Abstract
The double helical structure of DNA lends itself to topological constraints. Many DNA-based processes alter the topological state of DNA, generating torsional stress, which is efficiently relieved by topoisomerases. Maintaining this topological balance is crucial to cell survival, as excessive torsional strain risks DNA damage. Here, we review the mechanisms that generate and modulate DNA torsion within the cell. In particular, we discuss how transcription-generated torsional stress affects Pol II kinetics and chromatin dynamics, highlighting an emerging role of DNA torsion as a feedback mediator of torsion-generating processes.
Keywords: torsional stress, nucleosome turnover, nascent RNA, topoisomerase, RNA Polymerase II
Introduction
DNA is a highly ordered structure. It consists of two anti-parallel, complementary strands governed by base-pairing that follow a right-handed helical path about a central axis1 (Fig. 1A). This double helical structure presents an elegant solution to self-replication, but it also introduces a unique problem. DNA-based processes, such as replication, repair, recombination, and transcription, would have to overcome the topological constraints inherent in intertwined strands. To understand the many implications of such constraint, we first need to define a basic terminology for DNA topology.
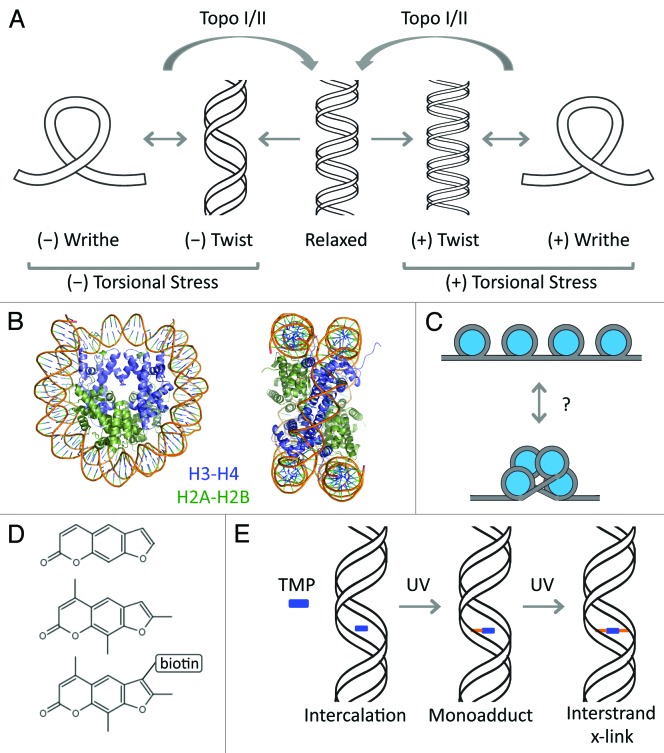
Figure 1. Effects of torsion on DNA and chromatin structure. (A) The relaxed state of the DNA double helix consists of ~10 bp per helical turn (center). When DNA-based processes exert torsional force on the DNA, it manifests as a change in twist or formation of writhe. (B) The nucleosome is composed of a left-handed wrapped DNA around an octameric core. H3-H4 tetramer is colored blue. H2A-H2B dimers are colored green. (C) Potential writhe in the context of a nucleosomal template. (D) Molecular structure of psoralen (top), tri-methyl psoralen (TMP) (middle), and biotin-TMP (bottom) (E) TMP intercalates into the DNA double-strand and forms monoadducts and interstrand crosslinks under UV light.
One complete helical turn of DNA about the central axis consists of ~10 base pairs. In topological terms, this is called the linking number (Lk), properly defined as the number of times the double-stranded DNA rotates around the axis in the right-handed direction.2 Therefore, a 20 bp fragment has a Lk of 2. In a relaxed DNA molecule, Lk is equal to the number of times the two strands wind around each other, which is called the twist (Tw). However, when the DNA is not relaxed, it can lead to altered Tw, or to coiling of the double strand about itself, resulting in formation of writhe (Wr) (Fig. 1A). The relationship between these three topological properties, Lk, Tw, and Wr, is summarized by the following equation: Lk = Tw + Wr.2,3 For closed, circular DNA of a given length where rotation cannot dissipate, this equation describes the direct relationship between linking number and the dynamic properties of twist and writhe. Although eukaryotic DNA is not circular, the basic principle largely applies because the genome is divided into large regions with fixed ends that prevent free rotation,4 giving rise to supercoiling domains.5 Instead of absolute numbers, the relationship becomes relative such that ΔLk = ΔTw + Wr.6 This equation suggests that when torsional force causes a change in Lk, it manifests as a change in Tw or a compensatory change in Wr, which is often referred to as supercoiling. Torsional force can result in over-twisting or under-twisting, forming positive or negative supercoiling, respectively (Fig. 1A).
