Abstract
Epichromatin, the surface of chromatin facing the nuclear envelope in an interphase nucleus, reveals a “rim” staining pattern with specific mouse monoclonal antibodies against histone H2A/H2B/DNA and phosphatidylserine epitopes. Employing a modified ChIP-Seq procedure on undifferentiated and differentiated human leukemic (HL-60/S4) cells, >95% of assembled epichromatin regions overlapped with Alu retrotransposons. They also exhibited enrichment of the AluS subfamily and of Alu oligomers. Furthermore, mapping epichromatin regions to the human chromosomes revealed highly similar localization patterns in the various cell states and with the different antibodies. Comparisons with available epigenetic databases suggested that epichromatin is neither “classical” heterochromatin nor highly expressing genes, implying another function at the surface of interphase chromatin. A modified chromatin immunoprecipitation procedure (xxChIP) was developed because the studied antibodies react generally with mononucleosomes and lysed chromatin. A second fixation is necessary to securely attach the antibodies to the epichromatin epitopes of the intact nucleus.
Keywords: nuclear architecture, retrotransposon Alu, epichromatin, epigenetics, myeloid leukemia, granulocytes, macrophage
Introduction
Current concepts of chromatin organization within the interphase nucleus include the nucleosomal “10 nm” and “30 nm” fibers, “megabase domains,” and “chromosome territories.”1-4 Interphase nuclear architecture is compartmentalized into more condensed, transcriptionally inactive heterochromatin, generally localized beneath the nuclear envelope (NE) and around the nucleolus, and less condensed, transcriptionally active euchromatin, located more centrally within the nucleus. Nuclear compartmentalization is disrupted during mitosis and rapidly reestablished during post-mitotic nuclear reformation.5-7 Beside the likely role of inner nuclear membrane proteins (e.g., lamin B receptor, LBR) in nuclear reformation, it has been suggested that properties of the NE-associated chromatin may facilitate the post-mitotic reconstruction of nuclear architecture.8,9 The surface of chromatin facing the nuclear envelope in an interphase nucleus (“epichromatin”) reveals a “rim” staining pattern with specific antibodies against histone H2A/H2B/DNA (mAb PL2-6) and phosphatidylserine (mAb 1H6) epitopes.8,9 High-resolution 3D-SIM images of PL2-6 and 1H6 immunostaining at the surface of interphase nuclei reveal a meshwork of chromatin fibers running parallel to the NE in tangential “grazing” views (Fig. 1). In the present study, we isolated the chromatin bound to PL2-6 and 1H6 in the three cell states of HL-60/S4 (undifferentiated, granulocyte, and macrophage) and analyzed the epichromatin DNA. We observed that epichromatin-associated DNA is very highly enriched in retrotransposon Alu, compared with its average content in the human genome.
