SUMMARY
Canonical primary microRNA transcripts (pri-miRNAs) are characterized by a ~30-bp hairpin flanked by single-stranded regions. These pri-miRNAs are recognized and cleaved by the Microprocessor complex consisting of the Drosha nuclease and its obligate RNA-binding partner DGCR8. It is not well understood how the Microprocessor specifically recognizes pri-miRNA substrates. Here we show that in addition to the well-known double-stranded RNA-binding domains, DGCR8 uses a dimeric heme-binding domain to directly contact pri-miRNAs. This RNA-binding heme domain (Rhed) directs two DGCR8 dimers to bind each pri-miRNA hairpin. The two Rhed-binding sites are located at both ends of the hairpin. The Rhed and its RNA-binding surface are important for pri-miRNA processing activity. Additionally, the heme cofactor is required for formation of processing-competent DGCR8-pri-miRNA complexes. Our study reveals a unique protein-RNA interaction central to pri-miRNA recognition. We propose a unifying model in which two DGCR8 dimers clamp a pri-miRNA hairpin using their Rheds.
Keywords: DiGeorge syndrome, miRNA maturation, pri-miRNA processing, protein-RNA interaction, ribonuclease III, iron, porphyrin
INTRODUCTION
In the canonical microRNA (miRNA) maturation pathway in animal cells, pri-miRNAs are specifically recognized and cleaved by the Microprocessor to produce precursor miRNAs (pre-miRNAs) in the nucleus (Kim et al., 2009; Guo, 2012). pre-miRNAs are exported to the cytoplasm where they are cleaved by the ribonuclease Dicer and mature miRNA strands are incorporated into the miRNA-induced silencing complexes. Previous studies have revealed fundamental features of pri-miRNAs, including a hairpin with mature miRNA strands located on either side of the stem (Lee et al., 2003) and unstructured regions flanking the hairpin (Zeng and Cullen, 2005; Han et al., 2006). The hairpin stems contain roughly 30 base pairs with internal loops and bulges at variable positions. Base pairing interactions in the stem, especially the bottom third, are clearly important for processing (Lee et al., 2003). The basal junction of a pri-miRNA, where the stem and the flanking unstructured regions join, is required for processing (Han et al., 2006). It has been proposed that the basal junction serves as an anchoring point for the Microprocessor to determine the cleavage sites ~11 bp away (the basal junction anchoring model). There have also been reports that apical region of the hairpin, including a ≥10nt terminal loop, is important for processing (Zeng et al., 2005; Zhang and Zeng, 2010). Recently, three short (2–4 nt) sequence motifs have been shown to be enriched in non-nematode pri-miRNAs and important for processing of some pri-miRNAs in human cells (Auyeung et al., 2013). Overall, pri-miRNAs are defined primarily by their structures, with some sequence elements involved.
The Microprocessor has to identify true pri-miRNA substrates out of the myriad of other RNAs and DGCR8 (DiGeorge Critical Region gene 8, called Pasha in flies and worms) (Lee et al., 2003; Denli et al., 2004; Gregory et al., 2004; Han et al., 2004; Landthaler et al., 2004) plays a major role in this recognition. The 773-residue DGCR8 contains a nuclear localization signal (NLS) in the N-terminal region (Yeom et al., 2006; Shiohama et al., 2007), a central heme-binding domain, two double-stranded RNA-binding domains (dsRBDs) and a C-terminal tail (CTT) (Figure 1A). Prior to this study, the dsRBDs were shown to bind RNAs (Han et al., 2006; Faller et al., 2007; Sohn et al., 2007) and were defined as the “DGCR8 core” (Sohn et al., 2007). However, the dsRBDs alone bind pri-miRNAs with varying stoichiometry (Roth et al., 2013), and thus are unlikely to be the sole specificity determinant. DGCR8 has been shown to crosslink to pri-miRNA basal junction fragments (Han et al., 2006). However, the crosslinked protein and RNA residues have not been identified. It remains unknown how DGCR8 recognizes pri-miRNAs.
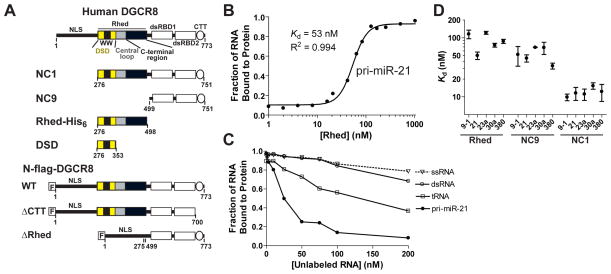
Figure 1. The Rhed contributes to pri-miRNA recognition by directly binding these RNAs and by collaborating with the dsRBDs.
(A) Recombinant human DGCR8 proteins used. “F” represents a FLAG tag. (B) A representative curve from filter binding assays showing that the Rhed binds pri-miRNAs. The data were fit using a cooperative binding model. The Kd is defined as the Rhed dimer concentration at which half maximal RNA binding is achieved. (C) Competition filter binding assays using unlabeled ssRNA, siRNA duplex, yeast tRNAs or pri-miR-21 to compete with a trace amount of 32P-labeled pri-miR-21 for association with 150 nM of Rhed dimer. An average molecular mass of 25 kDa was assumed in calculating molar concentrations of tRNAs. (D) Comparison of the Kd values of Rhed, NC9 and NC1 for a panel of five pri-miRNAs. The average Kd values and SD are summarized in Table 1. Purity of the recombinant proteins is shown in Figure S1. The sequences and MFOLD-predicted secondary structures (Zuker, 2003) of these pri-miRNAs are shown in Table S1 and Figure S2.
DGCR8 binds an essential heme (protoporphyrin IX in complex with iron) cofactor using a unique heme-binding domain. An active recombinant DGCR8 construct called NC1 (residues 276–751) and an isolated heme-binding domain (Figure 1A) both form constitutive dimers bound with one heme molecule (Faller et al., 2007; Senturia et al., 2010; Barr et al., 2011; Barr et al., 2012). Fe(III) heme directly binds the apo form of NC1 dimer and activates pri-miRNA processing in vitro (Barr et al., 2012). In HeLa cells, all known heme-binding-deficient DGCR8 mutants are inactive in pri-miRNA processing and heme availability affects processing efficiency (Weitz et al., 2014). A WW motif-containing dimerization sub-domain (DSD) resides in the heme-binding domain and contributes a surface for heme binding (Senturia et al., 2010). Dimerization and heme binding appear to be conserved features of DGCR8 homologs (Senturia et al., 2012). Despite the body of evidence supporting the importance of the DGCR8 heme-binding domain, its function in pri-miRNA processing remains unclear.
In this work, we show that the heme-binding domain of DGCR8 plays a key role in pri-miRNA substrate recognition. Therefore, we have renamed this domain the RNA-binding heme domain (Rhed). The Rhed directly binds pri-miRNAs at the basal and apical junctions of the hairpin. Using cellular and biochemical pri-miRNA processing assays, we show that the Rhed and the Rhed-RNA interaction are important for DGCR8 activity. By collaborating with other domains, the Rhed allows full structural features of pri-miRNAs to be recognized.
RESULTS
The Rhed of DGCR8 directly binds pri-miRNAs, contributing to affinity and specificity
Our biochemical analyses indicate a function of Fe(III) heme-bound Rhed in pri-miRNA recognition. Because the Fe(II) heme-bound and heme-free forms of the human Rhed are insoluble at pH 5–8, we exclusively use the Fe(III) heme-bound Rhed dimer in this study and refer to this form of the protein as the Rhed. Filter binding assays showed that the Rhed binds a panel of five pri-miRNAs (Figures S2) with Kd values ranging from 50–120 nM under equilibrium conditions (Figure 1B and Table 1). These interactions appear to be specific to pri-miRNAs, as neither a 21-nt single-stranded RNA (ssRNA) nor an siRNA duplex can compete with pri-miR-21 for binding the Rhed in competition filter binding assays (Figure 1C). tRNAs do compete but not as efficiently as unlabeled pri-miR-21 (Figure 1C). Therefore, it is likely that the Rhed contributes to the pri-miRNA-binding specificity of DGCR8.