As an added complexity, eukaryotic DNA is packaged into chromatin by wrapping 147 bp of DNA around eight histone proteins in a left-handed direction (Fig. 1B). This structure is called the nucleosome and forms the fundamental repeating unit of chromatin.7 The left-handed wrap of DNA around the nucleosome introduces negative Wr, thus forming constrained supercoiling. The structure of the nucleosome implies an intricate connection with DNA topology. In vitro, negatively supercoiled DNA templates readily form nucleosomes whereas positive supercoiling inhibits nucleosome formation,8 suggesting that torsional events generated in vivo would have a profound impact on nucleosome structure and chromatin organization (Fig. 1C).
In this extra view, we discuss how DNA topology is altered and managed within the cell, how we detect supercoiling states in vivo, and how generation of torsional stress, particularly during transcription, can re-organize chromatin structure, destabilize nucleosomes, feed back into Pol II regulation, and affect the affinity of other DNA-binding proteins.
Generating Torsion
Many DNA-based processes affect DNA topology primarily by changing the DNA Tw.9 Polymerases, in particular, are powerful torsion-generating motors.10 For example, during replication, the MCM helicase unwinds the two strands for use as templates by DNA Polymerase, which synthesizes the new copy. This unwinding event alters Tw and generates positive torsional stress ahead of the replication fork. Similarly, during transcription, the RNA Polymerase II (Pol II) machinery melts the promoter to access the transcription start site (TSS). Furthermore, as Pol II transcribes, the melted DNA bubble travels downstream, creating positive supercoiling ahead and negative supercoiling behind Pol II. This transcription-generated torsional effect is better known as the twin-supercoil domain model.11 The amount of torsional stress that Pol II generates can be inferred from single molecule experiments of bacterial RNAP, which shows that RNAP generates sufficient torque to distort DNA structure of arbitrary sequence.10 Modeling of supercoil dynamics reveals that supercoils propagate from Pol II at a rate of two orders of magnitude faster than the rate of Pol II itself12 in either 1D diffusion along the DNA or by a “hopping” mechanism.13 However, the diffusion of torsional stress is restricted within supercoiling domains.5 Although transcription-generated supercoiling constraints are not additive, if left unresolved, subsequent transcription events genome-wide would generate a significant amount of strain on DNA structure.
Non-polymerase based events also generate torsion. As discussed above, nucleosome assembly generates constrained negative supercoiling. Another chromatin-based process that generates torsion occurs during mitosis where ATP-dependent condensins generate positive supercoiling when condensing chromosomes.14 Furthermore, the activities of many DNA binding proteins also affect DNA topology. For example, in vitro, the binding of the general transcription factor TBP induces negative supercoiling.15 In yeast, the mediator component Hrs1 can induce plasmid supercoiling.16 Indeed, many components of the transcription machinery complex promote DNA looping of promoter sequence with enhancer regions often mediated by structural proteins such as cohesins and CTCF.17,18
Modulating Torsion
The ubiquity of torsion-generating processes poses a great risk for DNA damage unless torsion is relieved. Therefore, the activity of topoisomerases, enzymes that relieve torsional strain, is critical to cell survival. Virtually all life forms contain topoisomerases, including certain viruses, and the high degree of conservation across eukaryotes underlies their significance.19,20 Indeed, many antibiotics and anti-cancer drugs inhibit topoisomerases to effect cell death.21,22 Their primary function is to change DNA Lk through cleavage, torsion relief, and religation.23 There are two main types of topoisomerases as classified by the mode of DNA cleavage. Type I topoisomerases (Topo I) cleave one strand of the double helix whereas Type II topoisomerases (Topo II) cleave both strands. Although the end result of torsional relief is the same, the two types appear to have distinct and redundant functions within the cell (Table 1) (see ref. 23 for a thorough review on topoisomerases).
