Abstract
A significant proportion of warfarin dose variability is explained by variation in the genotypes of the cytochrome P450 CYP2C9 and the vitamin K epoxide reductase complex, VKORC1, enzymes that influence warfarin metabolism and sensitivity, respectively. We sought to develop an optimal pharmacogenetic warfarin dosing algorithm that incorporated clinical and genetic information. We enroled patients initiating warfarin therapy. Genotyping was performed of the VKORC1, –1639G>A, the CYP2C9*2, 430C>T, and the CYP2C9*3, 1075C>A genotypes. The initial warfarin dosing algorithm (Algorithm A) was based upon established clinical practice and published warfarin pharmacogenetic information. Subsequent dosing algorithms (Algorithms B and Algorithm C) were derived from pharmacokinetic / pharmacodynamic (PK/PD) modelling of warfarin dose, international normalised ratio (INR), clinical and genetic factors from patients treated by the preceding algorithm(s). The primary outcome was the time in the therapeutic range, considered an INR of 1.8 to 3.2. A total of 344 subjects are included in the study analyses. The mean percentage time within the therapeutic range for each subject increased progressively from Algorithm A to Algorithm C from 58.9 (22.0), to 59.7 (23.0), to 65.8 (16.9) percent (p = 0.04). Improvement also occurred in most secondary endpoints, which included the per-patient percentage of INRs outside of the therapeutic range (p = 0.004), the time to the first therapeutic INR (p = 0.07), and the time to achieve stable therapeutic anticoagulation (p < 0.001). In conclusion, warfarin pharmacogenetic dosing can be optimised in real time utilising observed PK/PD information in an adaptive fashion.
Clinical Trial Registration
ClinicalTrials.gov (NCT00401414)
Keywords: Pharmacogenetics, warfarin, clinical trial
Introduction
Warfarin is the most widely used anti-thrombotic agent. A narrow therapeutic window, however, limits its clinical utility, with excess thrombotic events occurring at subtherapeutic doses, and excess bleeding events occurring at supertherapeutic doses (1). Also, warfarin has about a 20-fold dose-response variability, with only one-fifth of this dose variability predicted by clinical factors (2, 3). Pharmacogenetics, the study of the effect of genetic variation on pharmacotherapy, may improve the efficacy and safety of drugs whose therapeutic response or metabolic disposition are affected by genetic variation; warfarin is one such drug (4-6).
The S-enantiomer of warfarin is principally metabolised by the hepatic cytochrome P450, family 2, subfamily C, polypeptide 9 (CYP2C9) enzyme. Genetic variation in this enzyme impairs the metabolism of the S-enantiomer and, consequently, reduces warfarin dose requirements (3, 7-10). Warfarin acts upon the hepatic vitamin K epoxide reductase complex, subunit 1 (VKORC1) enzyme, inhibiting recycling of vitamin K and therefore limiting the production of vitamin K-dependent clotting factors (11). Variation in the VKORC1 gene can enhance sensitivity to warfarin (12, 13). Genetic variation in the CYP2C9 and VKORC1 enzymes accounts for approximately 10% and 30% of warfarin dose variability, respectively, and in combination with clinical data allows for prediction of more than half of warfarin’s dose variability (2, 4).
A few studies have prospectively examined the utility of pharmacogenetic-guided warfarin dosing, and the results have been mixed (14-16). Mechanistic pharmacokinetic and pharmacodynamic (PK/PD) analyses that generally involve modeling and simulation have become an increasingly important component in drug dose selection (17-19). We sought to improve the performance of warfarin pharmacogenetic dosing by serially updating a dosing algorithm using a mechanistic PK/PD model as patient data accrued. Accordingly, the aim of the CROWN (Creating an Optimal Warfarin Nomogram) Trial was to develop an optimal dosing algorithm that incorporated clinical and genetic information to maintain patients within a therapeutic International Normalised Ratio (INR) range.
Methods
Study design
This was a prospective, single arm study in which pharmacogenetics was used for all patients. The initial dosing algorithm (Algorithm A) was based upon pre-existing clinical practice and historical warfarin pharmacogenetic data. Two subsequent dosing algorithms (Algorithms B and C, respectively) were generated based upon the observed warfarin pharmacogenetics, and then used in series (i.e. A, then B then C). Each dosing algorithm included an initiation dose that was given for four days, followed by a maintenance dose titration algorithm. The study was approved by the Partners Healthcare institutional review board and is registered on ClinicalTrials.gov (NCT00401414). Informed consent was obtained from all subjects prior to participation.
Inclusion and exclusion criteria
Inclusion criteria included a newly diagnosed condition that would require treatment and/or prevention of thrombosis with warfarin for at least three weeks, age 318 years, and written informed consent. Reasons for exclusion included warfarin treatment within the past three months, a contraindication to therapeutic anticoagulation, pregnancy or intent to become pregnant, life expectancy < 3 months, and a high likelihood of non-compliance.
Enrollment
Subjects were recruited from the outpatient and inpatient settings of six participating hospitals in the Partners Healthcare Network, Boston, Massachusetts, including Brigham and Women’s Hospital, Massachusetts General Hospital, Faulkner Hospital, Newton Wellesley Hospital, North Shore Medical Center, and Spaulding Rehabilitation Hospital. Participants were identified by review of the inpatient pharmacy record for warfarin or heparin orders, from among new referrals to the institutions’ anticoagulation services, from venous ultrasound studies performed in the vascular ultrasound laboratory, or from patients evaluated by the Vascular Medicine consultation service. Qualifying, consenting patients underwent genetic testing on the day of enrolment, and genotype data were usually available the same day. Patients who had received no more than two daily doses of warfarin were eligible to participate. Enrolment was from January 2007 to September 2009.
DNA extraction and genotyping
The Laboratory for Molecular Medicine (LMM, CLIA# 22D1-005-307) performed genetic testing. DNA was extracted from peripheral whole blood and was analysed by at least one of three methods validated by the laboratory: Third Wave Technologies (Madison, WI, USA) InvaderÒ assay, AutoGenomics (Vista, CA, USA) INFINITIÔ CYP450 2C9-VKORC1 assay, or a laboratory developed TaqmanÒ assay (20). Each assay was determined to have an accuracy of 100%, verified by DNA sequencing, and therefore test results from these methods were considered interchangeable (20). SNPs analysed were VKORC1, –1639G>A, rs9923231, CYP2C9*2, 430C>T (R144C), rs1799853, CYP2C9*3, 1075C>A (I359L), rs1057910.
Derivation of dosing algorithms
Three dosing algorithms (A, B, and C, respectively) were used in this investigation. The algorithms were developed sequentially to select both an initial warfarin dose and a titration scheme intended to maximise the likelihood of achieving and maintaining the target INR. The algorithms were refined using adaptive methods that allow for continual reassessment of patient-level data to optimise the predictive power of the algorithms. Monte-Carlo simulations using the concentration-INR relationship were then performed to identify the optimal algorithm. Details of each dosing algorithm and the warfarin PK/PD model can be found in the Appendix (available online at www.thrombosis-online.com).