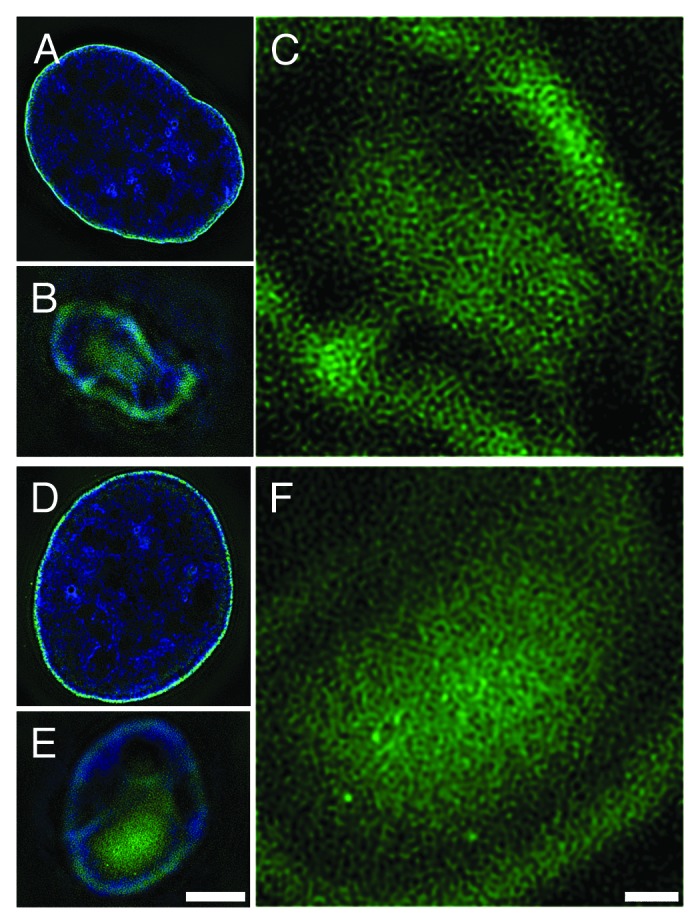
Figure 1. Immunostaining of epichromatin with monoclonal anti-H2A/H2B/DNA (PL2–6, A-C) and monoclonal anti-phosphatidylserine (1H6, D-F). 3-D SIM (Structured Illumination Microscopy) computed optical sections of immunostained U2OS cells, collected as described previously.9 (A, D) Mid-sections showing epichromatin staining (green) at the perimeter of DAPI stained interphase nuclei (blue). (B, E) Tangential “grazing” sections showing surface epichromatin staining. Scale bar (A, B, D, E) 5 µm. (C, F) Higher magnification views of the same tangential sections (B, E) illustrating the chromatin fiber mesh stained (green) by PL2–6 (C) or 1H6 (F). DAPI staining is not shown. Scale bar (C, F) 1 µm.
Results
Developing an appropriate chromatin immunoprecipitation (ChIP) protocol
Our original goal was to characterize epichromatin by conventional immunoprecipitation and DNA sequencing of formaldehyde-fixed and sonicated human tissue culture cells (“xChIP-Seq”), employing PL2-6 and 1H6. However, it became clear that these antibodies react with all mononucleosomes (Fig. 2), when binding was evaluated by an “electrophoretic mobility shift assay” (EMSA). Furthermore, PL2-6 reacts with most of the chromatin spilling out of hypotonically lysed erythroid cell nuclei (Fig. 3A and B). mAb 1H6 reacted with chromatin spilling from lysed erythroid nuclei in an identical manner (data not shown). We concluded that localized epitope accessibility and/or specific chromatin conformations, are preserved within fixed intact interphase nuclei, but readily destroyed when nuclear architecture is disrupted. We reasoned that sonication of cells after the first fixation, as in xChIP, would “open” more chromatin fragments to reaction with these antibodies. Therefore, we developed a “double-fixation” method (“xxChIP”) to identify specific antibody binding to the epichromatin region (see Materials and Methods for technical details). This method involves a second formaldehyde fixation after the primary antibody reaction and prior to sonication and immunoprecipitation. In this manner, the antibodies are fixed to the epichromatin sites and cannot bind to newly revealed epitopes following the sonication procedure. A human myeloid leukemic cell line (HL-60/S4) was selected for the xxChIP-Seq experiments, because it can be differentiated in vitro to granulocyte or macrophage forms by the addition of retinoic acid (RA) or phorbol ester (TPA), respectively.10,11 Validation of the protocol was observed by immunostaining with PL2-6 or 1H6 of “double-fixed” undifferentiated, RA and TPA treated HL-60/S4 cells, prior to sonication, in comparison to “single-fixed” cells. This experiment demonstrated that epichromatin localization of PL2-6 is unaffected by the additional fixation step (Fig. 3C–E). Convinced that we can maintain attachment of these two antibodies to epichromatin up to, and presumably, beyond the sonication process, we isolated the epichromatin fragments (~2 × 107 cells per experiment), purified the epichromatin-associated DNA pieces and performed Illumina sequencing.