Table 1. Comparison of the two types of topoisomerases.
| Topo I | Topo II | |
|---|---|---|
| # of cleavage | 1 | 2 |
| Mode of action | Twist reflief | Writhe relief |
| DNA replication | Required for cell proliferation in Drosophilia | Decatenation and strand separation |
| Chromatin remodeling | Hrp1 | ACF and CHRAC |
| Transcription | ||
| Site of function | Gene body | Promoters and gene ends |
| Main function | Relaxation of torsion during Pol II elongation | Secondary to Topo I: mostly on highly transcribed genes |
| Other functions | PIC formation Pol II pausing |
Pol II pausing Gene looping |
Topo I, acting as the main DNA “swivelase”, resolves topological issues by altering DNA twist. After cleaving one strand, the primary class of Topo I in eukaryotes swivels one end of the broken strand around the intact strand.24 Topo I is essential for viability in Drosophila with a critical role in all proliferating tissues, suggesting a function in DNA replication.25 In fission yeast, Topo I is also required for nucleosome disassembly through its interaction with the nucleosome remodeler Hrp1.26 Topo I, however, is most studied for its role as the primary reliever of transcription-generated torsional stress.27 It localizes to transcribed genes,28 has a preference for relieving positive supercoiling,29 and interacts with the Pol II C-terminal domain (CTD),30 suggesting that Topo I acts on gene bodies ahead of Pol II during active transcription. In addition to its action during elongation, Topo I has also been shown to regulate other steps in the transcription process. During initiation, Topo I facilitates the binding of TFIID-TFIIA to form the pre-initiation complex.31 Furthermore, genes containing a paused Pol II are hypersensitive to Topo I inhibition,32 suggesting that Topo I may also function in Pol II pause release.
In contrast to Topo I, Topo II is the main “writhase” of eukaryotic cells. It generates double-strand breaks on one DNA segment to create a gate and translocates another intact double-stranded segment through that gate.33,34 Aside from its writhase functions, Topo II also has the ability to decatenate sister chromatids during DNA replication and cell division as it toggles between writhase and decatenase functions.35 This toggle may be driven by the condensin-induced positive supercoiling,36 or by the tension on DNA as spindle forces pull on each sister chromatid.37 Topo II is more effective at relieving superhelical tension in nucleosomal templates than Topo I,38 and in both Drosophila and humans, it interacts with chromatin remodeling factors CHRAC and ACF,39 suggesting a functional role in chromatin remodeling. During transcription, Topo II has a largely secondary role to Topo I, mainly acting on promoters of highly transcribed genes.32 It has also been shown to bind at the 3′ end of genes, which, together with its function as a chromatin regulator, may indicate a role in DNA looping.
Together, these two types of topoisomerases effectively relieve torsional strain in vivo. The efficiency and redundancy of these enzymes seem to suggest that unconstrained supercoiling is largely absent within cells. However, recent advances in detecting supercoiling in vivo reveal that DNA topology is highly dynamic and well regulated.
Detecting Torsion
Several biochemical techniques are available to detect the effects of torsion on DNA. For example, excision and circularization of a DNA segments using Cre recombinase can trap supercoils ex vivo.40 However, to examine torsional effects in vivo, most methods rely on the basic properties of the molecule psoralen and its derivative tri-methyl psoralen (TMP). Psoralen (Fig. 1D) is a member of furocoumarins, a group of naturally occurring compounds in certain plant seeds that can freely cross lipid membranes and intercalate between the two strands of the DNA double helix.41 It has a preference for negatively supercoiled DNA,42 and when exposed to UV light, forms monoadducts and interstrand crosslinks between thymine residues43 (Fig. 1E). Taking advantage of this crosslinking property, several groups have used psoralen derivatives to map negatively supercoiled DNA in bacteria,44 yeast,45 Drosophila,44,46 and human cells.5,32,44 A comparison of the methods is shown in Table 2. For example, one group used a biotin-conjugated TMP (bTMP) to pull-down bTMP crosslinked DNA fragments.5 Another group enriched for TMP-crosslinked fragments by thermally denaturing all DNA fragments followed by gel electrophoresis.32 TMP-crosslinked fragments snap back into the double stranded form upon denaturation, and migrate slower in a gel. In the yeast study, the authors enriched for double-stranded crosslinks by a combination of denaturation and digestion with Exo I, a single-strand specific exonuclease.45 The TMP-crosslinked fragments re-nature efficiently and are thus protected from Exo I digestion. In all three variations, the crosslinked fragment was used as template for producing labeled probes to hybridize in microarrays, generating a global view of supercoiling states in yeast and human cells with varying resolution levels (Table 2).