The initial dosing algorithm (Algorithm A) was a dosing decision-tree that included both clinical and genetic factors. It was based upon optimal clinical practice at the Brigham and Women’s Hospital’s Anticoagulation Management Service as well as published literature that has utilised warfarin pharmacogenetics. Doses were subsequently adjusted based on serial INR measurements.
Dosing Algorithm B was generated from an analysis of warfarin dose, INR, genetic factors, demographic factors and concomitant drug therapy from an initial prospective group of 74 patients treated using Algorithm A. Using these data, a mechanistic concentration-INR model was constructed to refine the estimates of the effect of CYP2C9 genotypes, VKORC1 haplotypes, age, and concomitant medications. Actual plasma warfarin concentrations were not available, so plasma concentrations were imputed based upon established warfarin pharmacokinetics and study patients’ clinical and genetic characteristics (21). The concentration-INR model was then used to simulate the time course of the INR for different combinations of initial warfarin doses and titration schemes. We sought to identify a starting dose and titration scheme that would rapidly achieve and then maintain the INR in the target range of 2–3. The simulation conditions included 10,000 patients per dosing regimen for a duration of 90 days, where starting warfarin doses were given once a day for four days and then adjusted twice weekly based upon the INR results. The measurements evaluated in the simulation were: the percent of time within the target INR between days 5 and 90, the number of patients achieving the target INR on day 5 and on day 8 individually, the proportion of INR measurements within target INR range, the proportion of INRs outside of the target range, and the number of required dose adjustments. Results from the initial cohort (Algorithm A) for these endpoints were compared to simulated endpoint results to validate the performance of the model in identifying optimal starting and maintenance doses of warfarin.
Dosing Algorithm C was generated as an update of dosing Algorithm B and was based upon additional patient data, similar to what was described above for Algorithm B, from the prospective accrual of 203 patients in the CROWN trial. The major difference between Algorithm B and Algorithm C was an update of the half maximal inhibitory concentration (IC50) estimate for each VKORC1 haplotype in the model used to generate Algorithm B to reflect warfarin’s PD effect as evident in the acquired patient data. Simulations using Algorithm C were repeated using the clinical endpoints described above to derive the optimal starting warfarin doses and titration scheme that was tested prospectively in subsequent patients enrolled in the CROWN study. NONMEM 6 (ICON, MD) was used for the parameter estimation and Trial Simulator (Pharsight, Sunnyvale, CA, USA) was employed for the intensive simulation.
Study duration
Study duration was three months or to the end of warfarin therapy, if shorter. Orthopaedic surgery patients were generally treated for 21 days. The overall mean duration of time in the study for each participant was 75 days (SD=24 days).
Endpoints
The primary end point was the mean percentage of time that the INR was within the therapeutic range (TTR), using the method of linear interpolation introduced by Rosendaal et al. (22). This method assumes a linear increase or decrease between two consecutive INR determinations. TTR was calculated for the entire study period for each subject, and for the time periods 1–30 days and 31–60 days for each of the three algorithms. Though the target INR was 2.0 to 3.0, a therapeutic INR was considered 1.8 to 3.2 to allow for INR measurement error and avoid problems inherent in overcorrection. For this reason, and consistent with the design of the contemporaneous pharmacogenomics study by Anderson et al., the statistical analysis plan was amended during the course of the trial to use the therapeutic INR range of 1.8–3.2 as the primary endpoint, rather than the target INR of 2.0–3.0 (15).
Secondary endpoints were: 1) time to the first therapeutic INR, 2) the per-patient percentage of INRs out of the therapeutic range, 3) the time to stable anticoagulation, defined as two consecutive INRs within the therapeutic range 37 days apart and with no dose change during this time, and 4) the proportion of patients with serious adverse clinical events, defined as an INR 34.0, use of vitamin K, major bleeding events (as defined by the Thrombolysis in Myocardial Infarction [TIMI] criteria), thromboembolic events, stroke (all cause), myocardial infarction, and death (all cause) (15, 23). In addition, the accuracy of the initial recommended dose was calculated as the difference between the algorithm’s recommended dose and the actual dose at steady state. The stable maintenance dose was defined as the dose achieved on day 8 or later where two or more consecutive INR measurements measured 7 or more days apart varied by no more than 15% (15).
Genetic subtype analyses
For each endpoint, we repeated the analyses with subjects characterised by genetic subtype. The CYP2C9 alleles included *1, *2, and *3. The VKORC1 haplotypes included AA, AB, and BB. For these analyses, CYP2C9*1*1 / VKORC1 BB was designated wild type. Subjects were categorised as having 0 variants, 1 variant, or 2 or more variants from wild type. In this schema the wild-type genotype has the highest predicted dose requirement, and dose requirement declines with each additional variant. There were insufficient numbers of subjects within each algorithm to support separate analyses of persons with 3+ variants, and too few adverse events to support genetic subset analysis of adverse outcomes.
Statistical methods
Summaries of the baseline characteristics are presented as means, and standard deviations are presented for the continuous variables and percentages for the dichotomous variables. Comparisons of baseline characteristics among the subjects within the three algorithms were conducted using F-tests and chi-square tests. Analyses of the mean percentages of time in therapeutic range and the mean per-patient percentage of out-of-range INRs for the three algorithms were conducted using F-tests. Comparison of the times for individuals to reach therapeutic INR and the time to reach a stable INR among the three algorithms were made using a log-rank test to account for censoring of subjects if they transitioned into the next algorithm in series. Significance was set at p <0.05. Subjects were analysed according to the algorithm in place at the time of enrolment. If a subject transitioned to a newer algorithm, he or she was censored at that time.
Statistical analyses were performed using the statistical software package R (R foundation for Statistical Computing, Vienna, Austria, 2010) at the Massachusetts General Hospital Biostatistics Center. The authors had full access to and take full responsibility for the integrity of the data.
Results
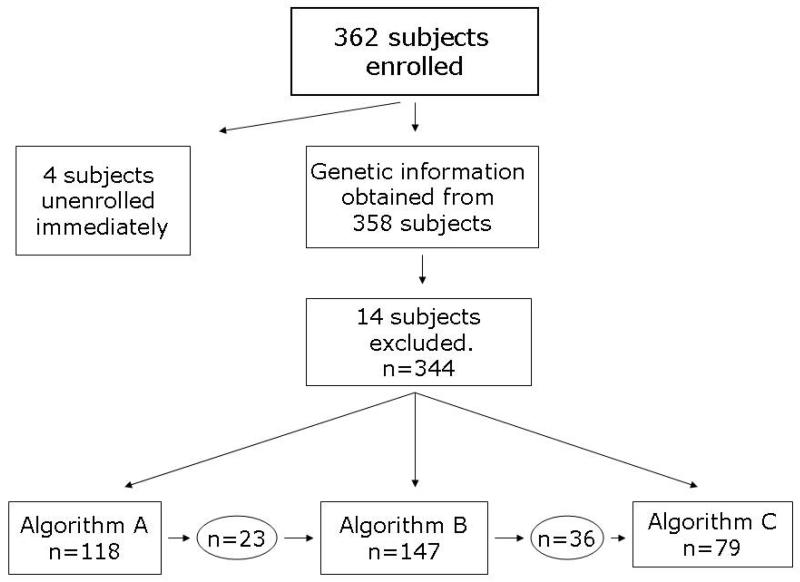
Patient enrolment and demographics A total of 362 patients were enrolled, as displayed in P Figure 1. Genetic information was obtained from 358 (99%) of the subjects. Eighteen subjects (5%) were excluded from the analyses because they withdrew prior to receiving any study drug, leaving 344 subjects who were included in the study analyses.