Table 1.
Summary of Kd values (nM) measured using filter binding assays.
| Rhed and dsRBDs work together to achieve high-affinity binding to pri-miRNA | ||||
|---|---|---|---|---|
| pri-miRNA | Rhed | NC9 | DSD | NC1 |
| pri-miR-380 | 87 ± 7 | 33 ± 4 | 143 ± 13 | 12 ± 4 |
| pri-miR-9-1 | 119 ± 18 | 52 ± 19 | 152 ± 51 | 10 ± 1 |
| pri-miR-21 | 50 ± 7 | 45 ± 6 | 158 ± 10 | 12 ± 4 |
| pri-miR-23a | 121 ± 7 | 69 ± 0.6 | 208 ± 23 | 10 ± 1 |
| pri-miR-30a | 75 ± 6 | 72 ± 19 | 204 ± 3 | 16 ± 1 |
| Kd of Rhed for truncated pri-miRNAs | |||||
|---|---|---|---|---|---|
| Apical Junctions | # bp in stem | Kd | Basal Junctions | # bp in stem | Kd |
| aj-miR-23a-C | 24 | 102 ± 6 | bj-miR-23a | 9 | 232 ± 15 |
| aj-miR-23a-D | 20 | 191 ± 24 | |||
| aj-miR-23a-E | 11 | FBmax=20–50% | |||
| aj-miR-23a-F | 7 | FBmax=20–50% | |||
| aj-miR-21-D | 18 | 178 ± 32 | bj-miR-21 | 8 | 326 ± 32 |
| aj-miR-21-E | 10 | FBmax=20–50% | |||
| Mutations in the Rhed region that directly or indirectly affect the affinity for pri-miRNAs | ||||||
|---|---|---|---|---|---|---|
| Mutants | R322A/R325A | R341A/K342A | K424A/K426A/K431A (G1) | |||
| Context | NC1 | DSD | NC1 | Rhed | NC1 | Rhed |
| pri-miR-380 | 25 ± 3 | 151 ± 16 | 32 ± 3 | N.B. | 50 ± 6* | 57.3 ± 20 |
| pri-miR-9-1 | 28 ± 5 | 116 ± 8 | 48 ± 5 | N.B. | 30 ± 5* | FBmax=20–50% |
| pri-miR-21 | 22 ± 1 | 240 ± 14 | 45 ± 3* | N.B. | 19 ± 1* | FBmax=20–50% |
| pri-miR-23a | 36 ± 20 | 128 ± 41 | 45 ± 5 | N.B. | 24 ± 5* | FBmax=20–50% |
Errors are S.D. from 3–9 repeats except those marked by “*”, which indicate ranges from two replicates. In the binding reactions, the fraction of RNA bound (FB) to protein generally plateaued to >0.9. Those reactions in which the maximal FB only reaches 20–50% are marked as “FBmax=20–50%”. “N.B.” indicates no binding. The highest protein concentrations used were 1 or 3 μM. See also Figures S2–S3 and Table S1.
To estimate the relative contribution of the Rhed and dsRBDs to pri-miRNA binding, we measured the affinity of Rhed, NC1 and NC9 (=NC1 ΔRhed) for pri-miRNAs. The NC1 protein we used in this study is a Fe(III) heme-bound dimer unless stated otherwise. With deletion of the Rhed that is also responsible for dimerization, NC9 is a monomer. The affinity of the Rhed for each of the five pri-miRNAs (Kd = 50–120 nM) is comparable to that of the dsRBDs-containing NC9 (Kd = 30–70 nM) (Figure 1D and Table 1), indicating the importance of Rhed in RNA binding. Both the Rhed and NC9 have lower affinities for each of the pri-miRNAs compared to NC1 (Kd = 10–16 nM) (Figure 1D and Table 1), suggesting that these domains work together to achieve tight binding. The Kd values for NC1 and NC9 are in qualitative agreement with other measurements for similar constructs as previously reported (Faller et al., 2007; Sohn et al., 2007; Faller et al., 2010; Roth et al., 2013).
Each pri-miRNA hairpin contains two binding sites for DGCR8
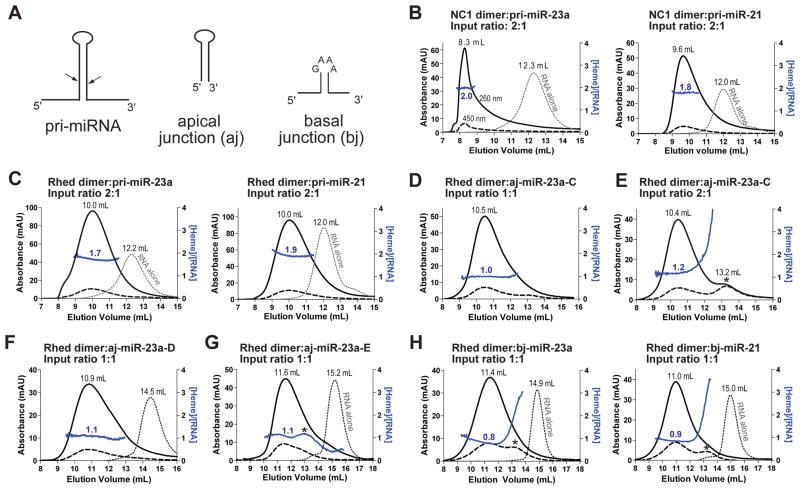
We analyzed the DGCR8-pri-miRNA binding stoichiometry using size exclusion chromatography (SEC), in which A450nm (the Söret peak of the DGCR8-bound heme) and A260nm (contributed mostly by RNA and to a lesser extent by the DGCR8-heme complex) were monitored simultaneously. Injection of NC1 mixed with pri-miR-23a or pri-miR-21 at 2:1 ratio resulted in single peaks (Figures 2A and 2B). Using a recently determined extinction coefficient (ε) of human DGCR8-bound heme (74 mM−1 cm−1 at 450 nm) (Senturia et al., 2012), we calculated the molar ratios of NC1 dimers and pri-miRNAs to be ~2:1 across the elution peaks (Figure 2B). These data suggest that there are two binding sites for DGCR8 dimers on each pri-miRNA hairpin.
Figure 2. RNA truncation and SEC analyses suggest that the Rhed binds to pri-miRNA junctions.
(A) Schematics of pri-miRNA fragments. The arrows indicate the Drosha cleavage sites. The sequences and secondary structures are shown in Table S1 and Figures S2 and S3. (B–H) Size exclusion chromatograms of NC1 in complex with 2 μM pri-miRNAs (B), the Rhed with 2 μM pri-miRNAs (C), the Rhed with 4 μM of aj-miR-23a-C at varying input ratios (D–E), the Rhed with 4 μM of aj-miR-23a-D (F), the Rhed with 4 μM of aj-miR-23a-E (G), and the Rhed with 4 μM of indicated basal junctions (H). Solid black lines indicate A260, dashed lines show A450 and dotted lines are A260 of the RNA-only runs. Solid blue lines represent heme:RNA ratios calculated from A450 and A260, following the scale on the right y axis. The asterisk in (E, G, H) marks a peak of free Rhed. See also Figures S1–S4 and Table S1.
