Abstract
Once overlooked as an evolutionary vestige, the primary cilium has recently been the focus of intensive studies. Mounting data show that this organelle is a hub for various signaling pathways during vertebrate embryonic development and pattern formation. However, how cilia form and how cilia execute the sensory function still remain poorly understood. Cilia dysfunction is correlated with a wide spectrum of human diseases, now termed ciliopathies. Various small GTPases, including the members in Arf/Arl, Rab, and Ran subfamilies, have been implicated in cilia formation and/or function. Here we review and discuss the role of one particular group of small GTPase, Arf/Arl, in the context of cilia and ciliopathy.
Keywords: Arf/Arl, cilia, ciliopathies
Cilia and Human Ciliopathies
Cilia emanate from the surface of most eukaryotic cells and perform diverse roles in motility and sensation. Cilia are microtubule-based organelles and serve as specialized subcellular compartments that harbor ciliary structural proteins, sensory receptors and signaling molecules. Evolutionarily conserved intraflagellar transport (IFT), a motor-mediated bidirectional transport along the axoneme, builds and maintains all cilia [Rosenbaum, 2002]. The IFT particle is composed of two subcomplexes (IFT-A and IFT-B). In a simple model, anterograde transport is regulated by kinesin-2, whereas dynein regulates retrograde transport. Sensory transduction capabilities of cilia are highly conserved in all ciliated organisms. Polarized cells utilize primary cilia to receive various environmental stimuli that are converted into physiological responses. For example, primary cilia of kidney nephrons and liver cholangiocytes use polycystins to sense liquid flow and convert this information into downstream signals [Masyuk et al., 2006; Nauli et al., 2003]. Primary cilia also regulate the transduction of various signals critical for embryonic development and pattern formation, such as Sonic hedgehog (Shh), Wnt, and PDGF pathways [Goetz and Anderson, 2010]. With rapid advancements during the past decade in the positional cloning of human disease genes, a wide variety of genetic disorders, such as autosomal dominant polycystic kidney disease (ADPKD), Bardet-Biedl syndrome (BBS), Joubert syndrome (JS), nephronophthisis (NPHP), Meckel-Gruber syndrome (MKS), and autosomal recessive polycystic kidney disease (ARPKD), have been characterized molecularly as cilia-related diseases, now known collectively as ciliopathies [Badano et al., 2006].
The Arf /Arl Family
Arf/Arls are enzymatically active small GTPases that belong to the Ras superfamily. The family of mammalian Arf/Arl GTPases has 29 members, including 6 Arfs, Sar1 and over 20 Arf-like proteins (Arls) [Gillingham and Munro, 2007]. Members of the Arf family were first identified as factors required for cholera toxin-dependent ADP-ribosylation of the heterotrimeric Gs protein [Kahn and Gilman, 1986]. The ADP-ribosylation factor (Arf) activity is shared by 6 closely related proteins Arf1–6, but not by Arls and Sar1, which were identified mainly by sequence alignment and structure similarity. The endogenous functions of most characterized Arf GTPases are involved in membrane traffic or cytoskeleton organization. Less is known about the in vivo role of Arls [Gillingham and Munro, 2007].
One shared, also unique, structural feature for the Arf/Arl family is the N-terminal amphipathic helix, which is indispensable for membrane insertion upon Arf/Arls binding to GTP. Posttranslational modifications are also critical for the membrane recruitment and biological activity of all Arfs and, at least, some Arls. For example, all Arfs and several Arls are found to be myristoylated at N-terminus during translation; and at least two Arls, ARFRP1 (ARF-related protein 1) and Arl8, are acetylated at N-terminus [Donaldson and Jackson, 2011]. Remarkably, the membrane insertion mechanism for Rabs and other small GTPases is the posttranslational modification at C-terminal cystines by prenyl groups. Thus, in contrast to Rabs and other small GTPases that contain a long linker between the C-terminal membrane-insertion domain and the N-terminal GTPase domain, Arf/Arls possess a short linker due to that the lipid modification or amphiphathic helix occurs near the GTPase domain. This structural characteristic suggests that the associated effectors of Arf/Arls are closer to the membrane than those of other small GTPases. The evidences that most identified Arf/Arl effectors are often coat proteins or lipid-modified enzymes further support this assumption [Gillingham and Munro, 2007].
Another feature for Arf/Arl family members is that they usually dissociate from membrane spontaneously upon GTP hydrolysis and do not require any GDP dissociation inhibitor (GDI) proteins [Donaldson and Jackson, 2011]. Interestingly, some Arfs can still bind to specific membrane in their GDP-bound forms [Donaldson and Jackson, 2011], indicating that GDP-bound Arf/Arls might have their specific effectors and play unknown roles in certain cellular processes.