Figure 1. CONSORT diagram.
A total of 362 subjects enrolled, 4 of which unenrolled prior to any study activity, and 14 of which were excluded because no study drug was administered. Of the 344 remaining subjects, 118 began on algorithm A, 147 on algorithm B, and 79 on algorithm C. If a subject transitioned to a newer algorithm, he was censored at that time.
Ninety-five subjects were treated using Algorithm A only, 111 using Algorithm B only, and 79 using Algorithm C only. Twenty-three subjects were begun on Algorithm A and were transitioned to Algorithm B, and 36 subjects were begun on Algorithm B and were transitioned to Algorithm C. When subjects were transitioned to a new algorithm they were excluded from further analyses.
Genotype distribution and indication for warfarin did not vary significantly among the three algorithm groups (P Table 1). There was a significant difference in the proportions of Caucasian subjects in each of the three algorithms, with a higher proportion in Algorithms B and C. There were more orthopaedic patients in Algorithm A compared with Algorithms B and C. The mean total time spent in the study was similar for subjects assigned to each of the three algorithms (p = 0.506). Since subjects were transitioned to a new algorithm in series if an algorithm change occurred during their participation, the data analysed included more than 10 days greater on average for participants in the Algorithm C group than those in the Algorithm A or B groups (p < 0.001).
Table 1. Baseline Subject Characteristics.
| Characteristic | All | Algorithm A | Algorithm B | Algorithm C | P-value |
|---|---|---|---|---|---|
| Number of Subjects | 344 | 118 | 147 | 79 | |
| Age | 60.1 (16.7) | 64.7 (16.6) | 57.9 (16.5) | 57.2 (15.6) | <0.001 |
| Male Gender (%) | 52.9% | 49.2% | 56.5% | 51.9% | 0.485 |
| Weight (kg) | 87.5 (25.5) | 85.9 (25.0) | 88.7 (24.9) | 87.5 (27.3) | 0.613 |
| Height (cm) | 171.9 (10.7) | 169.5 (10.7) | 172.6 (11.0) | 173.3 (9.94) | 0.018 |
| White Race | 83.1% | 75.4% | 85.0% | 91.1% | 0.003 |
| VKORC1 | |||||
| A/A | 16.9% | 13.6% | 19.7% | 16.5% | |
| A/B | 46.2% | 53.4% | 42.9% | 41.8% | 0.344 |
| B/B | 36.9% | 33.1% | 37.4% | 41.8% | |
| CYP 2C9: | |||||
| *1/*1 | 65.7% | 61.0% | 70.1% | 64.6% | |
| *1/*2 | 19.2% | 21.2% | 15.6% | 22.8% | |
| *1/*3 | 12.2% | 12.7% | 12.2% | 11.4% | 0.482 |
| *2/*2 | 1.5% | 1.7% | 1.4% | 1.3% | |
| *2/*3 | 1.5% | 3.4% | 0.7% | 0.0% | |
| Non-Orthopedic | 96.2% | 89.8% | 100.0% | 98.7% | <0.001 |
| Total Time in Study (Days) | 75.3 (23.7) | 74.7 (28.6) | 74.7 (64.4) | 74.8 (22.0) | 0.506 |
| Total Time in Study (Days) (after censoring) |
68.1 (28.0) | 66.5 (32.6) | 64.4 (27.5) | 77.2 (18.1) | <0.001 |
Comparisons of baseline characteristics among the subjects within the three algorithms were conducted using F-tests and chi-square tests
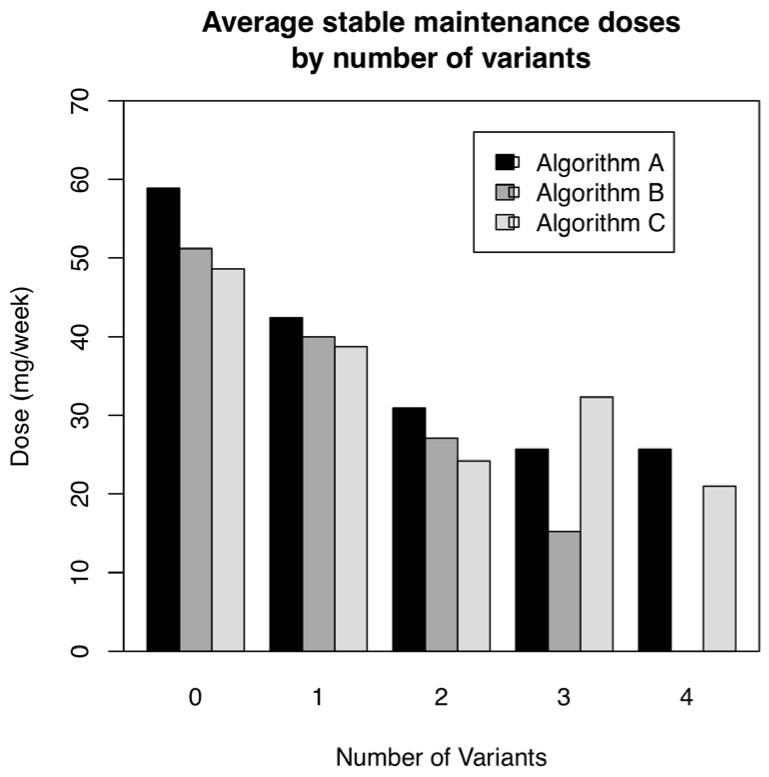
The mean weekly dose of warfarin at steady state decreased with increasing total number of CYP2C9 and VKORC1 variant alleles (p < 0.01), as expected (P Fig. 2). Subjects with 0 genetic variants, 1 genetic variant, and 2 genetic variants required an average of 50, 40, and 30 mg of warfarin weekly (p < 0.01). There were too few subjects with 3 or 4 variant alleles to have reliable estimates, but the trend toward lower warfarin doses appeared to continue.
Figure 2. The number of genetic variants and warfarin stable maintenance dose requirement.
The requirement for warfarin declined in a stepwise manner with increasing number of variant alleles, as would be expected.
Primary endpoint
The mean percentage time within the therapeutic range over the entire study period for each subject increased progressively from Algorithm A to Algorithm C from 58.9 (22.0), to 59.7 (23.0), to 65.8 (16.9) % (p = 0.04) (P Table 2). The improvement in time within the therapeutic range tended to be achieved primarily within the first 30 days compared with days 31–60; however, the improvement in the TTR across algorithms during the first 30 days was not different from the improvement in the TTR across algorithms between days 31-60.