The elution volume of the NC1-pri-miR-23a complex (8.3 mL) is close but not identical to the void volume (8.2 mL). We previously observed a similar elution volume for the NC1-pri-miR-30a complex (8.5 mL) (Figure 5D) (Faller et al., 2007). The 480-kDa apoferritin (one of the standard proteins used for calibration) also elutes in this region. These DGCR8-pri-miRNA complexes (including NC1-pri-miR-21) are expected to have molecular masses of ~260 kDa, but greatly deviate from globular shapes and contain peripheral RNA strands of various lengths and structures. Therefore, it is not surprising that these complexes elute as if with higher molecular masses. Importantly, the protein-RNA ratios determined from the A450 and A260 measurements are independent of the elution volumes and shapes of the complexes.
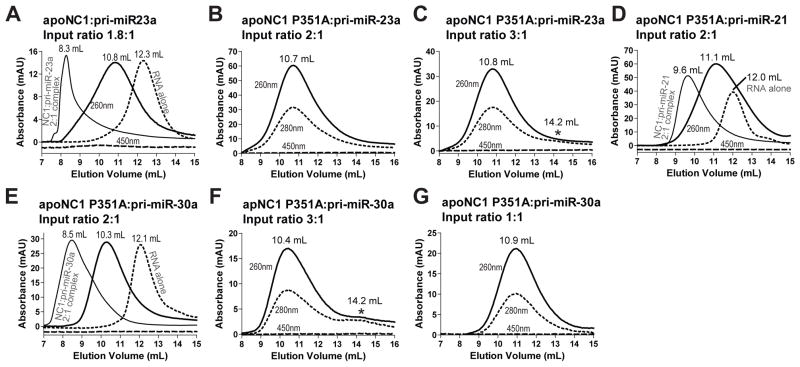
Figure 5. Fe(III) heme causes a large conformational change to DGCR8-pri-miRNA complexes.
Size exclusion chromatograms of (A) apoNC1 in complex with 0.45 μM pri-miR-23a and (B–G) apoNC1 P351A with 2 μM pri-miRNAs at the indicated input ratios. The asterisk in (C) and (F) marks a potential free protein peak.
The Rhed determines the stoichiometry of DGCR8-pri-miRNA interaction
We performed similar SEC analyses using the Rhed. An input containing the Rhed and pri-miRNA (pri-miR-23a, pri-miR-21 or pri-miR-30a) at 2:1 molar ratio elutes in a single peak that is about 2 mL earlier than that of the free RNA (Figures 2C and S4A). Based on the A450 and A260 in the chromatograms, we calculated the Rhed:RNA ratio to be ~2:1 across the elution peak. Therefore, we conclude that there are two Rhed-binding sites on a pri-miRNA and that the Rhed is responsible for determining the DGCR8-pri-miRNA binding stoichiometry. It is likely that the Rhed occupies similar pri-miRNA-binding sites whether it is in an isolated polypeptide or a part of processing-competent DGCR8 proteins.
We also analyzed the Rhed and pri-miR-30a complex at sub-stoichiometric (1:1) input ratio. We observed an SEC elution peak at 11.4 mL, between those of the 2:1 complex (10.1 mL) and the free RNA (~12 mL) (Figure S4B). The Rhed:RNA ratio gradually changed from 2:1 to 0 across the elution peak, indicating the presence of multiple species at 2:1, 1:1 and 0:1 ratios that were partially resolved. This result suggests that, at least in the absence of the dsRBDs and CTT, the Rhed does not strongly prefer to bind one site versus the other.
The Rhed binds both ends of a pri-miRNA hairpin—the apical and basal junctions
To locate the Rhed-binding sites on pri-miRNAs, we generated a series of truncated pri-miRNAs (Figures 2A and S3, Table S1) and analyzed their interactions with the Rhed using filter binding assays and SEC. A pri-miR-23a truncation contains the 10-nt hairpin loop and 24 bp of the upper stem, and thus includes the apical junction (aj-miR-23a-C, Figure S3A). The Rhed binds to aj-miR-23a-C with an affinity similar to that for pri-miR-23a (Table 1). Importantly, SEC analyses indicated that the Rhed dimer:aj-miR-23a-C molar ratio in their complex is reduced to ~1:1. When the Rhed:aj-miR-23a-C input ratio was 1:1, a single peak was observed; whereas increasing the input ratio to 2:1 resulted in a chromatogram containing both free-Rhed and complex peaks with nearly equal A450 (Figures 2D and 2E). We further truncated pri-miR-23a-C at its basal segments to generate aj-miR-23a-D, aj-miR-23a-E and aj-miR-23a-F, which contain 20 bp, 11 bp and 7 bp (Figures S3B–S3D), respectively. SEC analyses show that the Rhed forms ~1:1 complexes with all three RNAs (Figures 2F, 2G, S4C and S4D), suggesting that the apical junction region contains a core binding site for the Rhed. Filter binding assays showed that the affinity of Rhed for these RNAs decreases over the truncations, with Kd values increased to 191 nM for aj-miR-23a-D and the fractions of aj-miR-23a-E and aj-miR-23a-F bound to 1 μM Rhed only reached 0.25–0.30 and 0.15–0.20, respectively (Table 1). The large reduction in affinity caused by the deletion of 9 bp from aj-miR-23a-D suggests that the central segment of the pri-miR-23a stem is also important for the Rhed to bind the apical hairpin. Furthermore, to evaluate the importance of the hairpin loop, we replaced the 10-nt loop of aj-miR-23a-C with a GAAA tetraloop (aj-miR-23a-C-GAAA, Table S1) and found that the affinity of Rhed for this RNA decreased dramatically with the fraction of RNA bound to 1 μM Rhed only reached ~0.20. We also engineered pri-miR-21 to produce apical hairpins aj-miR-21-D and aj-miR-21-E that contain 18 bp and 10 bp in their stems (Figures S3E and S3F). The Rhed binds these RNAs with 1:1 stoichiometry (Figure S4E) and decreasing affinities over the truncations (Table 1), similar to the pri-miR-23a aj series. Therefore, we conclude that an Rhed-binding site is located in the apical junction area and a high-affinity interaction requires both the hairpin loop and ~20-bp of the upper stem.
To identify the second Rhed-binding site, we deleted the apical hairpins from the pri-miRNAs and thereby produced basal junction models that include 8–9 bp of the lower stem and 7–9 nt of single-stranded regions on both sides of the hairpin (Figures 2A and S3G–S3H). We linked the 5′ and 3′ strands using a GAAA tetraloop to stabilize the relatively short stem. The Rhed binds these bj-miRNAs, with affinities (Kd’s of 232–326 nM) modestly lower than those for pri-miRNAs (Kd’s of 50–120 nM) (Table 1), and the stoichiometry is ~1:1 (Figure 2H). As the bj RNAs are already quite short, we conclude that the second Rhed-binding site is located at the basal junction of a pri-miRNA.
Both the apical and basal Rhed-binding sites contain stem-ssRNA junctions, which are likely to be important features for Rhed to recognize. The stem and single-stranded regions of the junctions are all required for binding the Rhed, as neither ssRNA nor a duplex can compete with pri-miRNAs (Figure 1C) and substituting the hairpin loop of aj-miR-23a-C with GAAA disrupts the binding (see above). We also considered the possibility that certain pri-miRNA sequences drive the association with the Rhed. Three sequence motifs have recently been shown to be important for processing of some pri-miRNAs in human cells (Auyeung et al., 2013), including a “UG” at −14 position (14 nt upstream of the 5′ Drosha cleavage site), a “UGU” or “GUG” at P22–P44 positions (22–24 nt into the pre-miRNA) and a “CNNC” at positions 16–17 (16–17 nt downstream of the 3′ Drosha cleavage site). The −14 position is located at the basal junction, P22–P24 at the apical junction, and the 16–17 positions are close to the basal junction. We searched for these motifs in the five pri-miRNAs used in this study. pri-miR-30a has all three motifs, pri-miR-380 has none, and each of pri-miR-9-1, -21, and -23a contains two (Figures S2). There is no clear correlation between the presence of these motifs and the affinity for Rhed (Table 1). Therefore, it is likely that the Rhed recognizes the structures of pri-miRNA junctions.
