Abstract
We present a de novo high-resolution structure of the peptide Alanyl-Prolyl-Glycine using a combination of sensitive solid-state NMR techniques that each yield precise structural constraints. High-quality 13C–13C distance constraints are extracted by fitting rotational resonance width (R2W) experiments using Multimode Multipole Floquet Theory and experimental chemical shift anisotropy (CSA) orientations. In this strategy, a structure is first calculated using DANTE-REDOR and torsion angle measurements and the resulting relative CSA orientations are used as an input parameter in the 13C–13C distance calculations. Finally, a refined structure is calculated using all the constraints. We investigate the effect of different structural constraints on structure quality, as determined by comparison to the crystal structure and also self-consistency of the calculated structures. Inclusion of all or subsets of these constraints into CNS calculations resulted in high-quality structures (0.02 Å backbone RMSD using all 11 constraints).
Keywords: SSNMR, Structural determination
1. Introduction
Determining the high-resolution structures (backbone RMSD < 0.1 Å) of uniformly labeled polypeptides with solid-state NMR is often prohibitively difficult due to a lack of precision available with homonuclear broadbanded recoupling techniques such as RAD [1], DARR [2] and TSAR [3]. Although these experiments can define the secondary and tertiary structure and yield a clear global structure of the protein, the lack of high-precision measurements can leave detail of the active site and mechanisms that are of broader interest to the biological community still undefined. More precise techniques such as REDOR [4], RFDR [5], DRAWS [6], and local field correlations yield sub-angstrom precision, but are often limited to spin-pair labeled systems.
A previous study on the polypeptide, N-f-MLF-OH, demonstrated the use of FS-REDOR, and torsion angle constraints to determine a high-resolution de novo structure [7]. Since the publication of that structure, additional high-resolution techniques applicable to uniformly 13C, 15N labeled systems have been developed, such as rotational resonance width (R2W) [8,9] and DANTE-REDOR [10]. DANTE-REDOR can yield high-quality restrictions on torsion angles, in addition to heteronuclear distance constraints. R2W utilizes band-selective R2 to recouple homonuclear spin pairs and can separate weaker dipolar oscillations (from long distance, structurally relevant spin pairs) from relaxation processes using a constant mixing time strategy and has been recently applied to measure distances in uniformly labeled proteins [11]. However, Ramachandran et al. [12] discussed the effect of the relative CSA orientation of the two spins in extracting accurate distances from R2W experiments. To make R2W a truly de novo, precise, and accurate technique for making homonuclear distance measurements, one must obtain the relative CSA orientation of the spin pair experimentally.
We demonstrate the utility of an iterative approach to structure determination of uniformly labeled polypeptides using various solid-state NMR methods. Namely, ψ torsion angle and 13C–15N distances from NCCN [13] and DANTE-REDOR measurements are first used to establish a structure from which approximate relative CSA orientations are available. Those orientations are then utilized into the fitting routines of R2W profiles to also obtain precise and accurate 13C–13C distances. Finally all experimentally determined homonuclear, heteronuclear, and torsion angles constraints are used in a final energy minimization that yields a high-quality de novo structure. We further discuss the effect of each sub-group of structural constraints on the structural quality as judged by self-consistent backbone RMSD and comparison to the known crystal structure.
2. Results and discussion
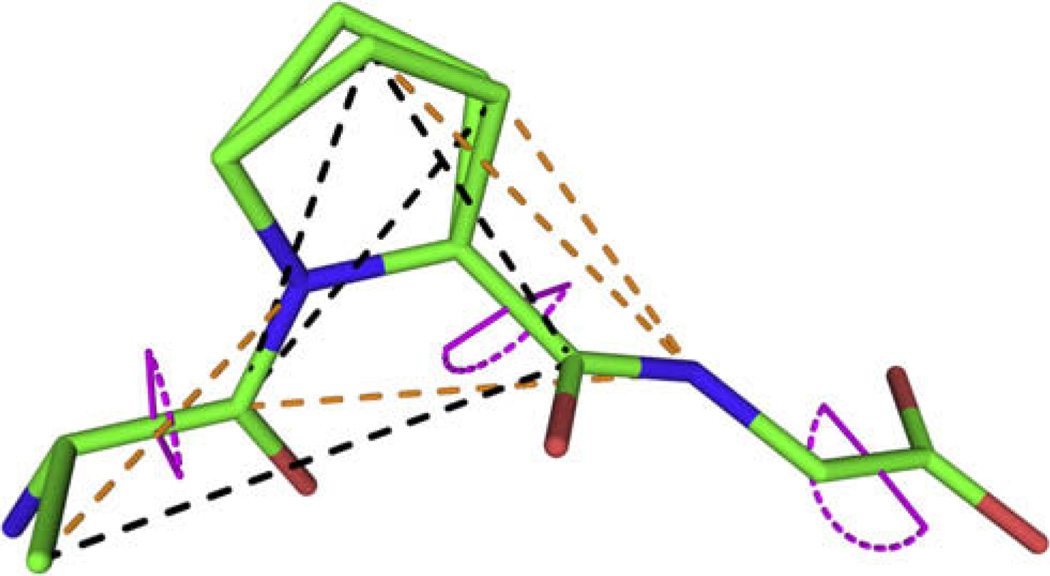
Fig. 1 shows the 11 constraints determined for APG listed in Table 1. Comparison with the X-ray crystal structure indicates the level of accuracy achieved in each measurement. Two of the restrictions on ψ dihedral angles were determined with a local field NCCN experiment [13]. DANTE-REDOR was used to determine the glycine ψ angle, and four 13C–15N internuclear distances. The four carbon internuclear distances were extracted using R2W.
Fig. 1.
Crystal structure of APG [14] showing the 11 SSNMR experimental constraints used in the structural refinement. Torsion angles are displayed in violet, REDOR distances in orange and R2W constraints in black. Note that the proline ring has a split occupancy between two conformers in the crystal structure. (For interpretation of colour mentioned in this figure, the reader is referred to the web version of this article.)