Table 2. Primary End Point Results Overall and by Genotypic Subsets.
| End Point | Algorithm A (n = 118) |
Mean (SD) A 1 gorithm B (n = 147) |
Algorithm C (n = 79) |
P-value |
|---|---|---|---|---|
| Time within therapeutic range (%), all subjects | 58.9 (22.0) | 59.7 (23.0) | 65.8 (16.9) | 0.039 |
| Time within therapeutic range (%),study days 1-30, all subjects |
47.3 (20.5) | 45.1 (21.1) | 52.9 (18.6) | 0.110 |
| Time within therapeutic range (%), study days 31-60, all subjects |
66.7 (28.3) | 71.4 (25.9) | 69.5 (22.7) | 0.440 |
| Time within therapeutic range (%), 0 genetic variants |
56.7 (20.9) | 59.2 (22.0) | 70.5 (14.2) | 0.026 |
| Time within therapeutic range (%) , 1 genetic variant | 57.0 (23.6) | 59.2 (22.0) | 63.1 (15.4) | 0.219 |
| Time within therapeutic range (%), 2+ genetic variants |
61.9 (21.4) | 60.8 (23.0) | 66.1 (21.2) | 0.589 |
Comparisons between the mean percentages of time spent in therapeutic range (INR = 1.8-3.2) in the three algorithms were conducted using F-tests.
Genetic subtype analyses of the primary endpoint
Wild-type (*1*1/B/B) subjects with 0 genetic variants appeared to derive the greatest benefit from the serial refinements in the pharmacogenetic algorithm (P Table 2). The TTR in wild-type subjects increased from 56.7 (20.9) % in Algorithm A, to 59.2 (22.0) % in (Algorithm B), to 70.5 (14.2) % in Algorithm C (p = 0.03). The TTR tended to improve across study algorithms in persons with 1 and 2+ variants, but these differences were not statistically significant.
Secondary endpoints
The time to the first therapeutic INR did not differ significantly among the three warfarin dosing algorithm groups (p = 0.07) (P Table 3). The per-patient percentage of INRs outside of the therapeutic range (INR 1.8–3.2) did improve over the three algorithms (p = 0.004) (P Table 3). The mean time to achieve stable therapeutic anticoagulation decreased across algorithms (p < 0.001) (P Table 3).
Table 3. Secondary Endpoints Overall and by Genetic Subsets.
| End Point | Algorithm A (n = 118) |
Mean (SD) Algorithm B (n = 147) |
Algorithm C (n = 79) |
P-value |
|---|---|---|---|---|
| Time to first therapeutic INR (days) | 9.1 (4.5) | 10.4 (4.9) | 9.7 (4.4) | 0.069 |
| Time to first therapeutic INR (days), 0 genetic variants |
9.5 (5.9) | 12.8 (16.2) | 9.4 (3.4) | 0.024 |
| Time to first therapeutic INR (days), 1 genetic variant |
8.2 (2.7) | 10.4 (4.3) | 9.7 (5.0) | 0.019 |
| Time to first therapeutic INR (days), 2+ genetic variants |
9.6 (4.6) | 8.7 (3.4) | 10.1 (4.2) | 0.335 |
| Per-patient percentage of INRs out of therapeutic range |
42.2 (21.6) | 37.7 (22.8) | 33.3 (16.8) | 0.004 |
| Per-patient percentage of INRs out of therapeutic range, 0 genetic variants |
44.7 (20.3) | 38.5 (21.9) | 30.0 (11.2) | 0.011 |
| Per-patient percentage of INRs out of therapeutic range, 1 genetic variant |
44.5 (23.1) | 37.6 (22.6) | 33.7 (16.7) | 0.025 |
| Per-patient percentage of INRs out of therapeutic range, 2+ genetic variants |
38.7 (21.1) | 37.2 (24.0) | 35.9 (21.3) | 0.616 |
| Time to stable therapeutic anticoagulation (days) |
50.8 (20.1) | 34.6 (14.9) | 31.5 (13.1) | <0.001 |
| Time to stable therapeutic anticoagulation (days), 0 genetic variants |
51.8 (23.2) | 38.6 (18.4) | 31.3 (12.1) | <0.001 |
| Time to stable therapeutic anticoagulation (days), 1 genetic variant |
47.9 (15.7) | 32.8 (13.1) | 31.8 (14.0) | <0.001 |
| Time to stable therapeutic anticoagulation (days), 2+ genetic variants |
52.3 (21.5) | 33.4 (13.4) | 31.8 (14.0) | <0.001 |
Genetic subtype analyses of secondary endpoints
The time to the first target INR (2-3) tended to vary by algorithm among subjects with both 0 and 2+ genetic variants, though no clear directional trend occurred (P Table 3). The per-patient percentage of INRs outside of the therapeutic range (1.8 to 3.2) decreased as the study progressed from Algorithm A to Algorithm C for each genetic variant group (P Table 3). The average number of days to stable therapeutic anticoagulation decreased in each genetic variant group (0, 1, 2+ genetic variants) as the study progressed from Algorithm A to Algorithm C (each p < 0.001) (P Table 3).
Dosing accuracy
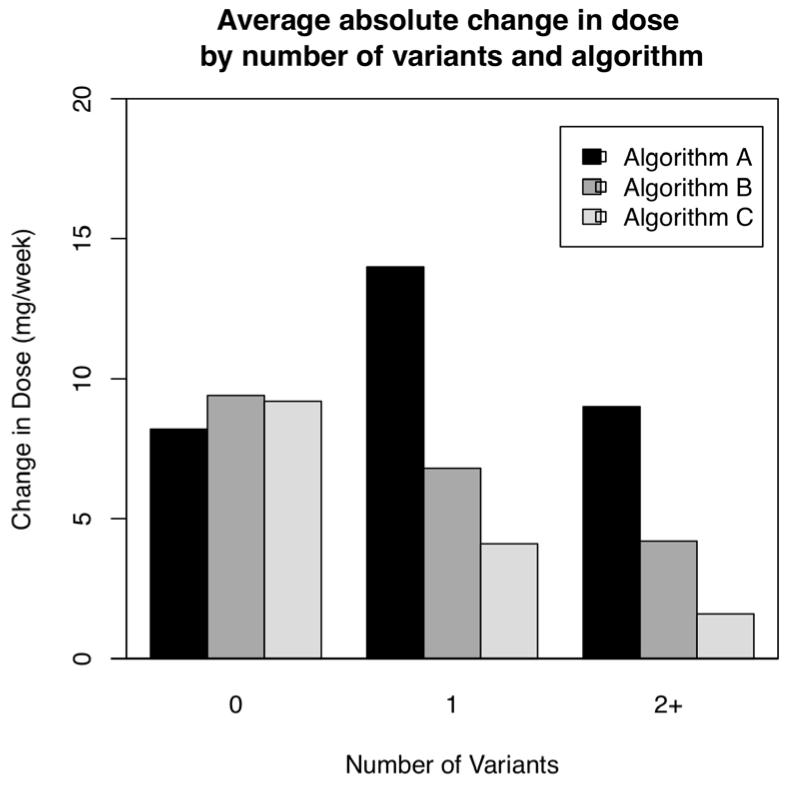
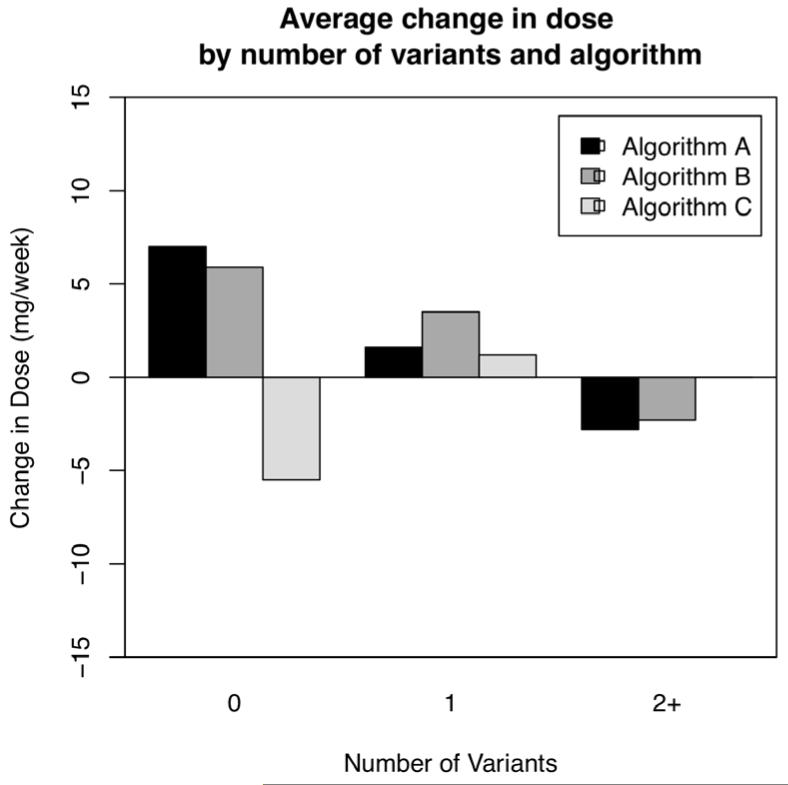
We examined algorithm dosing accuracy by comparing the mean absolute difference between the algorithm-recommended initial dose and the actual warfarin dose requirement at steady state (P Fig. 3). In subjects with 0 genetic variants, each algorithm had similar accuracy. Dosing accuracy improved across dosing iterations in subjects with 1 or 2+ genetic variants. Two-way analysis of variance demonstrated significant differences observed in the changes from initial to stable maintenance doses due to the algorithm used (p < 0.001) and genetic variant group (p = 0.001). To analyse the direction of algorithm dosing inaccuracy, we examined the mean difference between the algorithm-recommended and the actual warfarin dose requirement at steady state (P Fig. 4). In subjects with 0 genetic variants the first two algorithms tended to underestimate the required warfarin dose by approximately 7 mg per week, while the third algorithm tended to overestimate the required warfarin dose by about the same magnitude. In subjects with 1 and 2+ genetic variants, each algorithm was accurate within 5 mg/week.