Table 2. Comparison of genome-wide torsion studies (“supercoilinomics”).
| Bermudez et al. | Naughton et al. | Kouzine et al. | Teves and Henikoff | |
|---|---|---|---|---|
| Organism | Yeast | Human | Human | Drosophilia |
| Substrate | TMP | bTMP | TMP | TMP |
| Enrichment Method | Heat denuration and Exo I digestion | Streptavidin pulldown | Heat-glyoxal denaturation and gel electrophoresis | Heat denuration and Exo I digestion |
| Platform | Microarray | Microarray | Microarray | PE sequencing |
| Assay Resolution | 2 kb | Unknown | 250 bp | Nucleotide |
| Effective Resolution | 2 kb | ~10kb | ~1–5 kb | 150 bp |
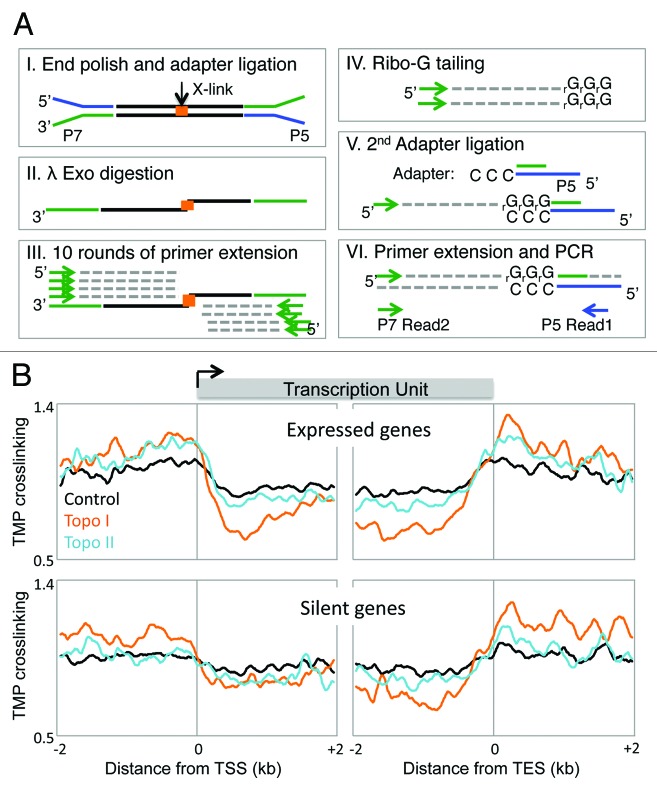
To achieve a higher degree of resolution, we had adapted the yeast method for next generation sequencing46 (Fig. 2A). Applying this method to Drosophila S2 cells, we observed high and low TMP crosslinking upstream and downstream, respectively, of TSSs. Furthermore, consistent with the twin-supercoil domain model, expressed genes were found to experience more torsional strain relative to silent genes (Fig. 2B). This torsional strain was exacerbated upon topoisomerase inhibition. Specifically, Topo I inhibition resulted in a greater change in supercoiling relative to Topo II (Fig. 2B), providing further evidence that Topo I is the major relaxer of transcription-generated torsional strain. Inhibition of Topo II primarily altered the supercoiling states of the highest expressed genes, also consistent with its role as a secondary relaxer when Topo I is outpaced by the rate of transcription. Indeed, Topo II is primarily localized at highly transcribed genes whereas Topo I is present in most genes.32
Figure 2. Transcribed gene regions experience torsional stress (A) Method for mapping DNA supercoiling genome-wide using next-generation sequencing.46 (I) Following Exo I-enrichment of crosslinked strands, Illumina paired-end (PE) adapters were ligated. (II) Lambda Exonuclease (Exo λ) was used to resect the 5′ strand until the crosslinking site. (III) Using a primer complementary to PE adapters, several rounds of primer extension were performed. (IV) The single-stranded products were appended with ribo-Gs at the 3′ ends using terminal transferase, (V) followed by ligation with a double-stranded PE adaptor with a CCC overhang. (VI) After a single round of primer extension to generate a double-stranded fragment, the products were amplified using PE sequencing primers. Sequencing from the CCC overhang allowed for the precise mapping of TMP-crosslinked sites throughout the whole genome. (B) Genes in Drosophila genome were split as expressed (top) or silent (bottom). High-resolution genome-wide maps of TMP crosslinking surrounding the transcription start site (TSS) and transcription end site (TES) are shown under normal conditions (black), Topo I inhibition (orange), and Topo II inhibition (teal). Expressed genes experience higher torsional stress, consistent with the twin-supercoil domain model.