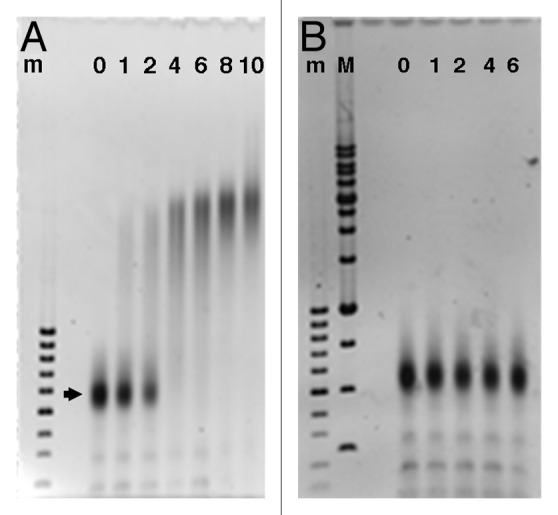
Figure 2. Electrophoretic mobility shift assay (EMSA) of Hela core mononucleosomes titrated with increasing amounts of PL2–6 (A) or normal mouse IgG (B). The lane numbers indicate the estimated molar ratios (IgG/nucleosome), based upon absorbance measurements of stock solutions prior to mixing, incubation and electrophoresis (see Materials and Methods). The arrowhead points to the position of monomer nucleosomes, which migrate at ~600 bp on 1% agarose gels. DNA size markers: m, 100 bp; M, 1 kb ladders.
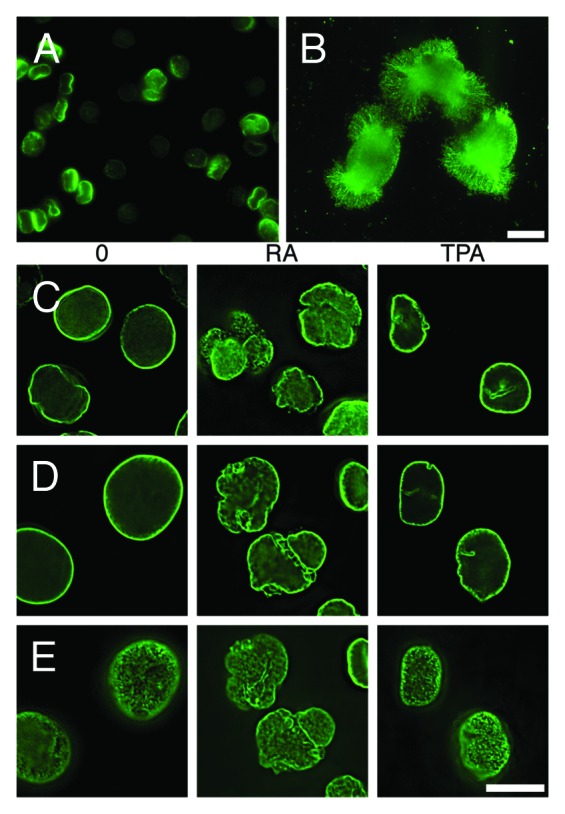
Figure 3. Necessity for and development of the “double-fixation” method (xxChIP). (A and B) Immunostaining by mAb PL2–6 of Xenopus nucleated erythrocytes (gift of C Cyr, College of Pharmacy, University of New England). Fresh RBCs were washed in PBS, allowed to settle on polylysine-coated slides, incubated in 1x (A) and 0.01x (B) PBS for 10 min at RT, fixed in 2% HCHO at the same buffer concentration (20 min, RT) and immunostained with PL2-6 as described previously.8 The nuclei remained intact in 1xPBS (A), exhibiting nuclear “rim” staining (green). In 0.01× PBS (B), swollen and exploded nuclei yielded “halos” (green) of immunostained chromatin fibers. Scale bar 10 µm. An identical set of images (not shown) was obtained from hypotonically lysed RBC stained with mAb 1H6 (anti-phosphatidyserine). (C-E) Deconvolved immunostaining patterns by PL2–6 of HL-60/S4 cells (O, undifferentiated; RA, granulocyte; TPA, macrophage) following single (x) or double (xx) fixation protocols. DAPI staining is not shown. (C) Mid-section images after single HCHO fixation prior to PL2–6 reaction. (D) Mid-section images after a second HCHO fixation following PL2–6, and prior to sonication. (E) Tangential “grazing” sections of the same cells as (D), showing epichromatin surface staining. Scale bar 10 µm.