Figure 3. Accuracy of pharmacogenetic dosing.
Shown is the average absolute difference between the algorithm prescribed initial dose and the actual steady-state warfarin dose. In subjects with 0 genetic variants each algorithm had similar accuracy. In subjects with 1 or more genetic variants, the dosing accuracy improved with each algorithm iteration.
Figure 4. Accuracy of pharmacogenetic dosing.
Shown is the mean difference between the algorithm-prescribed initial dose and the actual steady-state warfarin dose. Subjects with 0 genetic variants were tended to be under-dosed by the first 2 algorithms and over-dosed by the third. Subjects with 1 variant tended to be over-dosed by all 3 algorithms. Subjects with 2 variants tended to be over-dosed by the first 2 algorithms and appropriately dosed by the third.
Adverse events
There were no statistically significant differences in total or individual adverse event rates among the three dosing algorithms (P Table 4). The vast majority of adverse events were INR values 34.0, but without associated bleeding. There were few episodes of major bleeding, thrombosis, or vitamin K administration.
Table 4. Adverse Events Compared by Algorithm.
| Characteristic | All | Algorithm A | Algorithm B | Algorithm C | P-value* |
|---|---|---|---|---|---|
| Number of Subjects | 344 | 118 | 147 | 79 | |
| Total adverse events, number | 108 (31.4%) | 43 (36.4%) | 41 (27.9%) | 24 (30.4%) | 0.32 |
| Thrombotic events (Ml, Stroke, VTE)** | 2 (0.6%) | 0 (0.0%) | 2 (1.4%) | 0 (0.0%) | - |
| Major bleeding events (TIMI definition)** | 7 (2.0%) | 2 (1.7%) | 3 (2.0%) | 2 (2.5%) | - |
| Vitamin K administration** | 2 (0.6%) | 2 (1.7%) | 0 (0.0%) | 0 (0.0%) | - |
| INR ≥ 4, number events | 97 28.2%) | 39 (33.1%) | 36 (24.5%) | 22 (27.8%) | 0.35 |
Comparison by Chi-square test.
Note that numbers are too small for chi-square test
Discussion
We sought to improve the accuracy and efficiency of warfarin dosing through an adaptive pharmacogenetic dosing algorithm. We achieved our primary endpoint of an improvement in the time that the INR was within the therapeutic range, increasing from 57.7% with the first algorithm to 65.0% with the third and final algorithm. A secondary endpoint of lowering the per-patient percentage of INRs outside the therapeutic range also improved significantly during the course of our study, decreasing from 42.2% to 33.3%. Together these results suggest that an adaptive pharmacogenetic algorithm approach can be used to improve the accuracy and efficiency of warfarin dosing, and may ultimately improve the safety of warfarin therapy.
Previous prospective studies
Voora et al. demonstrated the feasibility of prospective application of point-of-care genotyping in patients initiating warfarin therapy in a small study of orthopaedic patients, and this was confirmed by Hillman et al. (10, 24). The Medco-Mayo Warfarin Effectiveness Study reported that pharmacogenetic dosing of patients initiating warfarin achieved a reduction in overall hospitalisation rates and in hospitalisations related to bleeding or thrombotic events, compared with a historical control group of conventionally dosed patients (25).
Three prospective randomised trials that compared pharmacogenetic-based dosing of warfarin with standard dosing and which included clinical endpoints have been reported. Caraco et al. reported improvement in the time to a first therapeutic INR (2.0 to 3.0), time to a stable warfarin dose, and time in the therapeutic range with a genotype dosing strategy that accommodated CYP2C9 variation (14). Huang et al. performed a prospective pharmacogenomics study utilising both the CYP2C9 and VKORC1 genotypes in patients undergoing heart valve replacement and reported significant improvement in time to reach a stable warfarin dose and time the INR was within the therapeutic range (1.8 to 3.0) (16). Anderson et al. also performed a controlled trial of genetic-guided warfarin dosing utilising both the CYP2C9 and VKORC1 genotypes. In that study, genotype-based dosing failed to significantly improve the percentage of INRs outside of the therapeutic range (1.8 to 3.2), the time to the first super-therapeutic INR, and the time within the therapeutic range compared with standard dosing (15). Thus, the available trial evidence is encouraging but has not convincingly demonstrated a benefit of pharmacogenetic-based dosing of warfarin compared with standard dosing.