Table 1.
Experimental solid-state NMR constraints used in the structure calculation and corresponding values from the crystal structure from all NMR experiments; DANTE-REDOR (orange), NCCN torsion angles and R2W distances (black).
| ψ Torsion Angles | X-ray (°) | SSNMR (°) |
|---|---|---|
| ψGly | 178 | 180 ± 15 |
| ψAla | 153 | 152 ± 5 |
| ψPro | 157 | 162 ± 5 |
| Distances | X-ray (Å) | SSNMR (Å) |
| Pro N–Ala Cβ | 3.2 | 3.2 ± 0.1 |
| Gly N–Pro Cβ | 3.2 | 3.2 ± 0.15 |
| Pro C′–Pro Cγ | 3.3/3.6 | 3.9 ± 0.39 |
| Ala C′–Pro Cβ | 3.6 | 3.8 ± 0.38 |
| Ala C′–Pro Cγ | 3.6 | 3.9 ± 0.39 |
| Gly N–Ala C′ | 4.1 | 4.1 +0.6/−0.3 |
| Gly N–Pro Cγ | 4.5/4.3 | 4.2 +0.6/−0.3 |
| Pro C′–Ala Cβ | 4.7 | 4.4 ± 0.44 |
2.1. CSA parameters for precise R2W fitting
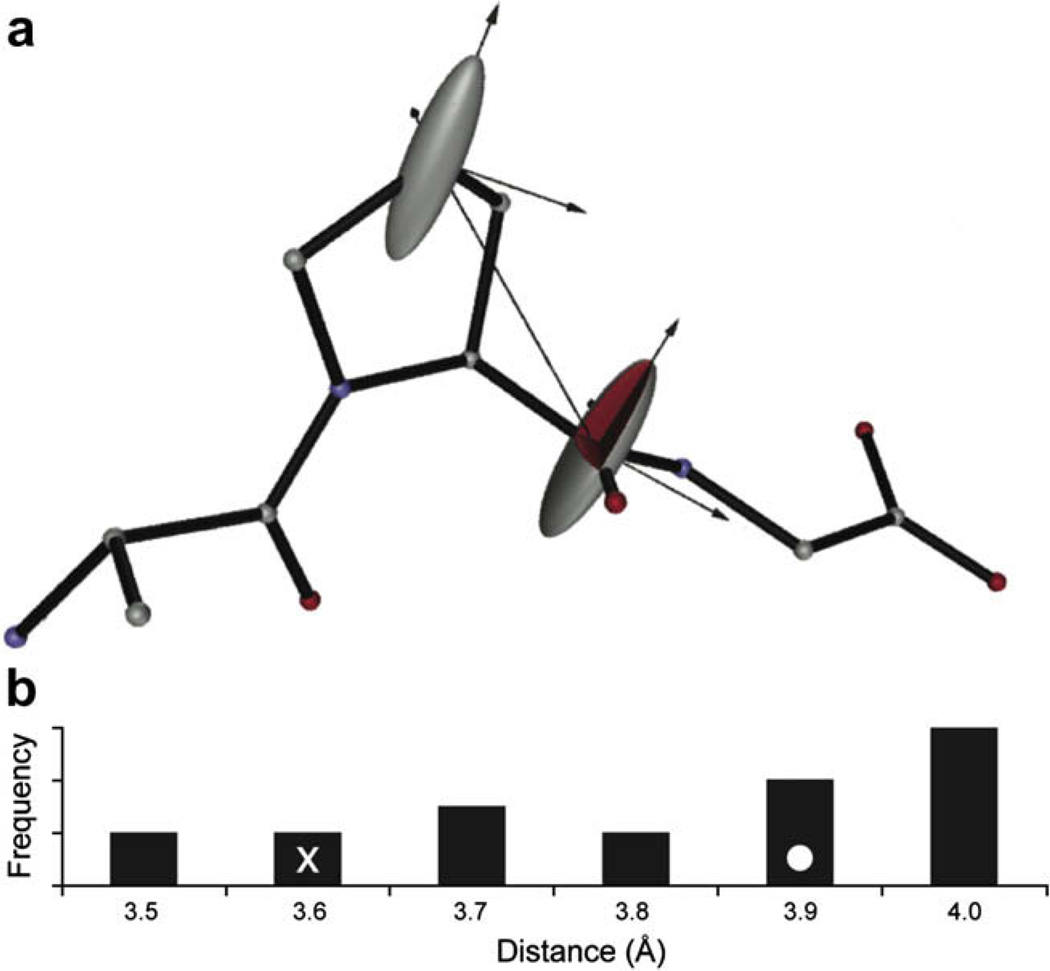
The CSA magnitude and relative orientation of the two spins being recoupled by R2 has a profound impact on the spin dynamics [15] and accuracy of the distances extracted using R2W [12]. For example Fig. 2b shows the distribution of distances resulting using different CSA orientations taken from an ensemble of structures generated from simulated annealing without experimental constraints. The range of motion displayed in unconstrained simulated annealing is large enough even in this small molecule to cause substantial deviation in the final R2 calculations, which demonstrates the importance of first calculating a lower resolution structure.
Fig. 2.
(a) Relative orientations of the Proline C′ and Cγ CSAs are shown superimposed on the initial structure calculated using REDOR and torsion angle constraints. (b) Histogram displaying the range of Pro C′-Pro Cγ distances calculated from an ensemble of accessible structures generated using simulated annealing (CNS) without any constraints. X-ray (x) and reported NMR (o) distances are indicated. This graphic was produced with SIMMOL [17].
As a starting point in the R2W fitting routine, an initial structure based on REDOR and torsion angle measurements was first calculated. The relative CSA orientation from this structure was then used as an input for the R2W fitting routine, which is derived from a Multimode Multipole Floquet Theory (MMFT) treatment of the two spins [16]. The 13C–13C distances from these R2W fits were then used together with the REDOR and torsion angle constraints to calculate a final structure. R2W fits generated with relative CSA orientations found in the final structure yielded the same distance as those generated from the initial REDOR-torsion angle structure. Thus we found it unnecessary to perform further iterations in which the structure generated with initial R2W distance constraints is used to refine these constraints.