Torsion and Chromatin
We have discussed how nucleosome assembly and chromosome condensation generate torsional stress, but, in a feedback manner, torsion can also affect chromatin structure and organization, despite the structural plasticity of chromatin fibers during torsional stress.47 In one study, the authors delineated large-scale supercoiling domains in human chromosome 11 with a median size of 100 kb that are dependent on transcriptional activity.5 Furthermore, the authors found that chromatin of underwound domains are more de-compacted than those of overwound domains. Regions of chromatin compaction, similar to supercoiling domains, are dependent on transcriptional activation.5 From these data, the authors proposed a model whereby transcription-generated supercoiling domains regulate chromatin compaction and organization. This is consistent with another study, which showed that large-scale chromatin movements are dependent on polymerases and Topo II,48 further implicating torsion in chromatin structure and dynamics.
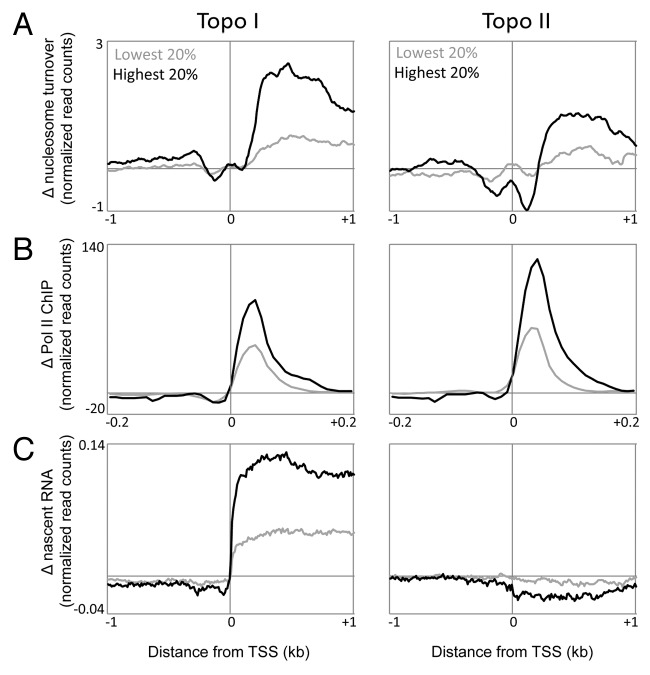
Changes in torsional states also affect fine-scale chromatin structure. Single molecule studies indicate that increased DNA torsion facilitates rapid H2A-H2B dimer exchange,49 further suggesting that DNA torsion mediates nucleosome structure and stability. We recently showed that in vivo, when topoisomerases are inhibited, the resulting accumulation of torsional strain results in increased nucleosome turnover within gene bodies.46 Nucleosome turnover is also dependent on transcription, as Pol II inhibition results in decreased nucleosome turnover for transcribed genes.50 When topoisomerases are inhibited, genes that experience the highest change in torsion also have the highest increase in nucleosome turnover (Fig. 3A). In contrast, genes that change the least in torsion also change the least in nucleosome turnover (Fig. 3A), further implicating DNA supercoiling in nucleosome dynamics.
Figure 3. Torsional stress affects nucleosome dynamics and Pol II kinetics. Genes were grouped based on change in torsional stress as measured by change in TMP crosslinking before and after Topo I or II inhibition. The highest (High TD) and lowest (Low TD) 20% of genes are shown. For these two groups, the change in nucleosome turnover (A), the change in Pol II pausing (B), and the change in Pol II elongation as measured by nascent RNA production (C) are plotted for regions surrounding the TSS.
Torsion and Transcription
Transcription-generated torsional stress has an inhibitory effect on polymerase activity. Indeed, one of the earliest indications of this relationship is that inhibition of both types of topoisomerases leads to an effective block of transcription of ribosomal genes in budding yeast.51 This feedback mechanism has likely evolved to prevent drastic accumulation of torsional strain. Recently, we showed that inhibition of individual topoisomerases affect specific aspects of Pol II kinetics.
Upon Topo I or II inhibition, Pol II pausing downstream of the TSS increased dramatically for all genes, although genes that experienced a greater change in torsional stress showed a greater increase in paused Pol II (Fig. 3B). Whereas a previous study has shown that genes regulated by a paused Pol II are more sensitive to Topo I inhibition,32 this result suggests that torsional relief by topoisomerases affects the kinetics of Pol II initiation and/or release from pause site. In contrast, Pol II elongation presents a different picture. Topo I inhibition affected Pol II elongation, as measured by nascent RNA production, much more strongly than Topo II inhibition (Fig. 3C), consistent with previous studies. Surprisingly, Topo I inhibition resulted in increased Pol II elongation, particularly in genes that experienced the most change in torsional stress. One possible explanation for this result is that the increase in nucleosome destabilization due to the accumulation of torsion transiently allows Pol II to proceed a short distance.