Ours is the first study, to our knowledge, to attempt to improve the accuracy and efficiency of a genetic dosing algorithm using an adaptive PK/PD modelling approach. Our study is different from the aforementioned prospective examinations of warfarin pharmacogenomics in that we did not have a control group that received standard, non-genetic dosing. It is therefore difficult to directly compare our results to those randomised trials. Nonetheless, our study suggests that further work employing the adaptive dosing algorithm approach we used might further improve upon existing pharmacogenetic algorithms.
Clinical implications
Much warfarin dose variability remains unexplained even after accounting for genetic variants. For example, in the International Warfarin Pharmacogenetics Consortium study of 5,052 subjects, an optimised pharmacogenetic algorithm predicted 49% and 43% of dose variability in the derivation and validation cohorts, respectively (4). In another large cohort, the Swedish Warfarin Genetics study of 1,677 subjects, a pharmacogenetic algorithm predicted 59% and 53% of dose variability in the derivation and validation cohorts (26). This suggests there may be a relatively small increment of potential benefit for warfarin pharmacogenetics using currently recognised genetic polymorphisms. The 65% time in the therapeutic range that we ultimately achieved, roughly the mean of that achieved in the randomised trials reviewed above, indicates that warfarin-treated persons spend about 35% of the time outside the therapeutic range, even when dosing utilises pharmacogenetics, and therefore these patients remain at risk for treatment-associated adverse events.
Further evidence suggests that a strategy of PK/PD modelling of warfarin dosing using the CYP2C9 and VKORC1 genotypes can provide modest incremental benefit in warfarin dosing beyond current high quality clinical practice. Salinger et al. used an existing PK/PD model to perform computer-run clinical trial simulations of pharmacogenomic-guided versus standard of care warfarin therapy (27). Pharmacogenomic guidance resulted in only a 3–4 percentage point absolute increase in time spent in therapeutic range compared with standard of care.
Employing pharmacogenetic-based dosing modifications beyond the initial dose may improve the performance of a pharmacogenetic warfarin algorithm. This could include loading doses in patients with a CYP2C9 variant, as well as allowing for more time before making warfarin dose adjustments in these same patients (21, 28). Indeed, the simulation of Salinger et al. found an absolute doubling of the improvement in warfarin dosing with concomitant use of loading and delayed-adjustment strategies (27).
The benefit of warfarin pharmacogenetics is likely greatest when INR measurement is least frequent (28). Salinger et al. found that the benefit of pharmacogenetic dosing was significantly greater when the simulated checking of INRs was assumed less frequent (27). While dedicated anticoagulation clinics perform frequent monitoring, more conventional practice is to measure the INR less frequently (29).
There are a few planned or active trials of warfarin pharmacogenetics that will provide greater clarity: The National Heart Lung and Blood Institute Clarification of Optimal Anticoagulation through Genetics (COAG) trial of 1,238 patients (ClinicalTrials.gov NCT00839657) and the European Commission FP7 Programme supported European Pharmacogenetics of Anticoagulant Therapy (EU-PACT) trial (ClinicalTrials.gov NCT01119300). Of note, COAG and the EU-PACT trials are powered to detect 5.49% and 5% improvements in the TTR, respectively, suggesting these investigators anticipate a relatively small incremental benefit from genetically based warfarin dosing. Also underway is The Genetics Informatics Trial (GIFT) of Warfarin to Prevent DVT (ClinicalTrials.gov NCT01006733), which is enroling 1,600 patients receiving warfarin prophylaxis following hip or knee arthroplasty and will compare clinical events (e.g. venous thromboembolism, haemorrhage, death) associated with pharmacogenetic versus conventional warfarin dosing.
Study strengths and limitations
Our study’s strengths include its prospective design and use of rapid genotyping to allow for pharmacogenetic dosing in clinical “real time”. The primary limitation is that the sample size may have been insufficient to fully realise the benefit of warfarin pharmacogenetics. For example, the results of the simulation by Salinger et al. suggested that a few thousand patients would be needed to have had 80% power to detect a benefit of warfarin pharmacogenetics (27). The sample size also was inadequate to detect differences in clinical event rates. Our study population was primarily Caucasian, therefore the results may not apply to other populations (30-33). Accounting for additional genotypic variants that influence warfarin metabolism or sensitivity may improve the predictive accuracy of a warfarin pharmacogenomic dosing algorithm (34, 35). Some subject characteristics, such as age, were not equally distributed between dosing algorithms, as would be expected from the non-randomised design.
Conclusions
This is the first clinical trial to prospectively utilise observed PK/PD information in an adaptive fashion to optimise warfarin pharmacogenetic dosing in real time. We observed an improvement in the time within the therapeutic INR range. These data support the conduct of larger trials of warfarin pharmacogenetics, with the ultimate goal of improving the efficacy and safety of warfarin therapy.
What is known on this topic
A pharmacogenetic dosing algorithm accounting for variation in the CYP2C9 and VKORC1 genes has the potential to improve prediction of required steady-state warfarin levels over an algorithm accounting for clinical factors alone.
Results of trials comparing the efficacy of a pharmacogenetic dosing algorithm with a standard clinical dosing algorithm in maintaining warfarin treated patients within the therapeutic range have been inconsistent.
What this paper adds
This is the first clinical trial to prospectively utilize observed PK/PD information in an adaptive fashion to optimize warfarin pharmacogenetic dosing in real time.
A strategy of modifying a warfarin pharmacogenetic dosing in real time in an adaptive fashion may have the potential to improve clinical outcomes in warfarin-treated patients.
Acknowledgements
We gratefully acknowledge the nurse coordinators, Regina Silver, Christine Harker, and Barbara Mahoney for their outstanding efforts in recruiting and managing the participants in this study.
Sources of Funding
This work was funded by Partners HealthCare through a grant to the Harvard Partners Center for Personalized Genetic Medicine. Dr Perlstein received support from the National Heart, Lung, and Blood Institute Research Career Development Award K12–HL083786. Dr. Creager is the Simon C. Fireman Scholar in Cardiovascular Medicine at Brigham and Women’s Hospital.
Appendices
Appendix 1. Algorithm A
Initiation dosing
| CYP2C9 Genotype | VKORC1 Haplotype | |||
| A/A | A/B | B/B | ||
| 4 | 6 | 8 | ||
| *1*2 | 3 | 5 | 7 | |
| *1*3 | 3 | 5 | 7 | |
| *2*2 | 2 | 4 | 6 | |
| *2*3 | 3 | 5 | 7 | |
| *3*3 | 2 | 4 | 6 | |
Dose adjustment:
(−) 1 mg if age > 65 years
(+) 1 mg if height > 175 cm
(−) 1 mg if height < 155 cm
(−) 1 mg if weight < 50 kg
(+) 1 mg if weight > 100 kg
(−) 2 mg if medication use*
(+) 2 mg if medication use**
(−) 2 mg if ALT > 110 IU/L
(+) 1 mg if ≤ 3 vegetable servings per day or enteral tube feeding Minimum allowable dose 1 mg, maximum allowable dose 10 mg
*Any of acetaminophen (> 1 g m/day), daily alcohol Intake, aspirin (> 325mg/d), non-steroidal anti-inflammatory drug (more than 4 tablets per day), amiodarone, cephalosporins, ciprofloxacin, clofibrate, clopidogrel, levofloxacin, erythromycin, azithromycin, clarithromycin, fluconazole, metronidazole, isoniazid, cimetidine, omeprazole, pantoprazole, sufamethoxazole/trimethoprim, tamoxifen, thyroid hormone, or fluoxetine.