Among all the Arf/Arl proteins, Arf4, Arl3, Arl6 and Arl13b have been implicated to play distinct roles in the context of cilia (Table 1). Here we will review them and their potential functional crosstalks within cilia.
Table 1.
Cilary Arf/Arl proteins and known effectors
| Small GTPases |
Organism | Gene Name | Localization | Function | GAP | Effectors |
|---|---|---|---|---|---|---|
| Arl13b |
C. elegans Zebrafish Human mouse |
ARL-13 arl13b ARL13B Arl13b |
Middle segment of cilia Cilia |
Cilia formation Ciliary protein targeting |
? | HDAC-6? |
| Arl6 |
C. elegans Human Mouse |
ARL-6/BBS-3 ARL6/BBS3 Arl6/Bbs3 |
Ciliated cell and cilia Cilia (ATP-bound form) |
Cilia formation Ciliary protein targeting |
? | BBSome |
| Arl3 |
Leishmania C. elegans Mouse |
Arl3 ARL-3 Arl3 |
Flagella Ciliated cell and cilia Cilia |
Flagella and cilia formation Ciliary protein targeting |
RP2 | UNC119 HDAC-6 |
| Arf4 |
X. laevis. Human Mouse |
arf4 ARF4 Arf4 |
Golgi | ciliary protein targetting | ASAP1 | ? |
Arf4 and TGN sorting of ciliary proteins
Arf4 is 90% identical to Arf5 and 80% identical to Arf1/2/3, and these five proteins are equally capable of recruiting COPI coats or the clathrin coat adaptor AP-1 to Golgi membrane [Nachury et al., 2011]. Of all 6 Arf proteins, Arf4 is the only reported one that shows cilia–related function. The physical interaction between Arf4 and the C-terminal VXPX motif of rhodopsin is required for the ciliary sorting of rhodopsin in photoreceptor cells [Deretic et al., 2005]. Further studies revealed that Arf4 forms a complex with Arf GTPase activating protein (GAP) ASAP1, another small GTPase Rab11, and Rab11 effector FIP3 to regulate the budding and sorting of rhodopsin-containing vesicles from Trans-Golgi-Network (TGN) to photoreceptor cilia [Mazelova et al., 2009]. As expected, expression of an Arf4 dominant-negative (DN) mutant resulted in rhodopsin mislocalization and retinal degeneration in Xenopus [Mazelova et al., 2009].
Interestingly, the VXPX sorting motif identified in rhodopsin exists in several other ciliary membrane sensory receptors, including polycystin-2 (PC2) [Geng et al., 2006] and polycystin-1 (PC1) [Ward et al., 2011], two proteins mainly affected in ADPKD disease. The VXPX domain of PC1 is required for the formation of a multimeric complex that contains Arf4, ASAP1, and PC1 [Ward et al., 2011]. Furthermore, a trafficking machinery consisting of Arf4, ASAP1, Rab6, and Rab11 mediates the transportation of PC1 from TGN to the ciliary base [Ward et al., 2011]. These data are consistent with rhodopsin sorting model and supportive for the assumption that a conserved Arf4-based transport machinery is used for sorting TGN cargoes into ciliary targeting vesicles.
Although the ciliary transportation of PC2 was reported to exit from cis-Golgi but not TGN for cilia targeting [Hoffmeister et al., 2011], the facts that the N-terminal ciliary targeting motif RVXP of PC-2 strongly interacts with Arf4 [Ward et al., 2011] and that Arf4 also presents on cis-Golgi surface [Volpicelli-Daley et al., 2005] hint the possibility that there might be another Arf4 mediated sorting pathway at cis-Golgi. Additionally, the clathrin adaptor AP-1, which is identified as one of Arf4 effectors, also plays a role in regulating the proper ciliary localization of odorant receptor ODR-10 in C. elegans [Dwyer et al., 2001]. No VXPX domain can be found in ODR-10 protein. It would be interesting to examine whether Arf4 might act through AP-1 and clathrin for the selective ciliary targeting of certain ciliary sensory receptors.