The Rhed is required for pri-miRNA processing
We next tested if the Rhed and its RNA-binding activity are important for pri-miRNA processing using cellular and biochemical assays. We first employed a recently developed fluorescent live-cell assay for pri-miRNA processing (Weitz et al., 2014). In this assay, a reporter plasmid inducibly expresses two fluorescent proteins, mCherry and eYFP (Figure 3A). A pri-miRNA sequence is inserted into the 3′-untranslated region of the mCherry expression cassette, so that cleavage of the pri-miRNA reduces mCherry expression. The eYFP and mCherry fluorescent signals for individual cells have a linear relationship and the slope faithfully indicates the efficiency of pri-miRNA processing but is not affected by subsequent steps of the miRNA maturation pathway (Weitz et al., 2014). Co-transfection of the reporter with the N-flag-DGCR8 expression plasmid (Figure 1A) increases pri-miRNA processing efficiency and the fluorescence slope (Figure 3B), thus provides a robust method for measuring the activity of DGCR8 mutants. Endogenous DGCR8 is expressed at a very low level in HeLa cells and does not seem to interfere with the measurements as the N-flag-DGCR8 expression is typically 30–100 folds higher (Weitz et al., 2014).
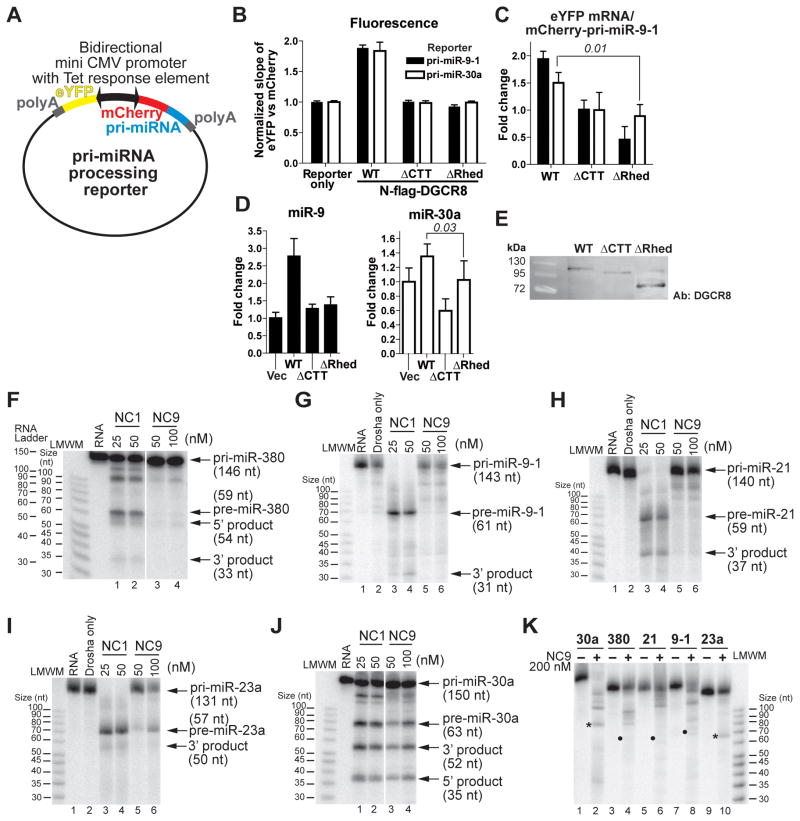
Figure 3. The DGCR8 Rhed is important for pri-miRNA processing.
(A) Schematic of the reporter plasmids. (B–E) The reporters were transfected into HeLa cells either alone or with the indicated N-flag-DGCR8 expression plasmids. (B) Slopes of the eYFP and mCherry fluorescence intensities, after normalization to that of the reporter-only transfection, are plotted. Error bars represent 95% CI. (C) Ratios of eYFP mRNA and mCherry-pri-miRNA. (D) Abundance of mature miR-9 and miR-30a normalized by that of β-actin (mean ± SD, n = 3). Select P values are indicated in italic. miR-30a is highly expressed endogenously in HeLa cells and thus the relative changes are modest. (E) An anti-DGCR8 immunoblot of nuclear extracts from the transfected cells. Equal amount of total proteins was loaded in each lane, as estimated using a Coomassie-stained SDS gel. (F–K) Reconstituted pri-miRNA processing assays. Low molecular weight marker, LMWM. Relationship between LMWM and a true RNA ladder in 15% gels is shown in panel (F). In panel (K), the asterisks mark a pre-miRNA band and the dots mark the position expected for a pre-miRNA product. See also Figure S1.
Using live-cell reporters containing either pri-miR-9-1 or pri-miR-30a, we found that deletion of the Rhed renders DGCR8 inactive. Unlike the wild type, expression of N-flag-DGCR8 ΔRhed (Figure 1A) fails to increase the eYFP vs mCherry slopes relative to the transfections either without exogenous DGCR8 expression or with an inactive DGCR8 mutant ΔCTT in which the CTT is deleted (Han et al., 2004; Faller et al., 2010) (Figure 3B). The lost activity of ΔRhed is further supported by quantitative RT-PCR measurements of the eYFP mRNA (for normalization), mCherry-pri-miRNA fusions and mature miRNAs (Figures 3C and 3D), and is not caused by reduced DGCR8 protein expression or lack of nuclear localization (Figure 3E). Altogether, our data suggest that the Rhed is required for pri-miRNA processing in human cells.
Two previous studies showed that recombinant DGCR8 proteins without the Rhed and NLS are active for processing pri-miR-16 and pri-miR-30a in vitro (Yeom et al., 2006; Faller et al., 2007). To clarify the functional importance of the Rhed, we compared the processing activity of NC1 and NC9 (Figure 1A) in vitro using four additional pri-miRNA substrates (Figures S2A–S2D; Table S1). Deletion of the Rhed from the highly active NC1 abolishes processing of pri-miR-380, pri-miR-9-1 and pri-miR-21 at all DGCR8 concentrations tested in vitro (Figures 3F–3H and 3K). The activity of NC9 is greatly reduced for pri-miR-23a compared to that of NC1, with some processing observed at high NC9 concentrations (100 nM in Figure 3I and 200 nM in Figure 3K). We also confirmed that NC9 has substantial pri-miR-30a processing activity (Figures 3J and 3K), similar to the previous report (Faller et al., 2007). The pri-miR-30a sequence used in in vitro processing assays is identical to the insert of the pri-miR-30a cellular reporter. It is possible that, in vitro, recombinant DGCR8 constructs with the Rhed deleted can assemble with certain pri-miRNAs into productive complexes that are not formed in vivo. Altogether, our cellular and biochemical results demonstrate that the Rhed is important for DGCR8 function. These observations also highlight the importance of using both biochemical and cellular assays in studying pri-miRNA processing mechanism.
The RNA-binding surface of Rhed is important for pri-miRNA processing
We next analyzed the RNA-binding surface of the Rhed using truncation and site-directed mutagenesis. The Rhed can be roughly divided into three regions: the N-terminal DSD, a central acidic loop, and a C-terminal region (Figure 1A). The DSD is soluble when expressed in E. coli without the rest of Rhed (Senturia et al., 2010). Filter binding assays show that the DSD binds pri-miRNAs, but does not bind the 21-nt ssRNA or the siRNA duplex. The affinities of the DSD for the five pri-miRNAs (Kd = 150–300 nM) are only modestly lower than those of the Rhed (Kd = 50–120 nM) (Table 1). However, unlike the Rhed, the DSD binds the junction-less aj-miR-23a-C-GAAA, with a Kd of 518 ± 45 nM (mean ± range, n = 2). These results suggest that the DSD makes an important contribution to the Rhed-pri-miRNA interaction, but it does not retain all the affinity or exactly the same binding specificity.