The default values from SIMMOL were used for relative CSA – molecular orientations, the span and skew of carbonyl carbons were extracted from a 4 kHz CP-MAS spectrum according to Herzfeld and Berger [18], and aliphatic carbon span and skew values were taken from Ye et al. [19].
2.2. Carboxyl terminus
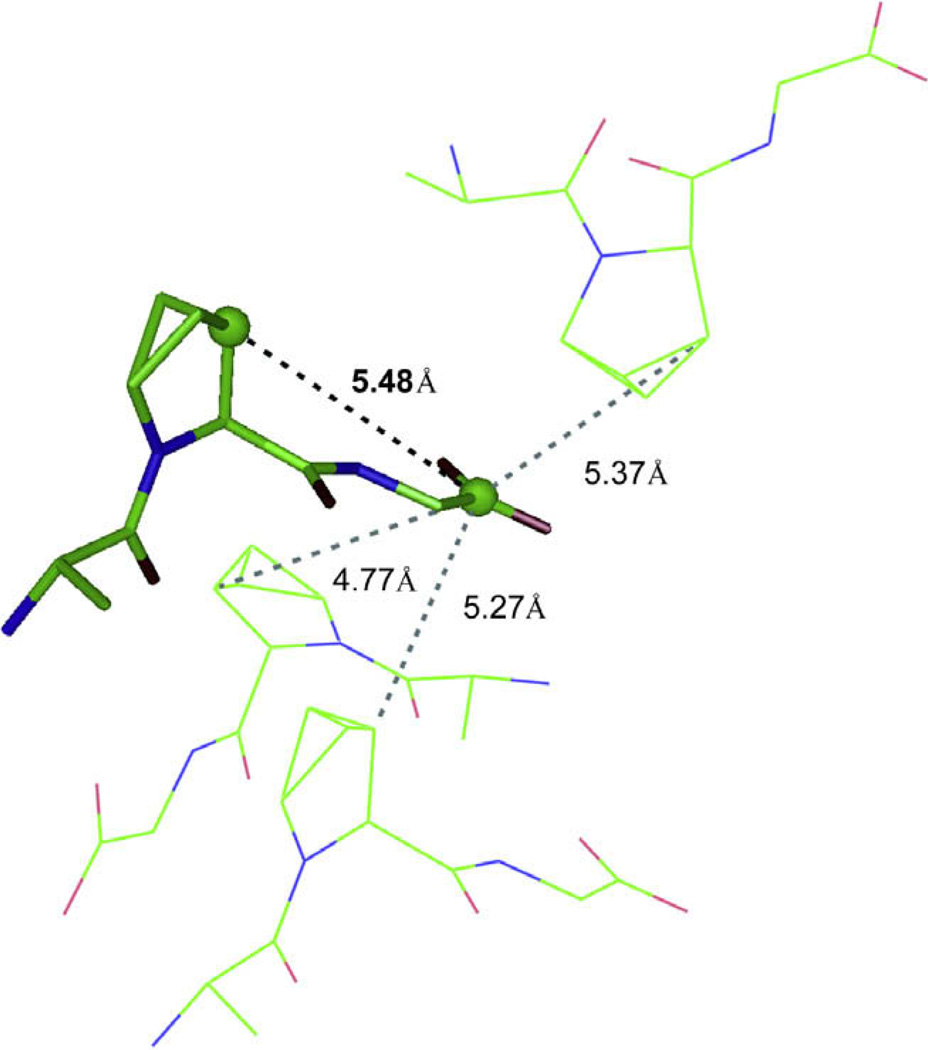
The carboxyl terminus is not well defined due to the absence of 13C–13C constraints to the Gly carboxyl carbon (see Fig. 1). This is predominantly due to intermolecular and dipolar truncation effects complicating any 13C–13C distance measurements between the Gly C′ and the Pro Cα and Pro Cβ. The Pro Cβ carbon to Gly C′ carbon intermolecular distance is 5.5 Å in the crystal structure, but there are three other intermolecular Pro Cβ to Gly C′ separations that are less than 5.4 Å (see Fig. 3). The crystals consisted of 10% 13C–labeled peptide diluted with natural abundance APG to reduce the number of intermolecular contacts, however there still appears to be enough coupling between intermolecular Pro Cβ–Gly C′ pairs to substantially effect the R2W profile and result in a fitting of the data to 4.7 Å rather than 5.5 Å seen in the crystal structure.
Fig. 3.
Crystal lattice of APG showing competing intermolecular distances to the long-range internuclear distance that could be used for structural refinement.
Furthermore, the one-bond Pro C′–Pro Cα dipolar coupling truncates the Pro C′–Gly Cα coupling which would also constrain the carboxyl terminus. Even though R2W is a band-selective recoupling technique and thus usually does not suffer from dipolar truncation [20], the single bond coupling is an order of magnitude stronger than the coupling of interest and is also recoupled when the spinning frequency is set to the N = 2 recoupling condition for the Pro C′ to Gly Cα interaction; the stronger coupling dominates and insufficient polarization is transferred to the Gly Cα. We note that the dipolar truncation should not be as pronounced at higher magnetic fields where the resonances are more separated and the recoupling condition is thus more selective.
2.3. Constraints effect on structural quality
The standard annealing protocol in the program CNS [21,22] was used to generate 40 APG structures in order to generate an ensemble that shows both an average geometry and demonstrates the degree of conformational variability allowed by the geometrical constraints.