Exactly how Arf4 gains the ability to specifically facilitate the ciliary sorting of the ciliary sensory receptors at the Golgi is unclear. However, the discovery that rhodopsin recruits Arf4 and initiates the formation of sorting complex indicates that this is a cargo-dependent event. The maintenance and function of cilia depend on the proper localization of hundreds of cilia-specific proteins, including structural proteins, membrane proteins and signaling molecules. After these proteins are synthesized and transported into the Golgi, little is known about how they gain access to cilia-targeting vesicles. Among a battery of small GTPases implicated in this particular sorting pathway [Li and Hu, 2011], Arf4 appears to act at the very beginning point, the budding step at TGN. Thus, it would be interesting to examine the protein profile in those Arf4-positive cilia-targeting vesicles to see whether the binding between transmembrane protein rhodopsin (or PC1) and Arf4-containing sorting machinery provides a mechanism for delivering other ciliary proteins in the same vesicles.
Arl3 in ciliogenesis and ciliary transport
A comparative genomic study indicated that Arl3 and Arl6 are present only in the ciliated organisms [Avidor-Reiss et al., 2004]. Arl3−/− mice were small, sick, and died by 3 weeks of age [Schrick et al., 2006]. Although Arl3 has not been identified as one of human ciliopathy loci, Arl3−/− mice exhibit a pleiotropic of ciliopathy phenotypes characterized by impaired photoreceptor development, cysts formation in the kidney, liver and pancreas. The Leishmania Arl3 ortholog targets to the flagellum and is essential for flagellum biogenesis [Cuvillier et al., 2000]. However, knockdown Arl3 by siRNA in mammalian systems only leads to defective ciliary transport but grossly normal cilia morphology [Lai et al., 2011].
Interestingly, arl-3 knockout C. elegans show normal cilia length but defective localization of ciliary sensory receptors ([Li et al., 2010] and unpublished data in our lab). As expected, overexpression of a dominant-active (DA) ARL-3 mutant compromises the normal ciliogenesis in C. elegans, indicating that ARL-3 might be a negative regulator for ciliogenesis [Li et al., 2010]. This also explains why depletion of Arl3 in either mammalian systems or C. elegans does not affect ciliogenesis. Further analyses suggested that ARL-3 is involved in regulating the association between IFT-B subcomplexes and IFT-B-associate kinesin motor OSM-3/KIF17. It is interesting that ARL-3 itself does not show any IFT movement in cilia. So the open question remains is how a non-IFT protein affects the IFT integrity and negatively regulates the ciliogenesis. We pinpointed one potential effector of ARL-3 in ciliogenesis to HDAC-6, but the mechanism behind this process is still poorly understood.
In mammalian photoreceptors, Arl3 localizes predominantly to the connecting cilium and interacts with the X-linked retinitis pigmentosa protein RP2, a GAP for Arl3 [Grayson et al., 2002]. RP2 is proposed to regulate Arl3 small GTPase activity and be involved in transporting and docking the vesicles from the Golgi to the base of the photoreceptor connecting cilia [Evans et al., 2010]. In primary cilium, the myristoylated ciliary cargo, such as NPHP3, is first transported by UNC119 to the ciliary base by unknown mechanism, and then inside the cilium, the binding between GTP-bound Arl3 and UNC119 somehow releases the myristoylated cargo into the appropriate ciliary membrane domain. Next, RP2 switches Arl3 from GTP-bound to GDP-bound and this leads to UNC119 dissociation and its exit from cilia [Wright et al., 2011].
Arl6/Bbs3 and ciliary receptors targeting
Arl6/Bbs3 was the first member of Arf/Arl family identified in human ciliopathy. It is one of a set of human genes that when mutated cause Bardet-Biedl syndrome (BBS) [Chiang et al., 2004; Fan et al., 2004]. BBS is a genetically heterogeneous human disorder characterized by obesity, polydactyly, mental retardation, retinal degeneration, renal cyst, and learning disabilities [Zaghloul and Katsanis, 2009]. Structural analyses demonstrated that the altered residues found in BBS3 patients cluster in or around Arl6 GTP binding domain and affect nucleotide binding capacity [Kobayashi et al., 2009; Wiens et al., 2010]. Arl6−/− mice develop BBS-associated phenotypes [Zhang et al., 2011]. In cultured mammalian cells, Arl6 GTPase activity is required for both ciliogenesis and Wnt signaling [Wiens et al., 2010]. Studies in C. elegans show that arl-6−/− worms still possess normal ciliogenesis but defective cilia sensory function. It’s unclear why other BBS proteins, but not ARL-6/BBS-3, are involved in regulating IFT integrity and ciliogenesis in C. elegans [Blacque et al., 2004]. However, recent studies in Arl6−/− knockout mice did suggest that Arl6/Bbs3 has both common BBS-associated and BBS3-unique functions [Zhang et al., 2011].