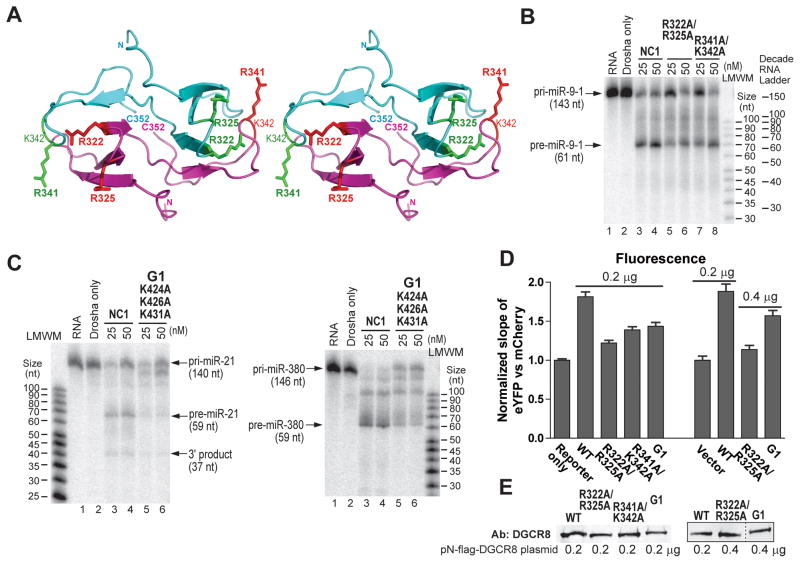
Site-directed mutagenesis demonstrated that both the DSD and C-terminal regions of the Rhed contribute to pri-miRNA binding. The central loop is the least conserved among DGCR8 homologs, thus was not explored here. We previously determined crystal structures of the DSDs from human and frog DGCR8 (Senturia et al., 2010; Senturia et al., 2012). Inspection of the structures identified four surface-exposed basic residues, R322, R325, R341 and K342, which we mutated to alanine in pairs of spatial proximity (Figure 4A). The C-terminal region (residues 413–498) is rich in conserved basic residues (Senturia et al., 2012). Because no structure is available for this region, we systematically mutated them to alanine in groups with each containing 2–3 nearby mutations. These mutations were introduced to DGCR8 in a variety of contexts for pri-miRNA binding and processing assays. Filter binding assays indicated that the R341A/K342A mutations reduce the affinities by 3–5 folds for pri-miRNAs in the context of the NC1 and abolish pri-miRNA binding in the context of the Rhed (Table 1). The lack of pri-miRNA binding of Rhed R341A/K342A was confirmed using SEC analyses (Figure S5A). These results clearly indicate that R341 and K342 make critical contribution to the Rhed-pri-miRNA interaction, most likely by directly participating at the RNA-binding interface. The modest affinity changes of the NC1 mutant are not surprising, as the dsRBDs are intact.
Figure 4. The pri-miRNA-binding surfaces of the Rhed are important for processing.
(A) Stereo diagram of the DSD crystal structure of human DGCR8 (PDB access code 3LE4) (Senturia et al., 2010), with the side chains of the mutated residues shown in sticks. The two subunits are drawn in cyan and magenta. (B, C) Reconstituted pri-miRNA processing assays. (D) Cellular assays using the pri-miR-9-1 reporter. The amounts of DGCR8 expression plasmids or the pCMV-Tag-2A vector are indicated on the graph. Error bars represent 95% CI. The presence of pCMV-Tag2A vector in the control transfection does not alter the fluorescence slope. (E). Anti-DGCR8 immunoblots of nuclear extracts from transfected cells. Equal amount of total proteins was loaded in each lane. See also Figures S1 and S5.
The R322A/R325A mutations reduce the affinity for pri-miRNAs by about 2–4 folds in the context of NC1, but make the Rhed protein insoluble and do not strongly alter the affinity for pri-miRNAs in the context of the DSD (Table 1). These observations suggest that R322 and R325 affect the Rhed-pri-miRNA interaction indirectly by partially disrupting the Rhed structure. The K424A/K426A/K431A mutations (G1) decreases the affinity for pri-miRNAs by 2–4 folds in the context of NC1 and render the Rhed protein incapable to plateauing to >50% occupancy of most pri-miRNAs in filter binding assays (Table 1). These RNA-binding defects were not caused by failure of the Rhed G1 mutant to bind the nitrocellulose membrane used in filter-binding assays, regardless whether pri-miRNAs are present (data not shown). However, SEC analyses of the Rhed G1-pri-miRNA complexes, assembled at higher protein and RNA concentrations (4 and 2 μM respectively), showed peaks similar to those of the complexes formed by the wild-type Rhed protein (Figure S5B). Overall, our data suggest that at least some of the mutated residues contribute to pri-miRNA binding but the RNA-binding defects of the mutant are not as severe as those of R341A/K342A.
The R341A/K342A, R322A/R325A and G1 mutants in the context of NC1 demonstrate reduced pri-miRNA processing activity in vitro (Figures 4B and 4C). Their electronic absorption spectra are indistinguishable from those of the wild type (Figures S5C–S5F), ruling out the possibility that the pri-miRNA processing defects are caused by a heme-binding deficiency. Finally, these mutations were tested in the context of N-flag-DGCR8 using the live-cell reporter assay. The normalized eYFP vs mCherry slopes were 1.22 ± 0.03, 1.39 ± 0.04 and 1.44 ± 0.05 (± 95% CI, same below) for R322/R325A, R341A/K342A and G1, respectively (Figure 4D). These slopes are significantly lower (p values < 0.0001) than the 1.81 ± 0.06 for the wide-type N-flag-DGCR8 but also significantly higher (p values < 0.0001) than the 1.00 ± 0.02 for the reporter-only transfections. Immunoblotting analyses indicated that the N-flag-DGCR8 mutants were expressed at levels either similar to (R341A/R325A) or slightly lower than (R322A/K325A and G1) that of the wild type (Figure 4E). We successfully compensated the lower expression levels of the N-flag-DGCR8 mutants by doubling the amounts of expression plasmids used in the transfections (Figure 4E), and observed no increase in the fluorescence slope for R322A/R325A (1.14 ± 0.05) and a slight increase of the fluorescence slope for G1 (1.57 ± 0.07) (Figure 4D). The G1 slope is still significantly lower than that of the wild-type control (1.88 ± 0.09) (p value < 0.0001). These results indicate that these mutations render the DGCR8 protein partially defective in cells. Altogether, our data demonstrate that the DSD and the C-terminal region of the Rhed contribute to pri-miRNA binding and that the RNA-binding surface is important for the pri-miRNA processing activity of DGCR8.
Fe(III) heme is required for formation of proper DGCR8-pri-miRNA complexes
To understand the role of heme in DGCR8-pri-miRNA interaction, we analyzed the interaction between the heme-free apoNC1 proteins and pri-miRNA. We previously showed that apoNC1 has affinity for pri-miRNAs similar to that of the Fe(III) heme-bound form (Barr et al., 2012). This is not surprising as the DSD and dsRBDs are still expected to be well folded. SEC analysis of the apoNC1 dimer with pri-miR-23a at an input ratio of 2:1 resulted in a peak at 10.8 mL (Figure 5A). This elution volume is between those of free pri-miR-23a (12.3 mL) and the Fe(III) heme-bound NC1-pri-miR-23a complex (8.3 mL), suggesting that the apoNC1 binds pri-miR-23a but in a conformation and/or stoichiometry different from those of the heme-bound NC1-pri-miR-23a complex.