Sequence analyses of the purified epichromatin-associated DNA
Purified epichromatin-associated DNA fragments (~200–400 bp) from HL-60/S4 cells (undifferentiated, RA- and TPA-treated) and corresponding input samples were subjected to paired-end sequencing (read length, 100 bp). Reads were aligned to the hg19 human genome assembly.12 Epichromatin regions were identified using ChIP-Seq peak calling software13 and annotated against genomic features (Genomatix Software GmbH). Table 1 summarizes the annotation statistics of enriched HL-60/S4 epichromatin regions (average region length ~1 kb). The most striking observation is that the vast majority (~95–99%) of epichromatin regions overlapped with repeat sequences, which were primarily retrotransposon Alu. The relative enrichment of Alu was ~10-fold, compared with the amount within the human genome (Alu ~11%); whereas L1 is enriched only ~2-fold (total genomic L1 ~17%).14,15 Furthermore, Table 1 shows that more than 50% of the epichromatin peak regions overlapped with introns (less than 10% overlapped with exons, promoters or transcription start sites), characteristic for Alu retrotransposons.15,16 The overlap statistics are quite similar comparing mAbs PL2-6 and 1H6, and comparing undifferentiated to granulocytic cell states. The minor exception was “TPA macro,” which exhibits a slightly lower association with Alu. It may be that scraping these attached fixed differentiated cells (prior to antibody addition) leads to some cell breakage, exposing general chromatin to reaction with PL2-6.
Table 1. Annotated epichromatin regions.
| mAb | Sample | Repeats (%) | Alu (%) | L1 (%) | Intergenic (%) | ≥1 locus (%) | ≥1 intron (%) | ≥1 exon (%) | Promoter (%) | TSS (%) |
Average region size (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PL2–6 | O un1 | 98.2 | 98 | 28 | 43.6 | 56.4 | 55.3 | 7.5 | 5.1 | 3.7 | 896.2 |
| PL2–6 | O un2 | 98.2 | 97 | 32 | 42.6 | 57.4 | 56.3 | 8.0 | 5.5 | 4.0 | 1008.9 |
| PL2–6 | RA gran1 | 99.0 | 97 | 32 | 44.0 | 56.0 | 54.9 | 7.2 | 4.9 | 3.6 | 975.0 |
| PL2–6 | RA gran2 | 98.2 | 98 | 30 | 45.1 | 54.9 | 53.9 | 7.1 | 4.8 | 3.6 | 961.0 |
| PL2–6 | TPA macro | 80.1 | 78 | 22 | 47.1 | 52.9 | 51.9 | 5.1 | 3.4 | 2.2 | 448.6 |
| 1H6 | O un | 94.8 | 93 | 28 | 45.0 | 55.0 | 54.1 | 5.9 | 4.1 | 2.8 | 727.3 |
| 1H6 | RA gran | 99.6 | 99 | 34 | 44.1 | 55.9 | 54.7 | 9.3 | 5.7 | 4.5 | 1192.1 |
Enriched epichromatin regions overlap more with retrotransposon Alu, than with L1. Samples were from HL-60/S4 cells incubated with PL2-6 (anti-H2A/H2B/DNA) or 1H6 (anti-phosphatidylserine): O un1 and O un2, duplicate experiments of undifferentiated cells; RA gran1 and RA gran2; duplicate experiments of granulocytes; TPA macro, macrophage. The columns list the percentage of overlap by the aligned “peak regions” with various elements within the human genome. Most of the data are derived from Genomatix RegionMiner Annotation and Statistics. Overlaps with Alu and L1 are derived from UCSC RepeatMasker annotation; mAb, monoclonal antibody; TSS, transcription start site.