Eight other BBS proteins, including BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, BBS9 and BBIP10 form a protein complex known as the BBSome [Loktev et al., 2008; Nachury et al., 2007]. The BBSome shares common structural features with COPI, COPII and clathrin coats and is the major effector of Arl6/Bbs3 in sorting membrane proteins to cilia [Jin et al., 2010]. Loss of Arl6 does not affect BBSome formation but disrupts normal localization of melanin concentrating hormone receptor 1 to ciliary membranes [Zhang et al., 2011]. Additionally, Arl6 and BBSome are required for the ciliary entry of somatostatin receptor 3 [Jin et al., 2010]. It was proposed that GTP-bound Arl6 recruits the BBSome and BBSome-associated cargo complex to membrane and assembles a polymerized coat-cargo complex that can be dragged through the periciliary diffusion barrier [Jin et al., 2010]. Interestingly, other than the ciliary entry of membrane receptors, loss of Arl6 can also affect the retrograde transport of sensory receptor Smoothened inside cilia, which leads to modest decrease in ciliary Shh signaling [Zhang et al., 2011]. Studies in our lab also confirmed that various sensory receptors consistently mislocalize in arl-6 knockout worms (unpublished results). Taken together, these evidences suggest that the role of Arl6 in the context of cilia is conserved and the variety of symptoms found in BBS patients is likely due to the compromised ciliary targeting or removal of various sensory receptors and/or signaling molecules.
A fact need to be pointed out is that Arl6 is the only Arf/Arl small GTPase that was found to move inside the cilium. C. elegans ARL-6 moves at typical IFT rates in both anterograde and retrograde directions [Fan et al., 2004]. However, our observations revealed that ARL-6 moves much less frequently when compared with IFT structural components or other BBSome components, indicating that ARL-6 might not be IFT integral protein but rather a cargo or cargo binding protein, which may just nonconstitutively attach to the IFT machinery. Why ARL-6 moves together with IFT machinery and whether this motility requires the GTPase activity need further investigations.
Arl13b in ciliogenesis, ciliary transport and signaling
Arl13b was initially cloned as the novel cystic kidney gene scorpion (sco) in zebrafish [Sun et al., 2004]. Knockdown of Arl13b leads to multiple cilia-associated phenotypes in zebrafish [Duldulao et al., 2009]. Unlike other small GTPases, Arl13b is an atypical small GTPase with an elongated C-terminus contains a coiled-coil domain and a proline-rich domain (PRD). Mutations in ARL13B has been identified in two families with the classical form of Joubert syndrome [Cantagrel et al., 2008], an autosomal recessive disorder characterized by congenital cerebellar ataxia, hypotonia, oculomotor apraxia, and mental retardation, cystic kidney, polydactyly. Arl13b null mice (hennin) mimic the mutant phenotypes in Joubert syndrome patients [Caspary et al., 2007], and show coupled defects in cilia structure and cilia signaling, including Shh signaling, BMB signaling, and abnormally expressed Wnt ligands [Caspary et al., 2007; Horner and Caspary, 2011; Larkins et al., 2011]. The cilia in Arl13b hennin mouse embryos are only one-half the length of wild-type and exhibit an unique mutant phenotype in that the B-tubules of the outer doublet microtubules are not closed. The unclosed B-tubule defect is also observed in arl-13 worm mutants, indicative of the conservation for Arl13b cilia-related function [Li et al., 2010].
Due to the lethality of hennin mice, C. elegans is used as an alternative model to gain mechanistic insights into the in vivo function of Arl13b protein. Studies from the Blacque’s laboratory and our laboratory revealed a role for ARL-13 at ciliary membranes, where it regulates ciliary transmembrane protein localizations and anterograde IFT assembly stability [Cevik et al., 2010; Li et al., 2010]. Moreover, our laboratory’s work showed that ARL-3 acts antagonistically with ARL-13 to regulate IFT integrity and ciliogenesis [Li et al., 2010]. Specifically, the absence of ARL-13 causes destabilized IFT complex and IFT-A and IFT-B tend to dissociate in anterograde IFT transport; whereas ARL-3 depletion can partially rescue ciliogenesis defects in arl-13 mutants by stabilizing the association between IFT-A and IFT-B [Li et al., 2010]. We will discuss this more in the following section.