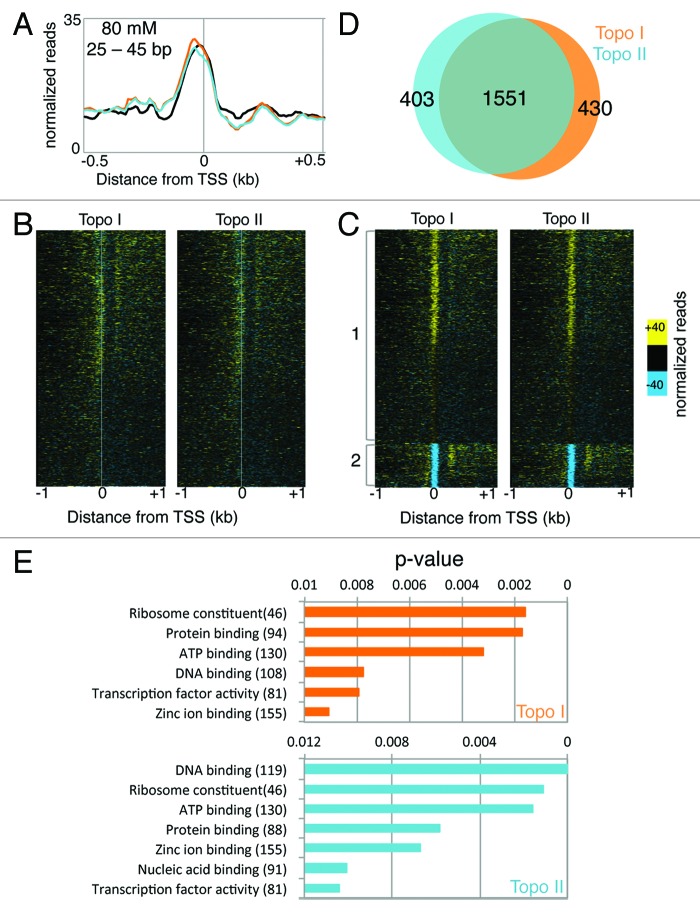
Some evidence also suggests that DNA supercoiling affects the affinity of DNA-binding proteins. For example, local melting of the c-myc promoter due to Pol II-generated negative supercoiling facilitates the binding of activators and repressors.40 DNA topology also influences the binding affinity of the tumor suppressor p53.52 In our recent study, we examined the effects of topoisomerase inhibition on the affinity of DNA binding proteins as measured by low-salt extraction. Low salt preferentially extracts euchromatic nucleosomes, but has also been used to map binding of sequence-specific and general transcription factors.46,53 Using this technique, we observed a strong peak of binding at the TSS, representing a cumulative view of DNA-binding proteins at the TSSs of all genes (Fig. 4A). Topo I or Topo II inhibition did not affect the averaged pattern of binding at the TSS (Fig. 4A). However, when the changes in binding were examined on a gene-by-gene basis using a heatmap, we observed variegated changes in binding due to topoisomerase inhibition (Fig. 4B). When we performed unbiased k-means clustering with k = 2, we detected two main groups of genes (Fig. 4C). Group 1 showed increased binding at the TSS whereas group 2 showed strong decrease in binding. Furthermore, genes in group 2 following Topo I inhibition strongly overlapped with those in group 2 of Topo II-inhibited samples (Fig. 4D), suggesting that TF binding at the TSS of these genes is hypersensitive to torsional stress. Gene ontology analysis of group 2 genes showed enrichment for ribosomal constituents and DNA binding proteins (Fig. 4E), consistent with previous studies. These data suggest that the torsional state of DNA affects the affinity of DNA binding factors on some promoter regions.