** Any of rifampin, dicloxacillin, nafcillin, griseofulvin, carbamazepine, oral contraceptives, barbiturates, haloperidol, sucralfate, antithyroid drugs, or cholestyramine.
Maintenance dosing
Coumadin dose for next 4 days* = (goal INR / current INR)1/3 × current 4-day dose
*With continued dose adjustments for medications and dietary intake as listed above for initial dosing.
Appendix 2. Algorithm B
Initiation dosing
| CYP2C9 Genotype | VKORC1 Haplotype | |||
| A/A | A/B | B/B | ||
| 3 | 4 | 5 | ||
| *1*2 | 3 | 4 | 5 | |
| *1*3 | 2.5 | 2.5 | 4 | |
| *2*2 | 2 | 2 | 2.5 | |
| *2*3 | 2 | 2 | 2.5 | |
| *3*3 | 0.5 | 1 | 1 | |
Dose adjustment:
(−) 1 mg if age > 65 years
(+) 1 mg if height > 175 cm
(−) 1 mg if height < 155 cm
Maintenance Dosing
| Achieved INR | Dose adjustment relative to previous dose |
| < 2 | 20% increase (10% increase for CYP2C9 *3*3) |
| 2 < INR < 3 | No adjustment |
| 3 < INR < 4 | 20% decrease |
| 4 < INR < 5 | 25% decrease |
| 5 < INR < 6 | 30% decrease |
| INR > 6 | 50% decrease |
Appendix 3. Algorithm C
Initiation dosing
| CYP2C9 Genotype | VKORC1 Haplotype | |||
| A/A | A/B | B/B | ||
| 3.5 | 5 | 7 | ||
| *1*2 | 3 | 4 | 4.5 | |
| *1*3 | 2.5 | 3 | 4 | |
| *2*2 | 1 | 1.5 | 2.5 | |
| *2*3 | 1 | 1.5 | 2.5 | |
| *3*3 | 1 | 1 | 1 | |
Maintenance Dosing
| Achieved INR | Dose adjustment relative to previous dose |
| < 1.8 | 20% increase (10% increase for CYP2C9 *3*3) |
| 1.8 < INR < 3.2 | No adjustment |
| 3.2 < INR < 4 | 20% decrease |
| 4 < INR < 5 | 25% decrease |
| 5 < INR < 6 | 30% decrease |
| INR > 6 | 50% decrease |
Appendix 4
Warfarin PK/PD model
A population model that describes the PK of S-warfarin and the PK/PD relationship between exposure and INR was applied. The model for pharmacokinetics of warfarin was derived from Hamberg and colleagues16, and the relationship between warfarin concentration and prothrombin complex activity was adapted from Chan and colleagues and modified to describe the relationship between warfarin concentration and INR.(16, 28)
Pharmacokinetic (PK) Modeling
Warfarin is a racemic mixture of two enantiomers: S-warfarin and R-warfarin. S-warfarin is known to be 3-5 times more potent as a vitamin K antagonist than R-warfarin and it is metabolized by CYP2C9. In this analysis, only S-warfarin was considered to describe the mechanism of action. A two-compartment model with first-order input and first-order elimination was applied (see below). Since the free concentrations of the drug are responsible for the anticoagulant effect of the drug, protein binding of warfarin was taken into account for expressing unbound plasma levels. The concentration was imputed given established PK parameters because the study did not measure warfarin levels.(16) CYP2C9 genotype status as well as clinical factors such as age and concomitant medications that induce or inhibit CYP2C9 were adjusted as covariates.
Absorption
Plasma
Tissue
where
ka First order rate constant for the absorption of S-Warfarin from the gut into the blood stream
CL Clearance of S-Warfarin
CL2 Inter-compartmental clearance of S-Warfarin
V1 Volume of distribution of S-Warfarin in the plasma compartment
V2 Volume of distribution of S-Warfarin in the peripheral tissue compartment
Since the free concentrations of the drug are responsible for the anticoagulant effect of the drug, protein-binding of warfarin was taken into account for expressing unbound plasma levels. Free plasma warfarin concentrations were determined as:
Pharmacodynamic (PD) Modeling
The clotting activity after warfarin exposure was defined as prothrombin complex activity (PCA) and described by an indirect response model with S-warfarin unbound concentration as an exposure.
Since the anticoagulant response generally obtained is INR, the response data can be transformed to PCA with the use of functional relationship between INR and prothrombin time (PT) and PCA(28, 29) described in the equations below:
where a=426; b=7.75
The CROWN trial measured INR rather than PCA; thus, the equations above were not well-suited to the observed study data. Hence, the original PD model was modified to describe the relationship between concentration and INR directly as follows:
Footnotes
Disclosures
None
References
- 1.Gallagher AM, Setakis E, Plumb JM, et al. Risks of stroke and mortality associated with subuptimal anticoagulation in atrial fibrillation patients. Thromb Haemost. 2011;106:968–977. doi: 10.1160/TH11-05-0353. [DOI] [PubMed] [Google Scholar]
- 2.Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008 Sep;84(3):326–31. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gage BF, Eby C, Milligan PE, et al. Use of pharmacogenetics and clinical factors to predict the mainenance dose of warfarin. Thromb Haemost. 2004;91:87–94. doi: 10.1160/TH03-06-0379. [DOI] [PubMed] [Google Scholar]
- 4.Klein TE, Altman RB, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009 Feb 19;360(8):753–64. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarz UI, Ritchie MD, Bradford Y, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008 Mar 6;358(10):999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodcock J, Lesko LJ. Pharmacogenetics--tailoring treatment for the outliers. N Engl J Med. 2009 Feb 19;360(8):811–3. doi: 10.1056/NEJMe0810630. [DOI] [PubMed] [Google Scholar]
- 7.Steward DJ, Haining RL, Henne KR, et al. Genetic association between sensitivity to warfarin and expression of CYP2C9*3. Pharmacogenetics. 1997 Oct;7(5):361–7. doi: 10.1097/00008571-199710000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Joffe HV, Xu R, Johnson FB, et al. Warfarin dosing and cytochrome P450 2C9 polymorphisms. Thromb Haemost. 2004 Jun;91(6):1123–8. doi: 10.1160/TH04-02-0083. [DOI] [PubMed] [Google Scholar]
- 9.Meckley LM, Wittkowsky AK, Rieder MJ, et al. An analysis of the relative effects of VKORC1 and CYP2C9 variants on anticoagulation related outcomes in warfarin-treated patients. Thromb Haemost. 2008;100:229–239. [PubMed] [Google Scholar]
- 10.Voora D, Eby C, Linder MW, et al. Prospective dosing of warfarin based on cytochrome P-450 2C9 genotype. Thromb Haemost. 2005 Apr;93(4):700–5. doi: 10.1160/TH04-08-0542. [DOI] [PubMed] [Google Scholar]