One unique feature for ARL-13 is its C-terminal PRD, which is indispensable for its specific targeting to the doublet segment of the cilium in C. elegans. PRD and its major interacting Src homology 3 (SH3) domain have been experimentally proven to be involved in various protein-protein interactions. Interestingly, PRD is found in many microtubule-associated proteins (MAPs), such as MAP2, Tau, MAP4, and required for the activities of MAPs in microtubule binding and assembly [Li et al., 2010]. Additionally, the PRD of the GTPase dynamin regulates its effector binding and microtubule association [Hamao et al., 2009]. In our studies, overexpression of ARL-13 PRD domain could induce a dominant negative effect on ciliogenesis [Li et al., 2010]. Overexpression of an ARL-13 truncation that lacks the PRD domain but preserves the whole GTPase domain could also cause ciliogenesis defects in wild-type animals [Li et al., 2010]. These two observations suggest that the PRD of ARL-13 is critical for not only its specific ciliary targeting but also its function in cilia formation. It would be of great interest to characterize the ciliary protein(s) that bind ARL-13 PRD domain.
Potential functional crosstalk among Arf/Arls in the context of cilia
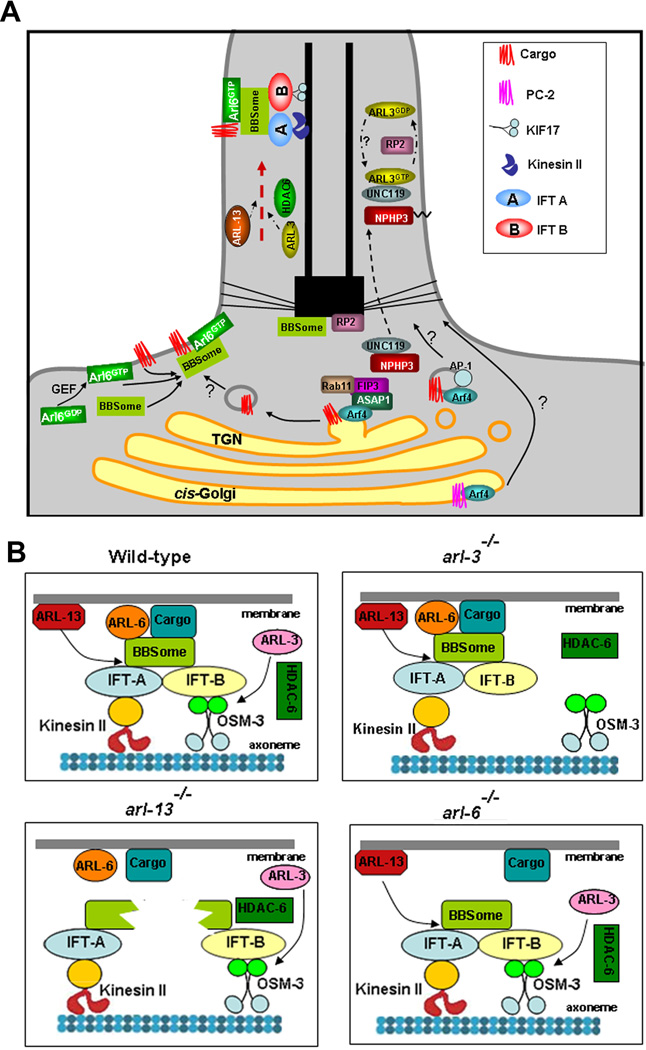
Numerous evidences show that Arf/Arl proteins often function in pair or as a network in one cellular process [Donaldson and Jackson, 2011]. Arf4, Arl3, Arl6 and Arl13b are all involved in targeted transport of membrane proteins to the cilia (Figure 1). The three Arl proteins also affect diverse signaling pathways. At least for Shh signaling pathways, the relative levels of Shh signaling components in cilia are regulated directly or indirectly by all three ciliary Arl proteins. It would be interesting to examine whether these Arf/Arls could possibly crosstalk in their functional pathways.
Figure 1.
Arl3, Arl6, and Arl13b are the only three Arl small GTPases implicated in human ciliopathies or vertebrate ciliopathy models. Not like Arf4, which mainly localizes on Golgi, Arl3, Arl6, and Arl13b all localize inside the cilium. Our previous work demonstrated that ARL-3 acts through an HDAC6-dependent pathway to partially rescue the ciliogenesis defect in arl-13 mutant [Li et al., 2010], indicating ARL-13 acts antagonistically with ARL-3 in cilia formation. HDAC6 belongs to histone deacetylase family and can deacetylates not only histone, but also α-tubulin, HSP90 and coractin. Activation of HDAC6 was found to promote cilia disassembly while loss of HDAC6 activity selectively stabilizes cilia in human epithelial cells [Pugacheva et al., 2007]. However, previous work and our data ruled out the possibilities of α-tubulin and HSP90 as the candidates in ARL-3-HDAC6 pathway in C. elegans [Fukushige et al., 1999; Li et al., 2010]. We propose that HDAC6 may affect the acetylation of an unknown adaptor protein in IFT complex. Interestingly, HDAC6 physically interacts with BBIP10, a subunit of the BBSome that binds to both IFT-A and IFT-B, suggesting a potential functional crosstalk between Arl3, Arl13, and Arl6-BBSome in cilia [Loktev et al., 2008].