Figure 4. Torsional stress affects the affinity of DNA-binding proteins at the TSS. (A) Micrococcal nuclease digested chromatin is extracted using low salt and sequenced using the paired-end Illumina platform. Reads were parsed computationally by size, with short fragments representing sites for DNA binding protein. The global pattern of short fragments from low-salt extraction of chromatin is analyzed for all genes surrounding the TSS before and after Topo I or II inhibition. (B) The change in short fragments after Topo I (left) or II (right) inhibition relative to control is displayed as a heat map for all genes arranged by expression level in untreated cells. (C) Unbiased k-means clustering of (B) with k = 2. (D) Venn diagram of group 2 genes in Topo I (orange) and Topo II (teal) inhibited samples. (E) Gene ontology analysis of group 2 genes performed as described previously.54,55
Conclusion
The discovery of the DNA double helix first introduced the concept of topological constraints. However, these constraints were generally overlooked in investigation of mechanisms behind many cellular processes such as DNA replication, transcription, and chromatin organization, because of the use of unconstrained DNA templates. Now, newly developed methods to study DNA topology in vivo have revealed the importance of DNA structural dynamics. As most DNA-based processes generate torsional stress, the resulting DNA strain in turn affects the same processes. This relationship creates a feedback loop based on DNA topology, with topoisomerases acting as regulatory modulators to fine-tune DNA structure.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- 1.Watson JD, Crick FHC. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–8. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Barbi M, Mozziconacci J, Victor JM, Wong H, Lavelle C. On the topology of chromatin fibres. Interface Focus. 2012;2:546–54. doi: 10.1098/rsfs.2011.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuller FB. The writhing number of a space curve. Proc Natl Acad Sci U S A. 1971;68:815–9. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, Cockroft SL, Bradley M, Ylstra B, Gilbert N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–95. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prunell A. A topological approach to nucleosome structure and dynamics: the linking number paradox and other issues. Biophys J. 1998;74:2531–44. doi: 10.1016/S0006-3495(98)77961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 8.Gupta P, Zlatanova J, Tomschik M. Nucleosome assembly depends on the torsion in the DNA molecule: a magnetic tweezers study. Biophys J. 2009;97:3150–7. doi: 10.1016/j.bpj.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavelle C. Pack, unpack, bend, twist, pull, push: the physical side of gene expression. Curr Opin Genet Dev. 2014;25C:74–84. doi: 10.1016/j.gde.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Ma J, Bai L, Wang MD. Transcription under torsion. Science. 2013;340:1580–3. doi: 10.1126/science.1235441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bécavin C, Barbi M, Victor J-M, Lesne A. Transcription within condensed chromatin: Steric hindrance facilitates elongation. Biophys J. 2010;98:824–33. doi: 10.1016/j.bpj.2009.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Loenhout MTJ, de Grunt MV, Dekker C. Dynamics of DNA supercoils. Science. 2012;338:94–7. doi: 10.1126/science.1225810. [DOI] [PubMed] [Google Scholar]
- 14.Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–34. doi: 10.1016/S0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 15.Kahn JD. Topological effects of the TATA box binding protein on minicircle DNA and a possible thermodynamic linkage to chromatin remodeling. Biochemistry. 2000;39:3520–4. doi: 10.1021/bi992263f. [DOI] [PubMed] [Google Scholar]
- 16.Piruat JI, Chávez S, Aguilera A. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics. 1997;147:1585–94. doi: 10.1093/genetics/147.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D. Transcription factors mediate long-range enhancer-promoter interactions. Proc Natl Acad Sci U S A. 2009;106:20222–7. doi: 10.1073/pnas.0902454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang WM. Bacterial diversity based on type II DNA topoisomerase genes. Annu Rev Genet. 1996;30:79–107. doi: 10.1146/annurev.genet.30.1.79. [DOI] [PubMed] [Google Scholar]
- 20.Viard T, de la Tour CB. Type IA topoisomerases: a simple puzzle? Biochimie. 2007;89:456–67. doi: 10.1016/j.biochi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Khadka DB, Cho WJ. Topoisomerase inhibitors as anticancer agents: a patent update. Expert opinion on therapeutic patents 2013; 23:1033-56. [DOI] [PubMed]