Other parameters of the epichromatin regions were estimated: (1) Most of the members of the AluS subfamily and a few AluY members are enriched within epichromatin; AluJ is considerably reduced (Table S1). (2) Epichromatin exhibits an enrichment of Alu dimers, trimers and tetramers, compared with the genomic background (Fig. S1; Tables S2–S5). (3) The percentage of total nuclear Alu present within epichromatin peak regions is estimated to be <30%. Therefore, the bulk of Alu is not represented within epichromatin and is considered by us to be “internal Alu.” (4) The percentage of the human genome enriched within epichromatin is ~4% (range, 2.5–6%), based upon total bp in epichromatin peaks/total bp in the human genome (~2.8 Gbp).
Mapping epichromatin regions on the human chromosomes
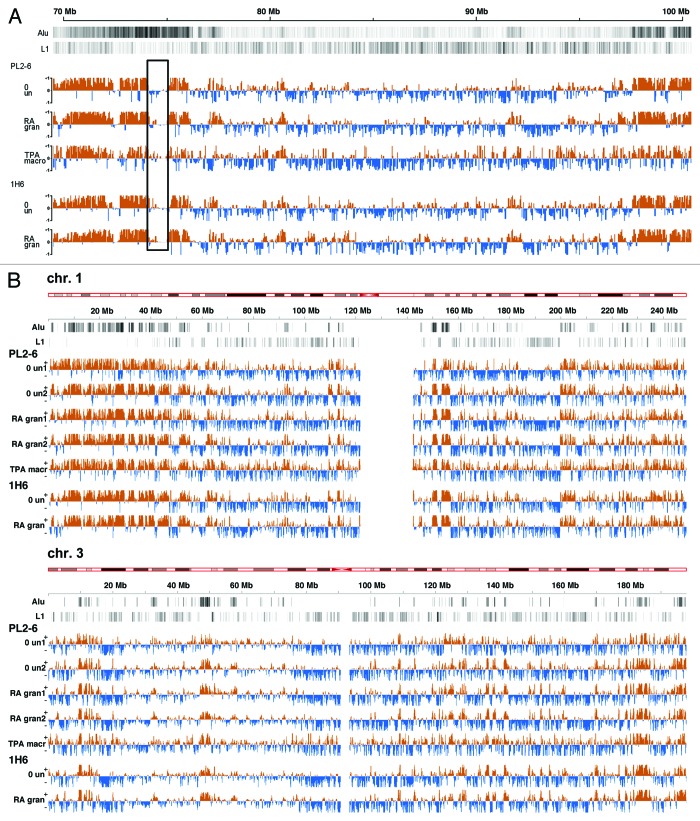
To examine the chromosome distribution of epichromatin regions and compare them with the distribution of Alu and L1 elements, we generated epichromatin maps (“epi-maps”) for the 22 human autosomes and the X chromosome (Fig. 4; Fig. S2). The epi-maps are normalized genome wide. They show the ChIP signal to input ratio, allowing different experiments and different chromosomes to be compared. Figure 4A presents an example of a small portion of a chromosome at high resolution (chromosome 7, “q” arm, 70–100 Mb). Epichromatin enriched regions (orange) are clearly not uniformly distributed on the human chromosomes. They seem to cover most, but not all, Alu-rich regions; e.g., an Alu-rich Chr. 7 region (74–75 Mbp, boxed area) is not enriched in epichromatin and, therefore, interpreted as “internal Alu.” Furthermore, much less association of epichromatin is observed to regions of high L1 density. These epi-maps demonstrate considerable resemblance along the same chromosome comparing different cell states (undifferentiated, granulocyte, and macrophage), suggesting “constitutive” (common) epichromatin regions, conserved even though the cells have diverged during differentiation. They also reveal considerable resemblance between PL2-6 (anti-H2A/H2B/DNA) and 1H6 (anti-phosphatidylserine). Two examples of epichromatin distribution on entire chromosomes are presented in Figure 4B; the entire set of epi-maps is shown in Figure S2.