We further analyzed pri-miRNA complexes with the apo form of a NC1 mutant P351A. The wild-type apoNC1 dimer is difficult to produce, usually has ~10% of residual heme associated and is not very soluble at pH 7 and above (Barr et al., 2012). In contrast, NC1 P351A is easily purified as a heme-free dimer and is soluble at pH 8 in which the binding assays are performed. apoNC1 P351A can bind Fe(III) heme to reconstitute a complex similar to the wild type (Barr et al., 2011). SEC of apoNC1 P351A dimer and pri-miR-23a at 2:1 input ratio resulted in a single peak at 10.7 mL, similar to the complex containing wild-type apoNC1 (Figure 5B). apoNC1 P351A with pri-miR-21 and pri-miR-30a also yielded elution peaks between those of the free RNAs and the heme-bound NC1-RNA complexes (Figures 5D and 5E). Increasing the protein:RNA input ratio to 3:1 did not shift the elution peaks, suggesting that the binding sites have been saturated (Figures 5C and 5F). Additionally, an increase of A280 was observed at ~14.2 mL, implying the presence of excess protein. Decreasing the input ratio to 1:1 shifted the elution peak to a larger volume but not as far as that of the free pri-miRNA (Figure 5G). These results suggest that apoNC1 binds a pri-miRNA with up to 2:1 stoichiometry. Overall, we conclude that the association of DGCR8 with Fe(III) heme causes a large conformational change in its complex with pri-miRNAs. It is also possible that the shift of elution volume is caused by partially disassembly of the apoNC1-pri-miRNA complexes during the SEC experiments. As heme-free DGCR8 is inactive in cells (Weitz et al., 2014), the conformation and/or stability induced by heme must be important for recognition and cleavage of pri-miRNAs by the Microprocessor.
The covalent linkage between the Rhed and dsRBDs is required for proper assembly of DGCR8-pri-miRNA complexes
To further dissect the relationship between the Rhed and dsRBDs in pri-miRNA recognition, we tested if the two components of NC1, namely the Rhed and NC9, can mediate pri-miRNA processing in trans or assemble into proper tertiary complexes with pri-miRNAs. As already shown in Figure 3, with NC9 alone, pri-miR-9-1 cannot be processed and pri-miR-23a is weakly processed in vitro. NC9 and Rhed together do not alter the pri-miRNA processing activity comparing to the NC9 alone (Figures S6A and S6B), indicating that the covalent linkage between the Rhed and dsRBDs is important for pri-miRNA processing. In SEC analyses with both the Rhed and NC9, the pri-miRNAs eluted at volumes smaller than those of the Rhed-pri-miRNA binary complexes but different from those of the NC1-pri-miRNA complexes (Figures S6C–S6E), suggesting that some (non-productive) tertiary complexes have formed. These observations suggest that the Rhed is responsible for properly anchoring the dsRBDs to pri-miRNAs for processing.
DISCUSSION
Our study identifies the DGCR8 Rhed as the junction-binding domain that anchors the Drosha-DGCR8 complex to pri-miRNAs. It is expected that dsRBDs of DGCR8 associate with the stems of pri-miRNA hairpins (Sohn et al., 2007). The Rhed and dsRBDs together should make extensive contacts with pri-miRNAs, allowing high-affinity binding and specific recognition.
The Rhed-junction interaction provides a physical basis for previously proposed models regarding pri-miRNA recognition. For the basal junction anchoring model (Han et al., 2006), the Rhed binding to the basal junction allows the Microprocessor complex to measure ~11 bp and thereby determine the Drosha cleavage sites (Figure 6A). It was also suggested that the apical junction may serve as an alternative anchoring point for the Microprocessor and this interaction leads to less efficient cleavages that are located in the middle of mature miRNA strands (called “abortive processing”) (Figure 6A) (Han et al., 2006). The capability of the Rhed to bind the apical junction can explain the abortive processing. In a second model, the Microprocessor binds the hairpin loop (approximately equivalent to the apical junction) and measures ~22 bp to determine the Drosha cleavage sites (Zeng et al., 2005; Zhang and Zeng, 2010) (Figure 6B). For this “apical junction anchoring model”, the Rhed-apical junction interaction results in productive processing. Common to both models is that association of DGCR8 with only one junction is sufficient to activate processing.
Figure 6. Models of how a pri-miRNA is recognized by the Microprocessor.
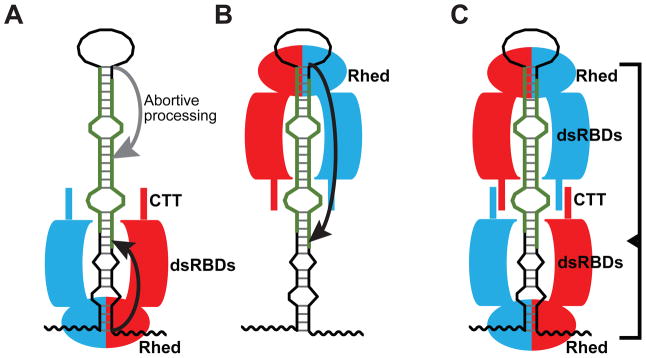
(A) The basal junction anchoring model (Han et al., 2006). (B) The apical junction anchoring model (Zeng et al., 2005). (C) Our proposed molecular clamp model. See Discussion for details. The DGCR8 subunits in a dimer are shown in red and cyan. The thick avocado strands represent 5′ and 3′ mature miRNAs.
The stoichiometry of DGCR8 and pri-miRNAs in processing competent complexes has been investigated in several studies, but no clear consensus has been reached. It has been previously reported that there is more than one copy of the DGCR8 protomer within the Microprocessor complex (Han et al., 2004). This observation may be explained by the dimerization of DGCR8 and/or by the formation of higher order structures of DGCR8 upon binding pri-miRNAs. The crystal structure of the monomeric DGCR8 core showed that the two domains adopt an α-β-β-β-α fold typical for dsRBDs and that these domains are held together by a relatively rigid interface involving an extra α-helix at the C-terminus of dsRBD2 (Sohn et al., 2007). The RNA-binding surfaces of dsRBD1 and dsRBD2 are expected to point to distinct directions. Based on this and other evidence, it was suggested that each DGCR8 core binds either one pri-miRNA hairpin that is severely bent or two separate hairpins. A recent NMR and biochemical study indicated that the DGCR8 core binds pri-miR-16 fragments with varying stoichiometry and that this interaction is reduced by competitor tRNAs (Roth et al., 2013). It was thereby concluded that the DGCR8-pri-miRNA interaction is non-specific.
In contrast to the DGCR8 core, DGCR8 proteins containing the Rhed do bind pri-miRNAs with specificity (Han et al., 2006; Faller et al., 2010) (Figure 1C) and well-defined stoichiometry. Based on biochemical analyses of DGCR8-pri-miRNA complexes, we previously proposed that DGCR8 dimers form a cooperative higher-order oligomer upon binding to a pri-miRNA (Faller et al., 2010). Using a previously estimated ε450 of 58 mM−1 cm−1 for DGCR8-bound Fe(III) heme, we derived NC1 dimer:pri-miRNA ratios of ~3:1 in SEC peaks of their complexes (Faller et al., 2007; Faller et al., 2010). The ε450 was recently revised to 74 mM−1 cm−1, using a widely accepted pyridine hemochromagen method (Senturia et al., 2012). Using this ε450 value, here we consistently obtained NC1 dimer:pri-miRNA ratios of ~2:1 for the previous and new data. Our study suggests that the Rhed is a major specificity and stoichiometry determinant. Any mechanism regarding pri-miRNA recognition must include the dimeric Rhed of DGCR8. Technically, it is important to use recombinant DGCR8 proteins with high heme content in the investigation (Barr and Guo, 2014).