- 22.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen SH, Chan NL, Hsieh TS. New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem. 2013;82:139–70. doi: 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 24.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–4. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 25.Lee MP, Brown SD, Chen A, Hsieh TS. DNA topoisomerase I is essential in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1993;90:6656–60. doi: 10.1073/pnas.90.14.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durand-Dubief M, Persson J, Norman U, Hartsuiker E, Ekwall K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010;29:2126–34. doi: 10.1038/emboj.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–11. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 28.Gilmour DS, Pflugfelder G, Wang JC, Lis JT. Topoisomerase I interacts with transcribed regions in Drosophila cells. Cell. 1986;44:401–7. doi: 10.1016/0092-8674(86)90461-7. [DOI] [PubMed] [Google Scholar]
- 29.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–7. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Phatnani HP, Hsieh TS, Greenleaf AL. The phosphoCTD-interacting domain of Topoisomerase I. Biochem Biophys Res Commun. 2010;397:117–9. doi: 10.1016/j.bbrc.2010.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shykind BM, Kim J, Stewart L, Champoux JJ, Sharp PA. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- 32.Kouzine F, Gupta A, Baranello L, Wojtowicz D, Ben-Aissa K, Liu J, Przytycka TM, Levens D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu LF, Liu CC, Alberts BM. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980;19:697–707. doi: 10.1016/S0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- 34.Laponogov I, Veselkov DA, Crevel IM, Pan XS, Fisher LM, Sanderson MR. Structure of an ‘open’ clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport. Nucleic Acids Res. 2013;41:9911–23. doi: 10.1093/nar/gkt749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roca J, Wang JC. The probabilities of supercoil removal and decatenation by yeast DNA topoisomerase II. Genes Cells. 1996;1:17–27. doi: 10.1046/j.1365-2443.1996.01001.x. [DOI] [PubMed] [Google Scholar]
- 36.Baxter J, Sen N, Martínez VL, De Carandini ME, Schvartzman JB, Diffley JF, Aragón L. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science. 2011;331:1328–32. doi: 10.1126/science.1201538. [DOI] [PubMed] [Google Scholar]
- 37.Farcas AM, Uluocak P, Helmhart W, Nasmyth K. Cohesin’s concatenation of sister DNAs maintains their intertwining. Mol Cell. 2011;44:97–107. doi: 10.1016/j.molcel.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salceda J, Fernández X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–83. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 40.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–54. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 41.Toussaint M, Levasseur G, Tremblay M, Paquette M, Conconi A. Psoralen photocrosslinking, a tool to study the chromatin structure of RNA polymerase I--transcribed ribosomal genes. Biochem Cell Biol. 2005;83:449–59. doi: 10.1139/o05-141. [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto K, Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J Cell Sci. 2004;117:3797–805. doi: 10.1242/jcs.01225. [DOI] [PubMed] [Google Scholar]
- 43.Dall’Acqua F, Marciani S, Rodighiero G. Inter-strand cross-linkages occurring in the photoreaction between psoralen and DNA. FEBS Lett. 1970;9:121–3. doi: 10.1016/0014-5793(70)80330-1. [DOI] [PubMed] [Google Scholar]
- 44.Sinden RR, Carlson JO, Pettijohn DE. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980;21:773–83. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- 45.Bermúdez I, García-Martínez J, Pérez-Ortín JE, Roca J. A method for genome-wide analysis of DNA helical tension by means of psoralen-DNA photobinding. Nucleic Acids Res. 2010;38:e182. doi: 10.1093/nar/gkq687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teves SS, Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bancaud A, Conde e Silva N, Barbi M, Wagner G, Allemand JF, Mozziconacci J, Lavelle C, Croquette V, Victor JM, Prunell A, et al. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat Struct Mol Biol. 2006;13:444–50. doi: 10.1038/nsmb1087. [DOI] [PubMed] [Google Scholar]
- 48.Zidovska A, Weitz DA, Mitchison TJ. Micron-scale coherence in interphase chromatin dynamics. Proc Natl Acad Sci U S A. 2013;110:15555–60. doi: 10.1073/pnas.1220313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheinin MY, Li M, Soltani M, Luger K, Wang MD. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat Commun. 2013;4:2579. doi: 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teves SS, Henikoff S. Heat shock reduces stalled RNA polymerase II and nucleosome turnover genome-wide. Genes Dev. 2011;25:2387–97. doi: 10.1101/gad.177675.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. DNA topoisomerase activity is required as a swivel for DNA replication and for ribosomal RNA transcription. NCI Monogr. 1987:11–5. [PubMed] [Google Scholar]
- 52.Jagelská EB, Brázda V, Pecinka P, Palecek E, Fojta M. DNA topology influences p53 sequence-specific DNA binding through structural transitions within the target sites. Biochem J. 2008;412:57–63. doi: 10.1042/BJ20071648. [DOI] [PubMed] [Google Scholar]
- 53.Kasinathan S, Orsi GA, Zentner GE, Ahmad K, Henikoff S. High-resolution mapping of transcription factor binding sites on native chromatin. Nat Methods. 2014;11:203–9. doi: 10.1038/nmeth.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317-22. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]