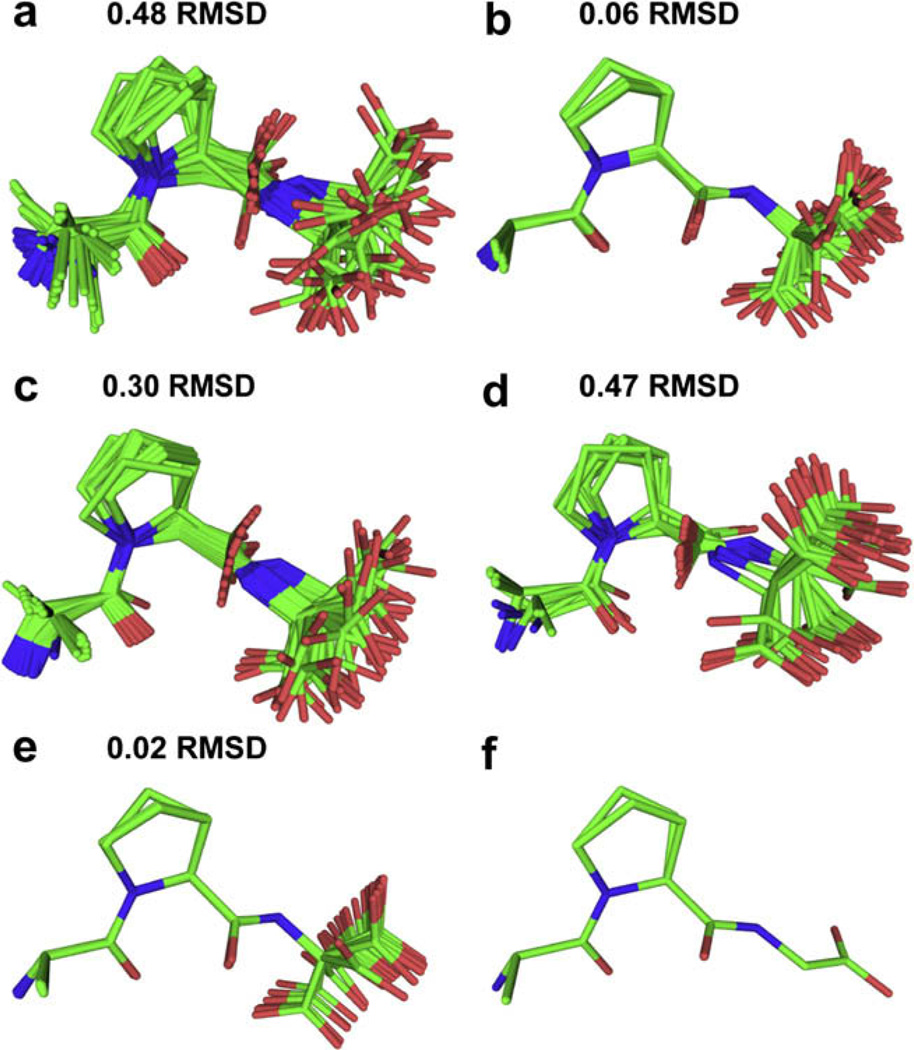
In order to compare the usefulness of different structural constraints, a backbone RMSD among the 40 structures of each ensemble was calculated. This shows the level of precision attained by the different NMR experiments, indicating their ability to define overall conformation in this peptide. The results are summarized in Fig. 4 and show excellent agreement with the crystal structure. Since we were not able to perform a measurement that constrains the ψGly angle, the Gly carboxyl group was omitted from the RMSD calculations. The final NMR structure and the crystal structure agree to within an RMSD of 0.09 Å.
Fig. 4.
Twenty lowest energy structures of APG using simulated annealing (CNS) (a) no constraints (b) 2 ψ angles from NCCN measurements (c) 4 13C–13C distance constraints (d) 4 13C–15N heteronuclear constraints (e) all 11 constraints (f) crystal structure.
3. Conclusions
Precise 13C–13C, 13C–15N, and ψ torsion angles were measured in a uniformly 13C, 15N labeled peptide using R2W in a de novo manner, DANTE-REDOR and local field NCCN experiments. Using these sub-angstrom structural constraints we were able to determine a high-quality structure of APG. The 0.02 Å backbone RMSD structure was sufficient to observe the same conformational heterogeneity of the proline ring that is seen in the X-ray structure at 100 K. The ability to discriminate the two conformers by using structural constraints with sub-angstrom precision and CNS could be applied to address biological questions that require high-resolution structural detail. Furthermore, the short intramolecular contacts present in the tripeptide lattice studied here should not be present in larger proteins and should enable the extension of this de novo R2W strategy to distances up to 8 Å.
Finally, the NCCN torsion and many of the internuclear distances that were experimentally determined with SSNMR are indicative of trans-proline. Although proline is almost always in the trans-isomer, the isomerization to the cis can play an important role in mitotic regulation [23], and we emphasize the strategies for structure determination presented here could be used to further investigate the biological function of proline isomerization.
4. Experimental methods
4.1. Sample preparation
Experiments were performed on a sample of uniformly 13C and 15N labeled Alanyl-Prolyl-Glycine (APG) that was diluted to 10% in the corresponding natural abundance tripeptide to attenuate intermolecular dipolar couplings. The tripeptide was synthesized by CS Bio (CA) with labeled amino acids from Cambridge Isotopes (Andover, MA) and natural abundance material from BaChem (Switzerland). A 9:1 natural abundance to labeled APG mixture was dissolved in a minimal amount of water (solubility was approximately 45 mg/ml). The aqueous solution was placed in a desiccator next to a container of excess ethylene glycol. After a week, crystals were collected, crushed and packed into a rotor.
4.2. Assignments
To assign the 13C resonances, a 2D 13C–13C SPC-5 [24] spectrum was recorded spinning at ωr/2π = 7.5 kHz using TPPM decoupling [25]. γB1 for 13C was set to 37.5 kHz required by the recoupling condition. The mixing time was 1.07 ms corresponding to 16 rotor cycles, during which time a 120 kHz continuous wave γB1 field was applied on the proton channel for decoupling. In the indirect dimension, 256 time points were recorded using 16 scans for each t1 point. The overall measurement time was 7 h. The direct and indirect dimensions were linearly predicted forward to 1024 points, zero-filled to 2048 points and Fourier transformed using NMRPIPE [26].