The observation of simultaneous and cooperative binding of two NC1 dimers to a pri-miRNA suggests a “molecular clamp” model. In this model, two DGCR8 dimers grip both ends of the hairpin using their Rheds and interact with each other, making extensive contacts with the pri-miRNA stem (Figure 6C). While it remains to be determined whether binding of both DGCR8 dimers to a pri-miRNA is required for assembly of active tertiary complexes with Drosha, this model is supported by recent cellular data showing that both the apical and basal junctions of pri-miRNAs are important for determining Drosha cleavage sites (Ma et al., 2013). Furthermore, a previous negative stain electron tomography study yielded a “fat butterfly” density for a DGCR8-pri-miR-30a complex (Faller et al., 2010). Compared to the trimer of dimers proposal, the molecular clamp model seems to be more consistent with this electron density in that the body of the butterfly may be the pri-miRNA hairpin and the four wings may be the DGCR8 subunits in the dimer of dimers.
The molecular clamp model allows the length of a full pri-miRNA stem to be measured through an inter-dimer interaction. The full stem length is one of the most important features of pri-miRNAs, but is measured in neither the basal junction anchoring model nor the apical junction anchoring model. We previously showed that the CTT of DGCR8 contains an amphipathic α-helix and that mutation of conserved hydrophobic resides on this α-helix reduces binding cooperativity and abolishes pri-miRNA processing (Faller et al., 2010). This helix may be involved in the communication between the two DGCR8 dimers. The CTT is also required for binding Drosha (Han et al., 2006), possibly linking assembly of DGCR8 dimer of dimers to activation of RNA cleavage.
A DGCR8-pri-miRNA complex seems to contain built-in asymmetry, which should be able to help Drosha identify the correct cleavage sites that is closer to the basal junction. The apical junction of a pri-miRNA is next to a closed hairpin loop, whereas the single-stranded regions of the basal junction lead to the open ends of the RNA. This topological difference may contribute to the asymmetry of the complex. In fact, our data show different features in the Rhed interactions with the apical and basal junctions—the central region of the miRNA stem appears to be more important for Rhed association with the apical junctions than with the basal junctions (Table 1). Overall, the molecular clamp model unifies features of previous proposals, is consistent with most available experimental data and explains how the full structural features of pri-miRNAs are recognized.
The mechanisms of substrate recognition by two ribonuclease III enzymes involved in miRNA maturation, Drosha and Dicer, bear interesting similarities and differences. Both Drosha and Dicer partner with dsRNA-binding proteins. However, Dicer-associated RNA-binding partners are not required for cleavage; instead they modulate substrate affinity and cleavage rates as well as loading of small RNAs to RNA-induced silencing complexes (Doyle et al., 2012). The PAZ domain of Dicer anchors the enzyme to the open end of a pre-miRNA hairpin, allowing the cleavage sites to be determined at a fixed distance (MacRae et al., 2006; Ma et al., 2012). The helicase domain of Dicer binds to the pre-miRNA hairpin loop and enhances the cleavage of pre-miRNAs over other Dicer substrates such as long dsRNAs (Tsutsumi et al., 2011; Ma et al., 2012). The interaction between the DGCR8 Rhed and pri-miRNA basal junctions seems functionally analogous to that between the Dicer PAZ domain and pre-miRNA open end. Similarly, the interaction between the Rhed and pri-miRNA apical junction may be comparable to the contact between the Dicer helicase domain and pre-miRNA hairpin loop. Thus, Drosha and Dicer systems appear to utilize distinct domains for the same purpose of recognizing the ends of substrate RNA helices.
To our knowledge, the Rhed is the first example of a heme-binding domain that directly binds nucleic acids. A number of transcription factors contain regulatory heme-binding domains, but these domains are separate from their DNA-binding domains (Gilles-Gonzalez and Gonzalez, 2005; Yin et al., 2007; Marvin et al., 2009). We believe that DGCR8 uses the heme cofactor for structural stabilization and/or for regulatory functions. Without heme, DGCR8 still binds pri-miRNAs but their complexes do not adopt processing-competent conformations.
The Rhed appears to have evolved together with animal miRNAs, consistent with an essential function in pri-miRNA recognition. Neither the Rhed nor canonical miRNAs are found in bacteria or archaea. Plants do not have the Rhed and their miRNAs are processed from primary transcripts with longer hairpins by Dicer-like enzymes (Axtell et al., 2011). The Rhed is unique to DGCR8 homologs, whereas the dsRBDs are distributed among a wide range of organisms and in proteins involved in diverse biological functions (Masliah et al., 2013). Most canonical animal miRNAs are thought to originate from unstructured RNA sequences. Emergence of new canonical miRNAs requires successful processing and thereby the formation of junction-containing hairpin structures. Thus the Rhed of DGCR8 imposes a strong constraint for a new miRNA gene and serves as a gatekeeper for miRNA maturation and subsequent gene regulation pathways.
DGCR8 has been shown to bind many other RNAs in mammalian cells, including mRNAs, small nucleolar RNAs (snoRNAs) and long noncoding RNAs (Macias et al., 2012; Heras et al., 2013). In the inherited neurodegenerative disorder Fragile X-associated tremor/ataxia syndrome, the expanded CGG repeats in the fragile X mental retardation 1 (FMR1) mRNA bind DGCR8, sequester the pri-miRNA processing machinery, decrease mature miRNA levels and cause neuronal cell dysfunction (Sellier et al., 2013). Furthermore, DGCR8 and Drosha are required for the function of a class of artificial pri-miRNAs called shRNAmir. As a DNA vector-based RNA interference technology, shRNAmir is widely used in biomedical research and is being explored for therapeutic potential (Silva et al., 2005; Ni et al., 2011). Our characterization of the previously unknown RNA-binding domain in DGCR8 should aid understanding of its role in both miRNA and non-miRNA pathways and enhance the rational design of artificial pri-miRNAs in the future.
EXPERIMENTAL PROCEDURES
Plasmids
Detailed are provided in the Supplemental Experimental Procedures.
Expression, purification and characterization of recombinant DGCR8 proteins
Same as previously described (Faller et al., 2007; Barr et al., 2011; Barr et al., 2012). See the Supplemental Experimental Procedures for details.
Transcription and purification of pri-miRNAs
Detailed are provided in the Supplemental Experimental Procedures.
pri-miRNA binding and processing assays
These assays were performed as described (Faller et al., 2007). Briefly, for filter binding assays, a trace amount of 32P-labeled pri-miRNA was incubated with DGCR8 proteins at room temperature for 30 min. For competition filter binding assays, unlabeled competitor RNAs were also included in the binding reactions (Faller et al., 2010). The mixtures were filtered through nitrocellulose (EMD Millipore, Billerica, MA) and positively changed nylon (GE Healthcare, Pittsburgh, PA) membranes. The autoradiography images of the membranes were analyzed using Quantity One (BIO-RAD, Hercules, CA; version 4.4.1). The data were fit and graphed using PRISM (GraphPad, La Jolla, CA; version 4).
SEC analyses were performed at room temperature. The NC1 or Rhed proteins were incubated with annealed pri-miRNAs at the indicated concentrations for >5 min; these binding reactions contained a total of 233 mM NaCl. The mixtures were analyzed using an KTA Purifier chromatography system and a Superdex 200 10/300 GL column (GE Healthcare), with a running buffer containing 20 mM Tris pH 8.0 and 80 mM NaCl. After baseline subtraction, A450 was used to calculate DGCR8-bound heme concentration. The contribution of heme-bound DGCR8 to A260 was calculated based on the A260/A450 ratio of the protein, and was subtracted from the A260 values in the chromatograms. The remaining A260 was used to calculate the RNA concentration. The chromatogram plots were generated using PRISM.