ARL-3 also has non-IFT related function in cilia. Overexpression of ARL-3 DA in arl-13 mutants produces a new defect in that some transition zones (TZs) at the ciliary base are totally missing [Li et al., 2010]. This striking phenotype is not due to the compromised IFT integrity in middle segments because previous TEM analyses showed that TZs can form normally in all IFT mutants examined in C. elegans [Perkins et al., 1986]. It’s known that ciliopathy NPHP and MKS proteins act together to establish the normal Y-link formation and the connection between the TZ and the ciliary membrane [Williams et al., 2011]. It would be interesting to examine if Arl-3 and Arl-13 function synergically in regulating the proper function and localization of NPHPs and MKSs at the TZ. Further understanding the crosstalk of the ciliary small Arls will definitely broaden our knowledge of cilia formation and function, as well as of ciliopathy pathogenesis.
Prospective
Our knowledge about the role of Arf/Arls in cilia is still in its infancy stage, with too many mechanistic insights missing. The fact that Arl3−/− and Arl13b−/− mice are embryonic lethal makes it difficult to study Arl function in living mammalian models. In contrast, worm arl-3, arl-6, and arl-13 single deletion mutants are all viable. Hence the simple model organisms will be invaluable in exploring the conserved pathways underlying ciliopathy Arls in their native ciliary environment.
Studies of Ras/Rho small GTPase families have demonstrated that numerous proteins bind to small GTPases to control their localization, activity, and downstream signaling pathways. The interactors include enzymes involved in posttranslational modifications, guanine nucleotide exchange factors (GEF), GTPase-activating factors (GAP), and effectors (such as adaptors, motors, kinases, and phosphatases) that bind specifically to the active forms of small GTPases [Bernards and Settleman, 2004; Cherfils and Chardin, 1999; Winter-Vann and Casey, 2005]. The BBSome serves as a major effector of Arl6 and mediates the ciliary cargo localization in an Arl6-GTP dependent way. HDAC-6 could be an effector for both ARL-13 and ARL-3. Functional identification and characterization of the binding partners of the three ciliary Arls as well as Arf4 would allow delineation of the poorly defined Arf/Arl signaling networks and would significantly enhance our understanding of their physiological roles in cilia biogenesis and function. In addition, the fact that small GTPases and many of their effectors are enzymatically active molecules makes these proteins promising targets for the therapeutic invention in future ciliopathy treatment.
Among all Arl effectors, the most important, also the least defined, ones are GEFs and GAPs. All small GTPases are regulated spatially and temporally through a cycle of GTP binding and hydrolysis, which mediates by GEFs and GAPs. However, no GEFs were identified for Arls and only two GAPs (RP2 and ELMOD2) are specific for Arl3 and Arl2/3, respectively. Arf GEF/GAPs are defined by the conserved signature domains and this has facilitated the identification process for other Arf GEF/GAPs from yeast to human. However, evidences are lacking in demonstrating that the GEF/GAPs of Arfs also work on Arl proteins. In addition, at least one GAP for Arl2, ELMOD2, which is also reported to possess GAP activity against Arf1 and Arf6, has no homology to typical Arf GAPs [Bowzard et al., 2007]. Since all three ciliary Arls are expressed in ciliated organisms, and most importantly, only in ciliated cells in model organisms, there is a good reason to speculate that the effectors of these ciliary Arls might also be cilia-specific. Focusing on the candidates in the existing ciliary proteome library could be a shortcut for the future characterization process.
Acknowledgement
Research in the authors’ laboratory is supported by the NIH/NIDDK, Mayo translation center for PKD, Mayo Clinic Center for Cell Signaling in Gastroenterology, and PKD Foundation.
Contract grant sponsor: NIH/NIDDK; Contract grant number: RO1-DK090038
Contract grant sponsor: NIH/NIDDK; Contract grant number: P30-DK084567-03
Contract grant sponsor: PKD Foundation; Contract grant number: 04YI09b
Footnotes
We apologize to the many individuals whose valuable contributions were unable to be cited due to space restrictions.