- 11.Rost S, Fregin A, Ivaskevicius V, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004 Feb 5;427(6974):537–41. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 12.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005 Jun 2;352(22):2285–93. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 13.Harrington DJ, Underwood S, Morse C, et al. Pharmacodynamic resistance to wararin associated with a Va166Met substitution in vitamin K epoxide reductase complex subunit 1 Thromb Haemost 2005. 93:23–26. doi: 10.1160/TH04-08-0540. [DOI] [PubMed] [Google Scholar]
- 14.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008 Mar;83(3):460–70. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 15.Anderson JL, Horne BD, Stevens SM, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007 Nov 27;116(22):2563–70. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 16.Huang SW, Chen HS, Wang XQ, et al. Validation of VKORC1 and CYP2C9 genotypes on interindividual warfarin maintenance dose: a prospective study in Chinese patients. Pharmacogenet Genomics. 2009 Mar;19(3):226–34. doi: 10.1097/FPC.0b013e328326e0c7. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Bhattaram AV, Jadhav PR, et al. Leveraging prior quantitative knowledge to guide drug development decisions and regulatory science recommendations: impact of FDA pharmacometrics during 2004-2006. J Clin Pharmacol. 2008 Feb;48(2):146–56. doi: 10.1177/0091270007311111. [DOI] [PubMed] [Google Scholar]
- 18.Bhattaram VA, Bonapace C, Chilukuri DM, et al. Impact of pharmacometric reviews on new drug approval and labeling decisions--a survey of 31 new drug applications submitted between 2005 and 2006. Clin Pharmacol Ther. 2007 Feb;81(2):213–21. doi: 10.1038/sj.clpt.6100051. [DOI] [PubMed] [Google Scholar]
- 19.Bhattaram VA, Booth BP, Ramchandani RP, et al. Impact of pharmacometrics on drug approval and labeling decisions: a survey of 42 new drug applications. AAPS J. 2005;7(3):E503–12. doi: 10.1208/aapsj070351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi VA, Duffy E, Funke BH, et al. Platform evaluation for rapid genotyping of CYP2C9 and VKORC1 alleles. Per Med. 2009;6(4):449–57. doi: 10.2217/pme.09.8. [DOI] [PubMed] [Google Scholar]
- 21.Hamberg AK, Dahl ML, Barban M, et al. A PK-PD model for predicting the impact of age, CYP2C9, and VKORC1 genotype on individualization of warfarin therapy. Clin Pharmacol Ther. 2007 Apr;81(4):529–38. doi: 10.1038/sj.clpt.6100084. [DOI] [PubMed] [Google Scholar]
- 22.Rosendaal FR, Cannegieter SC, van der Meer FJ, et al. A method to determine the optimal intensity of oral anticoagulant therapy. Thromb Haemost. 1993 Mar 1;69(3):236–9. [PubMed] [Google Scholar]
- 23.Antman EM. Hirudin in acute myocardial infarction. Thrombolysis and Thrombin Inhibition in Myocardial Infarction (TIMI) 9B trial. Circulation. 1996 Sep 1;94(5):911–21. doi: 10.1161/01.cir.94.5.911. [DOI] [PubMed] [Google Scholar]
- 24.Hillman MA, Wilke RA, Yale SH, et al. A prospective, randomized pilot trial of model-based warfarin dose initiation using CYP2C9 genotype and clinical data. Clin Med Res. 2005 Aug;3(3):137–45. doi: 10.3121/cmr.3.3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Epstein RS, Moyer TP, Aubert RE, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study) J Am Coll Cardiol. 2010 Jun 22;55(25):2804–12. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Wadelius M, Chen LY, Lindh JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009 Jan 22;113(4):784–92. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salinger DH, Shen DD, Thummel K, et al. Pharmacogenomic trial design: use of a PK/PD model to explore warfarin dosing interventions through clinical trial simulation. Pharmacogenet Genomics. 2009 Dec;19(12):965–71. doi: 10.1097/FPC.0b013e3283333b80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds KK, Valdes R, Jr., Hartung BR, et al. Individualizing warfarin therapy. Personalized Medicine. 2007;4(1):11–31. doi: 10.2217/17410541.4.1.11. [DOI] [PubMed] [Google Scholar]
- 29.Ansell J, Hollowell J, Pengo V, et al. Descriptive analysis of the process and quality of oral anticoagulation management in real-life practice in patients with chronic non-valvular atrial fibrillation: the international study of anticoagulation management (ISAM) J Thromb Thrombolysis. 2007 Apr;23(2):83–91. doi: 10.1007/s11239-006-9022-7. [DOI] [PubMed] [Google Scholar]
- 30.Yuen E, Gueorguieva I, Wise S, et al. Ethnic differences in the population pharmacokinetics and pharmacodynamics of warfarin. J Pharmacokinet Pharmacodyn. 2010 Feb;37(1):3, 24. doi: 10.1007/s10928-009-9138-4. [DOI] [PubMed] [Google Scholar]
- 31.Wu AH, Wang P, Smith A, et al. Dosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multi-ethnic population: comparison with other equations. Pharmacogenomics. 2008 Feb;9(2):169–78. doi: 10.2217/14622416.9.2.169. [DOI] [PubMed] [Google Scholar]
- 32.You JH, Zuo Z, Lo CM, et al. Any effect of CYP2C9 variants on warfarin clearance in Chinese patients? Thromb Haemost. 2007;97:866–868. [PubMed] [Google Scholar]
- 33.Geisen C, Watzka M, Sittinger K, et al. VKORC1 haplotypes and their impact on the inter-individual and inter-ethnical variability of oral anticoagulation. Thromb Haemost. 2005;94:773–779. doi: 10.1160/TH05-04-0290. [DOI] [PubMed] [Google Scholar]
- 34.King CR, Deych E, Milligan P, et al. Gamma-glutamyl carboxylase and its influence on warfarin dose. Thromb Haemost. 2010;104:750–754. doi: 10.1160/TH09-11-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vecsler M, Loebstein R, Almog S, et al. Combined genetic profiles of components and regulators of the vitamin K-dependent gamma-carboxylation system affect individual sensivity to warfarin. Thromb Haemost. 2006;95:205–211. doi: 10.1160/TH05-06-0446. [DOI] [PubMed] [Google Scholar]
- 36.Chan E, McLachlan A, O’Reilly R, et al. Stereochemical aspects of warfarin drug interactions: use of a combined pharmacokinetic-pharmacodynamic model. Clin Pharmacol Ther. 1994 Sep;56(3):286–94. doi: 10.1038/clpt.1994.139. [DOI] [PubMed] [Google Scholar]
- 37.Adcock DM, Duff S. Enhanced standardization of the International Normalized Ratio through the use of plasma calibrants: a concise review. Blood Coagul Fibrinolysis. 2000 Oct;11(7):583–90. doi: 10.1097/00001721-200010000-00001. [DOI] [PubMed] [Google Scholar]