4.3. NCCN local field correlations
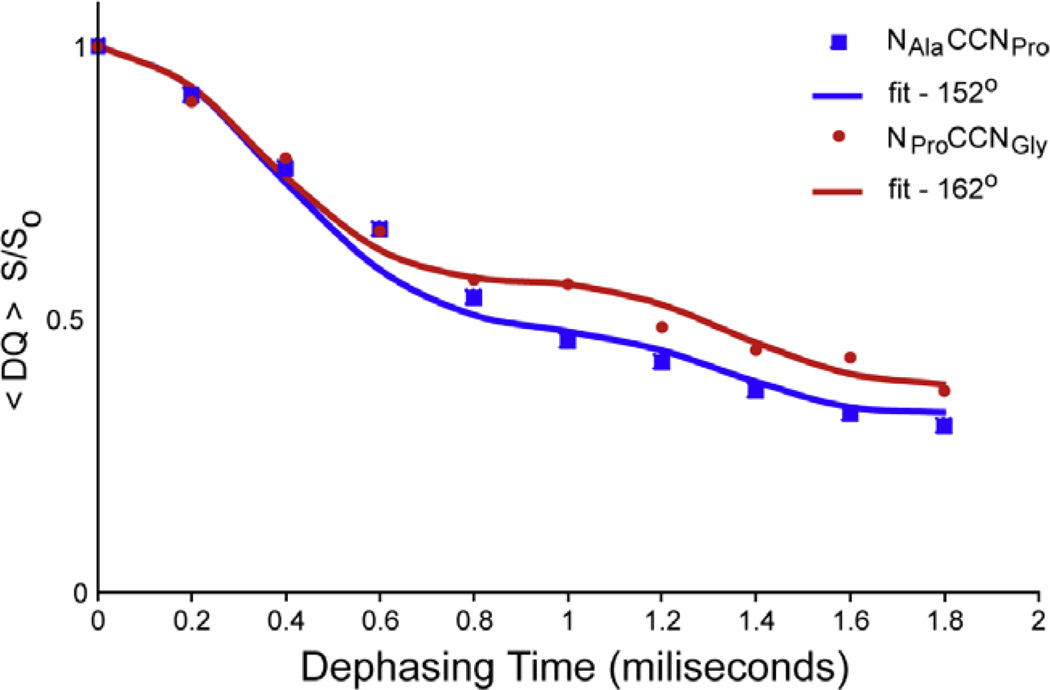
The torsion angles, ψAla and ψPro were determined with the double-quantum heteronuclear local field experiment previously described [13]. SPC-5 [24] was used to create the 13C–13C double-quantum coherence. The homonuclear mixing time was 800 µs, corresponding to eight rotor periods and the 13C carrier was set precisely between the two resonances targeted for recoupling. γB1 equal to 50 kHz was used for both 13C and 15N during homonuclear mixing and the REDOR [4] dephasing pulses. The spinning frequency was 10 and 110 kHz continuous wave γB1 decoupling field on protons was applied during SPC-5 mixing and REDOR periods; 136 kHz TPPM [25] decoupling was used during the acquisition period. About 512 scans were taken per dephasing time, and a 3 s recycle delay was used between scans. Each dephasing point was normalized to the intensity of the corresponding resonances absent of REDOR pulses during the dephasing period. The resonances were integrated and the intensities fit (see Fig. 8) to extract the torsion angle with SPINEVOLUTION [27].
Fig. 8.
Dephasing curves for extraction of Ψ torsion angle. An NGlyCCNPro Ψ angle of 152° (blue) was extracted compared to 153° from the X-ray data and NAlaCCNGly Y angle was found to be 162° (red) versus 157° from the X-ray data. SPINEVOLUTION [27] was used to fit the data. (For interpretation of colour mentioned in this figure, the reader is referred to the web version of this article.)
4.4. Rotational resonance width
NMR spectra were recorded on a 360 MHz Spectrometer (courtesy of Dr. D.J. Ruben) with a commercial Chemagnetics triple resonance MAS probe equipped with a 4.0 mm Chemagnetics spinning module. Spinning frequencies were regulated with a Bruker MAS controller.
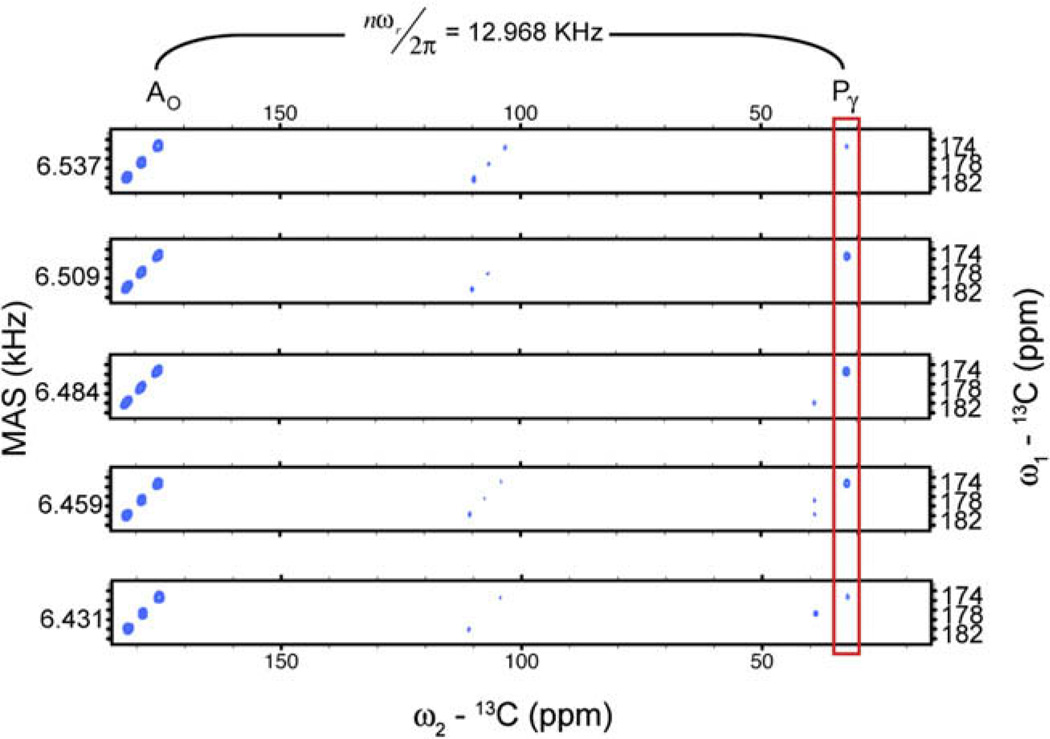
The crosspeak volumes (see Fig. 7) were extracted by automated fitting to two-dimensional Gaussians using NMRPIPE [26]. All data points in each 2D-slice were normalized to the carbonyl intensities of reference experiments conducted at identical spinning frequencies with zero mixing time.