References
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Bernards A, Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 2004;14:377–385. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Blacque OE, Reardon MJ, Li C, McCarthy J, Mahjoub MR, Ansley SJ, Badano JL, Mah AK, Beales PL, Davidson WS, Johnsen RC, Audeh M, Plasterk RH, Baillie DL, Katsanis N, Quarmby LM, Wicks SR, Leroux MR. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowzard JB, Cheng D, Peng J, Kahn RA. ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J Biol Chem. 2007;282:17568–17580. doi: 10.1074/jbc.M701347200. [DOI] [PubMed] [Google Scholar]
- Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, Bertrand JY, Audollent S, Attie-Bitach T, Holden KR, Dobyns WB, Traver D, Al-Gazali L, Ali BR, Lindner TH, Caspary T, Otto EA, Hildebrandt F, Glass IA, Logan CV, Johnson CA, Bennett C, Brancati F, Valente EM, Woods CG, Gleeson JG. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Cevik S, Hori Y, Kaplan OI, Kida K, Toivenon T, Foley-Fisher C, Cottell D, Katada T, Kontani K, Blacque OE. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J Cell Biol. 2010;188:953–969. doi: 10.1083/jcb.200908133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Chardin P. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, Ferguson AL, Secrist J, Braun T, Casavant T, Stone EM, Sheffield VC. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3) Am J Hum Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvillier A, Redon F, Antoine JC, Chardin P, DeVos T, Merlin G. LdARL-3A, a Leishmania promastigote-specific ADP-ribosylation factor-like protein, is essential for flagellum integrity. J Cell Sci. 2000;113(Pt 11):2065–2074. doi: 10.1242/jcs.113.11.2065. [DOI] [PubMed] [Google Scholar]
- Deretic D, Williams AH, Ransom N, Morel V, Hargrave PA, Arendt A. Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4) Proc Natl Acad Sci U S A. 2005;102:3301–3306. doi: 10.1073/pnas.0500095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duldulao NA, Lee S, Sun Z. Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development. 2009;136:4033–4042. doi: 10.1242/dev.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer ND, Adler CE, Crump JG, L'Etoile ND, Bargmann CI. Polarized dendritic transport and the AP-1 mu1 clathrin adaptor UNC-101 localize odorant receptors to olfactory cilia. Neuron. 2001;31:277–287. doi: 10.1016/s0896-6273(01)00361-0. [DOI] [PubMed] [Google Scholar]
- Evans RJ, Schwarz N, Nagel-Wolfrum K, Wolfrum U, Hardcastle AJ, Cheetham ME. The retinitis pigmentosa protein RP2 links pericentriolar vesicle transport between the Golgi and the primary cilium. Hum Mol Genet. 2010;19:1358–1367. doi: 10.1093/hmg/ddq012. [DOI] [PubMed] [Google Scholar]
- Fan Y, Esmail MA, Ansley SJ, Blacque OE, Boroevich K, Ross AJ, Moore SJ, Badano JL, May-Simera H, Compton DS, Green JS, Lewis RA, van Haelst MM, Parfrey PS, Baillie DL, Beales PL, Katsanis N, Davidson WS, Leroux MR. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat Genet. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Siddiqui ZK, Chou M, Culotti JG, Gogonea CB, Siddiqui SS, Hamelin M. MEC-12, an alpha-tubulin required for touch sensitivity in C. elegans. J Cell Sci. 1999;112:395–403. doi: 10.1242/jcs.112.3.395. [DOI] [PubMed] [Google Scholar]
- Geng L, Okuhara D, Yu Z, Tian X, Cai Y, Shibazaki S, Somlo S. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J Cell Sci. 2006;119:1383–1395. doi: 10.1242/jcs.02818. [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson C, Bartolini F, Chapple JP, Willison KR, Bhamidipati A, Lewis SA, Luthert PJ, Hardcastle AJ, Cowan NJ, Cheetham ME. Localization in the human retina of the X-linked retinitis pigmentosa protein RP2, its homologue cofactor C and the RP2 interacting protein Arl3. Hum Mol Genet. 2002;11:3065–3074. doi: 10.1093/hmg/11.24.3065. [DOI] [PubMed] [Google Scholar]
- Hamao K, Morita M, Hosoya H. New function of the proline rich domain in dynamin-2 to negatively regulate its interaction with microtubules in mammalian cells. Exp Cell Res. 2009;315:1336–1345. doi: 10.1016/j.yexcr.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Hoffmeister H, Babinger K, Gurster S, Cedzich A, Meese C, Schadendorf K, Osten L, de Vries U, Rascle A, Witzgall R. Polycystin-2 takes different routes to the somatic and ciliary plasma membrane. J Cell Biol. 2011;192:631–645. doi: 10.1083/jcb.201007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner VL, Caspary T. Disrupted dorsal neural tube BMP signaling in the cilia mutant Arl13b hnn stems from abnormal Shh signaling. Dev Biol. 2011;355:43–54. doi: 10.1016/j.ydbio.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Gilman AG. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. J Biol Chem. 1986;261:7906–7911. [PubMed] [Google Scholar]