For reconstituted pri-miRNA processing assays (Barr and Guo, 2014), uniformly 32P-labeled pri-miRNAs were annealed and incubated at 37°C for 30 or 45 min with purified recombinant His6-Drosha390–1374 and DGCR8 proteins. The reactions were analyzed using 7 M-urea 15% PAGE and autoradiography.
Live-cell pri-miRNA processing assays
Detailed procedures have been described (Weitz et al., 2014). Briefly, HeLa Tet-On cells (Clontech, Mountain View, CA) were transfected with reporter and/or N-flag-DGCR8-expression plasmid. Cells were immediately induced with 2 μg/mL doxycycline and imaged 18–24 hr later. Total eYFP and mCherry intensities for individual cells were fit by linear regression (y = slope * x) and slopes were obtained. P-values were determined using the linear regression function of PRISM. Expression levels of N-flag-DGCR8 were analyzed using immunoblotting (Gong et al., 2012). The mCherry-pri-miRNA fusion and the eYFP mRNA levels were determined using qRT-PCR as described (Weitz et al., 2014). The miRNA levels were measured using Taqman assays (Life Technologies, Grand Island, NY).
Supplementary Material
HIGHLIGHTS.
A unique RNA-binding domain is discovered in the pri-miRNA processing factor DGCR8
The RNA-binding heme domain binds both ends of a pri-miRNA hairpin
The Rhed and its RNA-binding surface are important for pri-miRNA processing
The molecular clamp model allows recognition of full structural features of pri-miRNAs
Acknowledgments
We thank R. Johnson, D. Black, K. Yeom and I. Barr for critical comments of the manuscript, S. Weiss for support, Y. Chen, M. Gong, S. Griner and N. Prakash for technical assistance. This work was partially supported by NIH grants GM080563 (to F.G.), Ruth L. Kirschstein National Research Service Award GM007185 (to J.Q. and G.S.), T32GM008496 (to S.H.W.), and by the Stein Oppenheimer Endowment Award (to F.G.).
Footnotes
AUTHOR CONTRIBUTIONS
J.Q. performed most protein-RNA binding assays (filter-binding and SEC), and mutagenesis analyses. J.P.J. did most in vitro pri-miRNA processing assays. S.H.W. performed the live-cell assays. G.S. initiated the SEC analyses and purified the RNA truncations. R.S. initiated the project. F.G., J.Q., J.P.J. and S.H.W. designed most experiments. F.G., J.Q., and J.P.J. wrote the manuscript with input from other authors.
Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Westholm JO, Lai EC. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr I, Guo F. Primary microRNA processing assay reconstituted using recombinant Drosha and DGCR8. Methods Mol Biol. 2014;1095:73–86. doi: 10.1007/978-1-62703-703-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr I, Smith AT, Chen Y, Senturia R, Burstyn JN, Guo F. Ferric, not ferrous, heme activates RNA-binding protein DGCR8 for primary microRNA processing. Proc Natl Acad Sci USA. 2012;109:1919–1924. doi: 10.1073/pnas.1114514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr I, Smith AT, Senturia R, Chen Y, Scheidemantle BD, Burstyn JN, Guo F. DiGeorge Critical Region 8 (DGCR8) is a double-cysteine-ligated heme protein. J Biol Chem. 2011;286:16716–16725. doi: 10.1074/jbc.M110.180844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Doyle M, Jaskiewicz L, Filipowicz W. Dicer proteins and their role in gene silencing pathways. In: Part B, Guo F, Tamanoi F, editors. The Enzymes: Eukaryotic RNases and their partners in RNA degradation and biogenesis. Amsterdam, Netherlands: Academic Press; 2012. pp. 1–35. [Google Scholar]
- Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat Struct Mol Biol. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- Faller M, Toso D, Matsunaga M, Atanasov I, Senturia R, Chen Y, Zhou ZH, Guo F. DGCR8 recognizes primary transcripts of microRNAs through highly cooperative binding and formation of higher-order structures. RNA. 2010;16:1570–1583. doi: 10.1261/rna.2111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles-Gonzalez MA, Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem. 2005;99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Gong M, Chen Y, Senturia R, Ulgherait M, Faller M, Guo F. Caspases cleave and inhibit the microRNA processing protein DiGeorge Critical Region 8. Protein Sci. 2012;21:797–808. doi: 10.1002/pro.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Guo F. Drosha and DGCR8 in microRNA biogenesis. In: Part B, Guo F, Tamanoi F, editors. The Enzymes: Eukaryotic RNases and their partners in RNA degradation and biogenesis. Amsterdam, Netherlands: Elsevier Academic Press; 2012. pp. 101–121. [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, Garcia-Perez JL, Caceres JF. The Microprocessor controls the activity of mammalian retrotransposons. Nat Struct Mol Biol. 2013;20:1173–1181. doi: 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Ma E, Zhou K, Kidwell MA, Doudna JA. Coordinated activities of human dicer domains in regulatory RNA processing. J Mol Biol. 2012;422:466–476. doi: 10.1016/j.jmb.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Wu Y, Choi JG, Wu H. Lower and upper stem-single-stranded RNA junctions together determine the Drosha cleavage site. Proc Natl Acad Sci U S A. 2013;110:20687–20692. doi: 10.1073/pnas.1311639110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias S, Plass M, Stajuda A, Michlewski G, Eyras E, Caceres JF. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol. 2012;19:760–766. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Marvin KA, Reinking JL, Lee AJ, Pardee K, Krause HM, Burstyn JN. Nuclear receptors homo sapiens Rev-erbbeta and Drosophila melanogaster E75 are thiolate-ligated heme proteins which undergo redox-mediated ligand switching and bind CO and NO. Biochemistry. 2009;48:7056–7071. doi: 10.1021/bi900697c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah G, Barraud P, Allain FH. RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence. Cell Mol Life Sci. 2013;70:1875–1895. doi: 10.1007/s00018-012-1119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BM, Ishimaru D, Hennig M. The core Microprocessor component DiGeorge syndrome critical region 8 (DGCR8) is a non-specific RNA-binding protein. J Biol Chem. 2013;288:26785–26799. doi: 10.1074/jbc.M112.446880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, Alunni V, Moine H, Thibault C, Page A, et al. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–880. doi: 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senturia R, Faller M, Yin S, Loo JA, Cascio D, Sawaya MR, Hwang D, Clubb RT, Guo F. Structure of the dimerization domain of DiGeorge Critical Region 8. Protein Sci. 2010;19:1354–1365. doi: 10.1002/pro.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senturia R, Laganowsky A, Barr I, Scheidemantle BD, Guo F. Dimerization and heme binding are conserved in amphibian and starfish homologues of the microRNA processing protein DGCR8. PLoS One. 2012;7:e39688. doi: 10.1371/journal.pone.0039688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Sohn SY, Bae WJ, Kim JJ, Yeom KH, Kim VN, Cho Y. Crystal structure of human DGCR8 core. Nat Struct Mol Biol. 2007;14:847–853. doi: 10.1038/nsmb1294. [DOI] [PubMed] [Google Scholar]
- Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol. 2011;18:1153–1158. doi: 10.1038/nsmb.2125. [DOI] [PubMed] [Google Scholar]
- Weitz SH, Gong M, Barr I, Weiss S, Guo F. Processing of microRNA primary transcripts requires heme in mammalian cells. Proc Natl Acad Sci U S A. 2014;111:1861–1866. doi: 10.1073/pnas.1309915111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010;38:7689–7697. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.