Fig. 7.
R2W spectra at five spinning frequencies. The Pγ crosspeak intensifies as the spinning frequency is swept the R2 condition between the spin pair.
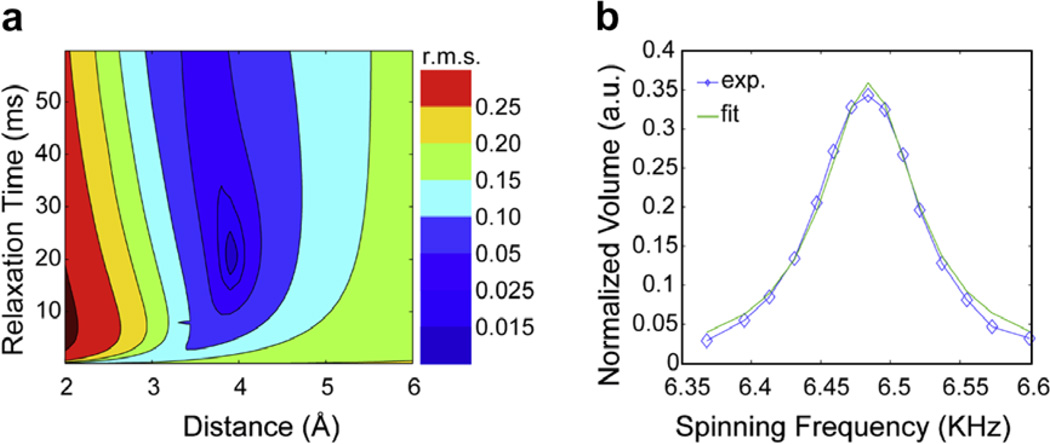
A Gaussian pulse of 250 µs duration was used, followed by a z-filter of 750 µs to select the carbonyl region of the spectrum. 32 t1 points were recorded with an increment of 250 µs and 16 scans per point with a recycle delay of 3.1 s leading to a total experimental time of about 50 min per two-dimensional slice. A series of experiments were performed as a function of the sample spinning frequency under a constant mixing time of 30 ms (see Fig. 5).
Fig. 5.
(a) Contour plot of the root mean square deviation between experimental and simulated R2W profiles for PO to Pγ as a function of distance and relaxation. (b) Magnetization transfer from Po to Pγ in R2W as a function of spinning frequency. The experimental data is shown in blue, and the fit in green. (For interpretation of colour mentioned in this figure, the reader is referred to the web version of this article.)
4.5. DANTE-REDOR
DANTE-REDOR measurements were carried out on a custom assembled NMR spectrometer with a Discovery console (Tecmag; Houston, TX), 14.1 T magnet (Magnex; Oxford, England), and a 39-channel matrix shim system (Resonance Research, Inc.; Billerica, MA), operating at a proton frequency of 600.377 MHz (150.987 MHz for 13C). A doubly tuned 4 mm magic angle spinning probe (Doty Scientific; Columbia, South Carolina) was employed.
The DANTE-REDOR pulse sequence is described in [10] and follows after DANTE-REDOR based work of Kaustov [28] and Schaefer [29]. This sequence adapts the orientation information available in a specifically labeled sample to a uniformly labeled system. By employing a DANTE pulse train as the selective pulse in FS-REDOR, the orientation information in REDOR sidebands is retained. Thus at the end of the REDOR evolution time, the 13C spin-echo intensity is modulated only by dipolar couplings to the selected 15N and the resulting REDOR curve can be fit to the analytical expression for an isolated spin pair. The effects of proton couplings are attenuated with Small Phase Incremental Alternation (SPINAL-64) decoupling [25]. The proton decoupling field was 100 kHz, the 15N REDOR π-pulse width was 16 us and the DANTE π-pulse trains consisted of 33 2 µs rotor synchronized pulses.
Fourier transforms were applied with no apodization and zero filling to twice the acquisition time of 15.36 ms. Only zero order phase correction was used. Dephasing (S) and full-echo (S0) peak areas were determined by integrating the center band and all observed sidebands. For distance determinations, the sum of all bands in the S and S0 spectra were used. For extraction of orientation information, the dephasing of each individual band was compared.
Distances were determined by fitting the experimentally observed REDOR curve to the analytical expression, ΔS/S0(τ) = λ[1 − 〈cos(ωCNτ)〉]. ΔS/S0 = 1 − S/S0, where S and S0 are the dipolar dephasing and reference intensities, respectively. The coupling ωCN is a function of the dipolar coupling constant, bCN, and 〈〉 indicates the powder average over a uniform distribution of crystallite orientations. The scaling factor λ accounts for 13C spins without a neighboring 15N spin, which is a result of imperfect labeling and dilution of the labeled compound in natural abundance material. In the DANTE-REDOR experiment, λ also accounts for imperfect inversion of the 15N spin magnetization by the selective pulse and decay of coherence due to insufficient decoupling.
