how much one would have to add to each sample to achieve 150 nM biosensor final concentration.
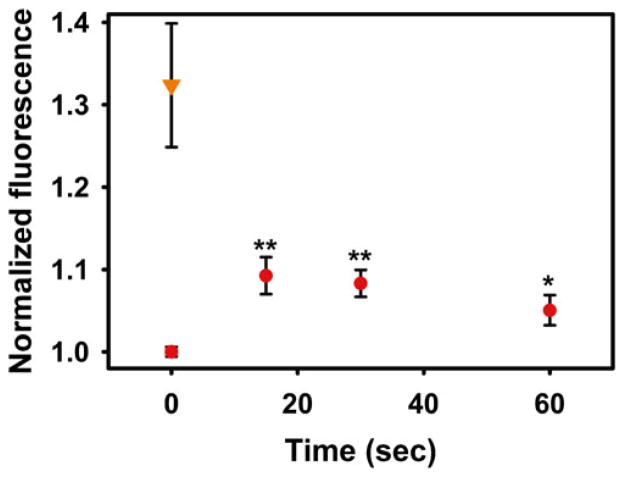
Just prior to the measurement, mix appropriate volume of meroCBD (may require dilution) with the lysate sample, load into the cuvette, and measure the emission spectral profile. This procedure is repeated for all samples. Wash the cuvette in between each sample to minimize cross-contamination. The representative result is shown in Fig. 2.
Fig. 2.
Fluorometric measurements of lysates of RAW cells stimulated with Fractalkine (50 ng/mL) at indicated times (red circles). **P < 0.01, *P < 0.05, n = 6–7. At T = 0 s, samples of lysates were also pre-equilibrated with 100 μM GTPγS as positive controls (Orange inverted triangle)
Acknowledgments
This work was supported by grants GM093121 (D.S., L.H.), T32GM007491 (V.M.), and GM071828 (D.C.).
Footnotes
Choice of bacteria is very important. BL21(DE3) must be used. Do not use the BL21(DE3)pLysS, as this will severely reduce the protein yield.
pET-CBD-EGFP will produce a fusion protein of the biosensor CBD with EGFP. This will be useful for determining the amount of biosensor pipetted into the cuvette during the fluorometric measurements. If the pipetting of biosensor is done precisely, this step can be omitted and the pET-CBD-MBP can be used. This version does not contain the EGFP. Additionally, EGFP contains two cysteines which will interfere with the stoichiometric labeling of the mero-87-IAA dye, whereas the CBD-MBP only contains one reactive Cysteine in the whole molecule, eliminating this problem.
Starting from the plated colonies accelerates the bacterial growth in the flasks as they are still in the log-growth phase. Overnight liquid starter cultures can be used instead, but this will significantly slow down the large-scale bacterial growth as they have reached the plateau-phase in the liquid starter culture.
NaCl must be added to the Talon buffer fresh, just prior to use.
PMSF is only useful for approximately 30 min after addition into an aqueous solution, so add this just prior to bacterial resuspension.
When the ultrasonication starts, the consistency of the bacterial lysis buffer changes from watery, to viscous, to again watery. This indicates the various stages of bacterial lysis. When the liquid becomes viscous, the bacterial outer membrane is lysed but the genomic DNA is not yet sheared. When the viscosity turns to thin again, it indicates that the DNA has been sheared and the lysis is more or less complete.
The samples collected throughout the protein purification steps should be run on an SDS-PAGE gel, and Coomassie stained to determine the extent of protein purification.
Also collect some amount of the bulk dialysis buffer, as this will be useful for correctly producing the blanking solution during concentration measurement of the dialyzed protein. Trace amount of Imidazole present in the protein sample will affect the absorbance if not properly corrected for by using the bulk dialysis buffer also containing identical amount of imidazole.
In the case of CBD-EGFP, prolonged reaction times or increased dye concentrations will result in overlabeling due to the presence of two additional cysteines on EGFP. This effect will reduce the EGFP stability and reduce the fluorescence as well as reducing the fluorescence emission intensity from the mero-87. In the case of CBD-MBP, there is only one Cysteine, but overlabeling is still possible due to reactions with other residues including lysines and histidines if the reaction times/dye concentrations are increased unreasonably.
Here, the manner in which pipetting is performed is absolutely critical to remove errors. Use fresh pipette tips all the time, do not pipette into a fresh pipette tip more than one time as the surface wetting and viscosity effects will slightly change the amount being pipetted.
Here, choose the lowest concentrated lysate and use that as the baseline. One can always dilute more concentrated lysates to make it the same concentration, never the reverse.
The volume of lysates could be more or less than the suggested amount. Use the appropriate cuvettes that are optimized for various volumes.
References
- 1.Benard V, Bokoch GM. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 2002;345:349–359. doi: 10.1016/s0076-6879(02)45028-8. [DOI] [PubMed] [Google Scholar]
- 2.Nalbant P, Hodgson L, Kraynov V, et al. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 3.Toutchkine A, Nguyen DV, Hahn KM. Merocyanine dyes with improved photostability. Org Lett. 2007;9:2775–2777. doi: 10.1021/ol070780h. [DOI] [PubMed] [Google Scholar]
- 4.Toutchkine A, Kraynov V, Hahn K. Solvent-sensitive dyes to report protein conformational changes in living cells. J Am Chem Soc. 2003;125:4132–4145. doi: 10.1021/ja0290882. [DOI] [PubMed] [Google Scholar]
- 5.Cox D, Chang P, Zhang Q, et al. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J Exp Med. 1997;186:1487–1494. doi: 10.1084/jem.186.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]