- Kobayashi T, Hori Y, Ueda N, Kajiho H, Muraoka S, Shima F, Kataoka T, Kontani K, Katada T. Biochemical characterization of missense mutations in the Arf/Arl-family small GTPase Arl6 causing Bardet-Biedl syndrome. Biochem Biophys Res Commun. 2009;381:439–442. doi: 10.1016/j.bbrc.2009.02.087. [DOI] [PubMed] [Google Scholar]
- Lai CK, Gupta N, Wen X, Rangell L, Chih B, Peterson AS, Bazan JF, Li L, Scales SJ. Functional characterization of putative cilia genes by high-content analysis. Mol Biol Cell. 2011;22:1104–1119. doi: 10.1091/mbc.E10-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins CE, Gonzalez Aviles GD, East MP, Kahn RA, Caspary T. Arl13b Regulates Ciliogenesis and the Dynamic Localization of Shh Signaling Proteins. Mol Biol Cell. 2011 doi: 10.1091/mbc.E10-12-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hu J. Small GTPases and cilia. Protein Cell. 2011;2:13–25. doi: 10.1007/s13238-011-1004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wei Q, Zhang Y, Ling K, Hu J. The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J Cell Biol. 2010;189:1039–1051. doi: 10.1083/jcb.200912001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loktev AV, Zhang Q, Beck JS, Searby CC, Scheetz TE, Bazan JF, Slusarski DC, Sheffield VC, Jackson PK, Nachury MV. A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell. 2008;15:854–865. doi: 10.1016/j.devcel.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Masyuk AI, Masyuk TV, Splinter PL, Huang BQ, Stroope AJ, LaRusso NF. Cholangiocyte cilia detect changes in luminal fluid flow and transmit them into intracellular Ca2+ and cAMP signaling. Gastroenterology. 2006;131:911–920. doi: 10.1053/j.gastro.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazelova J, Astuto-Gribble L, Inoue H, Tam BM, Schonteich E, Prekeris R, Moritz OL, Randazzo PA, Deretic D. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. Embo J. 2009;28:183–192. doi: 10.1038/emboj.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2011;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AE, Lu W, Brown EM, Quinn SJ, Ingber DE, Zhou J. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J. Intraflagellar transport. Curr Biol. 2002;12:R125. doi: 10.1016/s0960-9822(02)00703-0. [DOI] [PubMed] [Google Scholar]
- Schrick JJ, Vogel P, Abuin A, Hampton B, Rice DS. ADP-ribosylation factor-like 3 is involved in kidney and photoreceptor development. Am J Pathol. 2006;168:1288–1298. doi: 10.2353/ajpath.2006.050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- Volpicelli-Daley LA, Li Y, Zhang CJ, Kahn RA. Isoform-selective effects of the depletion of ADP-ribosylation factors 1–5 on membrane traffic. Mol Biol Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward HH, Brown-Glaberman U, Wang J, Morita Y, Alper SL, Bedrick EJ, Gattone VH, 2nd, Deretic D, Wandinger-Ness A. A conserved signal and GTPase complex are required for the ciliary transport of polycystin-1. Mol Biol Cell. 2011;22:3289–3305. doi: 10.1091/mbc.E11-01-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens CJ, Tong Y, Esmail MA, Oh E, Gerdes JM, Wang J, Tempel W, Rattner JB, Katsanis N, Park HW, Leroux MR. Bardet-Biedl syndrome-associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signaling. J Biol Chem. 2010;285:16218–16230. doi: 10.1074/jbc.M109.070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CL, Li C, Kida K, Inglis PN, Mohan S, Semenec L, Bialas NJ, Stupay RM, Chen N, Blacque OE, Yoder BK, Leroux MR. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol. 2011;192:1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer. 2005;5:405–412. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, Kwong M, Wang W, Pretorius PR, Sheffield VC, Sengupta P, Slusarski DC, Jackson PK. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011;25:2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J Clin Invest. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Nishimura D, Seo S, Vogel T, Morgan DA, Searby C, Bugge K, Stone EM, Rahmouni K, Sheffield VC. Bardet-Biedl syndrome 3 (Bbs3) knockout mouse model reveals common BBS-associated phenotypes and Bbs3 unique phenotypes. Proc Natl Acad Sci U S A. 2011;108:20678–20683. doi: 10.1073/pnas.1113220108. [DOI] [PMC free article] [PubMed] [Google Scholar]