For each distance measurement, the weighted χ2, , was minimized to fit the data. . In this expression, wi is the inverse uncertainty in each point squared (σ−2). Since the noise in each data point is approximately constant, wi was easily determined by propagation of error. In the minimization of , the coupling constant, bCN, was varied freely and the amplitude scaling factor λ was varied within the narrow range of values determined for strong couplings, for which this parameter can be easily determined. The reported uncertainties are at the 95% confidence level according to the following procedure [30]. First, bCN and λ are varied to determine the best-fit dipolar coupling, . Next, several trial bCN values are selected about the best-fit, and the data are re-fit by optimizing λ within the previously defined range. The uncertainties are represented by values that differ from by where F(ν) is a constant that depends on the number of degrees of freedom, ν, and the confidence level. For the case of 95% confidence, and 2 degrees of freedom, F(ν) = 19.
Each best-fit bCN value was linearly scaled by dividing by 0.95 in order to account for thermal motion at 298 K. Internuclear C–N distances are related to the dipolar coupling constants by the equation [31].
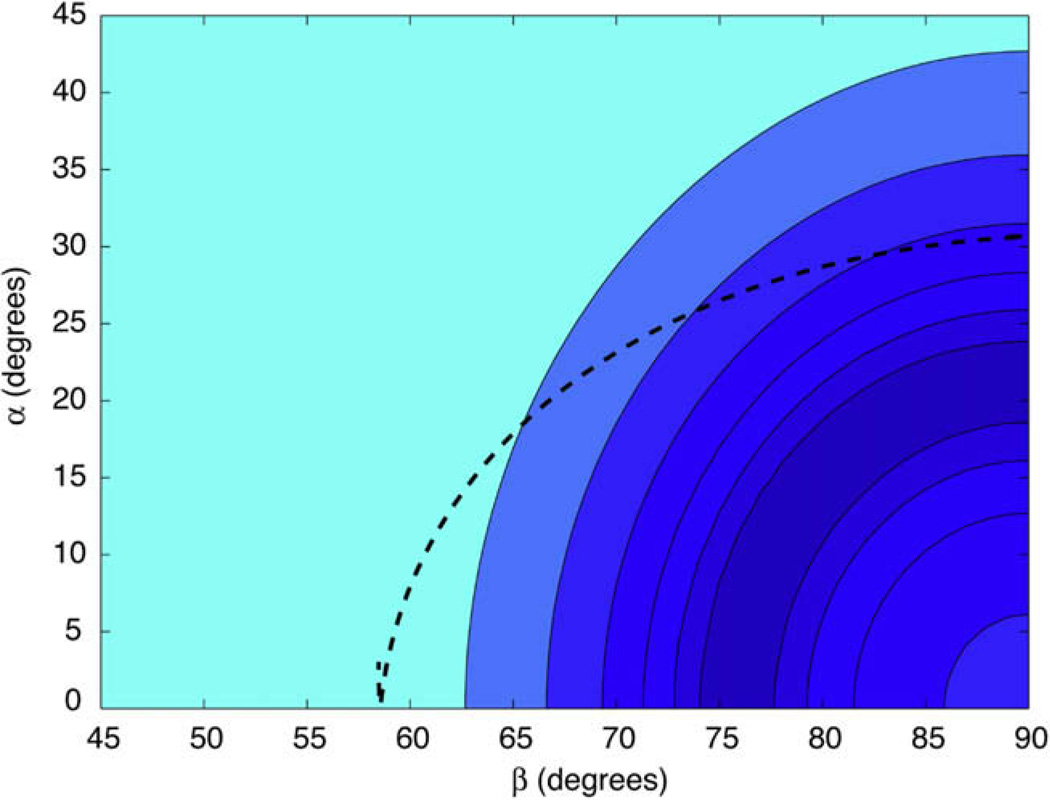
The orientation of the nitrogen in the CSA principle axis system was determined by fitting the experimentally observed REDOR curves (see Fig. 6) of each sideband to numerical simulations explained previously [32]. The weighted χ2, , was minimized to fit the data. , where the sum is taken over spectral bands N, and mixing times i. As before, wi is the inverse uncertainty in each point squared (σ−2). The uncertainties in the dipolar and azimuthal angles alpha and beta are represented by values that differ from by where F(ν) is a constant that depends on the number of degrees of freedom, ν, and the confidence level. The best-fit dipolar coupling is used in this determination, so this is really a three-parameter fit. However, since the REDOR dephasing is approximately linear in the beginning of the curve (before the first extremum in the REDOR curve, where we fit the data) a small error in the dipolar coupling does not significantly alter the position of the best-fit orientation, and thus can be treated as a two parameter fit.
Fig. 6.
The χ2 surface for azimuthal (α) and polar (β) angles of the CSA-dipole orientation with each contour level two times the previous level. Labeled APG was diluted to 10% in natural abundance APG and data were collected at 14.1 Tesla with an MAS spinning frequency of 8929 Hz. The superimposed dashed line shows the allowed azimuthal and polar angles corresponding to a rotation about the terminal ψ dihedral angle, with the best-fit at α = 31°, β = 90°.
Acknowledgments
A.B.B. was supported through an NSF graduate research fellowship. We thank Marvin Bayro for supplying the pulse sequence for the NCCN experiment and Jozef Lewandowski and Matthew Eddy for helpful discussions, and Matthias Zeller for the crystal structure. This research was supported by the National Institutes of Health (EB003151 and EB002026).
References
- 1.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J. Am. Chem. Soc. 2004;126:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 2.Takegoshi K, Nakamura S, Terao T. C-13–H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 3.Lewandowski JR, de Paepe G, Griffin RG. Proton assisted insensitive nuclei cross polarization. J. Am. Chem. Soc. 2007;129:728–729. doi: 10.1021/ja0650394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J. Magn. Res. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Bennett AE, Rienstra CM, Griffiths JM, Zhen WG, Lansbury PT, Griffin RG. Homonuclear radio frequency-driven recoupling in rotating solids. J. Chem. Phys. 1998;108:9463–9479. [Google Scholar]
- 6.Gregory DM, Mitchell DJ, Stringer JA, Kiihne S, Shiels JC, Callahan J, Mehta MA, Drobny GP. Windowless dipolar recoupling – the detection of weak dipolar couplings between spin-1/2 nucleic with large chemical-shift anisotropies. Chem. Phys. Lett. 1995;246:654. [Google Scholar]
- 7.Rienstra CM, Tucker-Kellogg L, Jaroniec CP, Hohwy M, Reif B, McMahon MT, Tidor B, Lozano-Perez T, Griffin RG. De novo determination of peptide structure with solid-state magic-angle spinning NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 2002;99:10260–10265. doi: 10.1073/pnas.152346599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramachandran R, Ladizhansky V, Bajaj VS, Griffin RG. 13C–13C rotational resonance width distance measurements in uniformly 13C-labeled peptides. J. Am. Chem. Soc. 2003;125:15623–15629. doi: 10.1021/ja037761x. [DOI] [PubMed] [Google Scholar]
- 9.Costa P, Sun B, Griffin RG. Rotational resonance NMR: separation of dipolar coupling and zero quantum relaxation. J. Magn. Res. 2003;164:92–103. doi: 10.1016/s1090-7807(03)00083-1. [DOI] [PubMed] [Google Scholar]
- 10.Andreas LB, Mehta AK, Mehta MA. Determination of global structure from distance and orientation constraints in biological solids using solid-state NMR spectroscopy. J. Am. Chem. Soc. 2007;129:15233–15239. doi: 10.1021/ja074789q. [DOI] [PubMed] [Google Scholar]
- 11.van der Wel PCA, Eddy MT, Ramachandran R, Griffin RG. Targed 13C-13C distance measurements in a microcrystalline protein via J-decoupled rotational resonance width measurements. Phys. Chem. Chem. Phys. 2009;10 doi: 10.1002/cphc.200900102. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran R, Lewandowski JR, van der Wel PCA, Griffin RG. Multipole-multimode Floquet Theory of rotational resonance width experiments: 13C–13C distance measurements in uniformly labeled solids. J. Chem. Phys. 2006;124:214107. doi: 10.1063/1.2194905. [DOI] [PubMed] [Google Scholar]
- 13.Costa PR, Gross JD, Hong M, Griffin RG. Solid-state NMR measurement of Psi in peptides: a NCCN 2Q-heteronuclear local field experiment. Chem. Phys. Lett. 1997;280:95–103. [Google Scholar]
- 14.Wu S, Declercq JP, Tinant B, Vanmeerssche M. Crystal-structure and conformation of short linear peptides.6. l-Alanyl-l-prolyl-glycine monohydrate. Bull. Soc. Chim. Belg. 1987;96:515–520. [Google Scholar]
- 15.Levitt MH, Raleigh DP, Creuzet F, Griffin RG. Theory and simulations of homonuclear spin pair systems in rotating solids. J. Chem. Phys. 1990;92:6347–6364. [Google Scholar]
- 16.Ramachandran R, Griffin R. Multipole-multimode Floquet Theory in nuclear magnetic resonance. J. Chem. Phys. 2005;122:164502. doi: 10.1063/1.1875092. [DOI] [PubMed] [Google Scholar]
- 17.Bak M, Schultz R, Vosegaard T, Nielsen NC. Specification and visualization of anisotropic interaction tensors in polypeptides and numerical simulations in biological solid-state NMR. J. Magn. Res. 2002;154:28–45. doi: 10.1006/jmre.2001.2454. [DOI] [PubMed] [Google Scholar]
- 18.Herzfeld J, Berger AE. Sideband intensities in NMR spectra of samples spinning at the magic angle. J. Chem. Phys. 1980;73:6021–6030. [Google Scholar]
- 19.Ye C, Fu R, Hu J, Hou L, Ding S. Carbon-13 chemical shift anisotropies of solid amino acids. Magn. Reson. Chem. 1993;31:699–704. [Google Scholar]
- 20.Bayro MJ, Huber M, Ramachandran R, Davenport TC, Meier BH, Ernst M, Griffin RG. Dipolar truncation in magic-angle spinning NMR recoupling experiments. J. Chem. Phys. 2009;130:114506. doi: 10.1063/1.3089370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse- Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 23.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld J-U, Xu J, Kuang J, Kirschner MW, Fischer G, Cantley LC, Lu KP. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 24.Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG. Fivefold symmetric homonuclear dipolar recoupling in rotating solids: application to double quantum spectroscopy. J. Chem. Phys. 1999;110:7983–7992. [Google Scholar]
- 25.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 1995;103:6951. [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe – a multidimensional spectral processing system based on unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Veshtort M, Griffin RG. SPINEVOLUTION: a powerful tool for simulations of solid and liquid state NMR experiments. J. Magn. Res. 2006;178:248–282. doi: 10.1016/j.jmr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Kaustov L, Kababya S, Belakhov V, Baasov T, Shoham Y, Schmidt A. Inhibition mode of a bisubstrate inhibitor of KDO8P synthase: a frequency-selective REDOR solid-state and solution NMR characterization. J. Am. Chem. Soc. 2003;125:4662–4669. doi: 10.1021/ja028688y. [DOI] [PubMed] [Google Scholar]
- 29.Klug CA, Zhu W, Tasaki K, Schaefer J. Orientational order of locally parallel chain segments in glassy polycarbonate from 13C–13C dipolar couplings. Macromolecules. 1997;30:1734–1740. [Google Scholar]
- 30.Shoemaker D, Garland C, Steinfeld J, Nibler J. Least-Squares fitting procedures. Experiments in Physical Chemistry. 1989 [Google Scholar]
- 31.Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Frequency selective heteronuclear dipolar recoupling in rotating solids: accurate C-13–N-15 distance measurements in uniformly C-13, N-15-labeled peptides. J. Am. Chem. Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- 32.O’connor RD, Schaefer J. Relative CSA-dipolar orientation from REDOR sidebands. J. Magn. Res. 2002;154:46–52. doi: 10.1006/jmre.2001.2461. [DOI] [PubMed] [Google Scholar]