Figure 4. Epichromatin maps. (A) Normalized epichromatin ChIP-Seq read enrichments (orange) across a section of chromosome 7 (“q” arm, 70‒100 Mb). These “epi-maps” illustrate: epichromatin association with some Alu and lack of association with L1 retrotransposons; the locations of “internal Alu” and the existence of “constitutive” (common) epichromatin peak regions in undifferentiated, granulocytic and macrophage cells. Alu and L1 tracks (black and white lines near the top of the map) depict the density of elements per 10 kb window, where darker regions have greater density than lighter regions. Read enrichment tracks (orange) for PL2-6 pooled the duplicate experiments for “un” and for “gran.” All tracks depict the density of elements per 10 kb window and have input subtracted. The orange (+) sections of the tracks show regions with increased epichromatin read densities compared with input control; the blue (-) sections of the tracks show regions enriched in input control compared with the epichromatin read densities. The box highlights a chromosomal segment (74–75 Mb) which is Alu-rich, but epichromatin-poor. (B) Two entire chromosomes (Chr. 1 and 3) illustrating the constancy of “constitutive” regions and the variation between different chromosomes in their epichromatin distribution. These “epi-maps” are displayed with window step size of 25 kb and a smoothing bandwidth of 50 kb. Maps of all human autosomes plus the X chromosome are presented in Figure S2.
We suggest that each epi-map represents an average over a large ensemble of nuclear architectural arrangements, illustrating the proportion of cells with any portion of a chromosome to be represented within epichromatin. Interphase chromosome 3D arrangements are considered stable, with some level of rearrangement during mitosis (for a review, see ref. 4). Chromosomal rearrangements during each cell division would generate large variations in nuclear architecture within a single cell population.
Comparison of epichromatin regions to data on epigenetic modifications
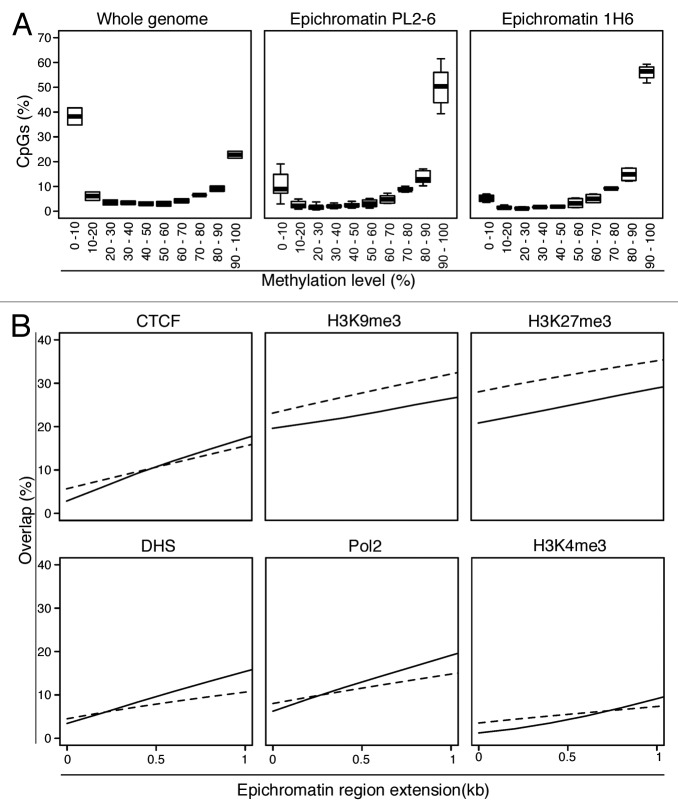
We attempted to predict properties of epichromatin by examining available chromatin epigenetic features available for HL-60 cells. Reduced representation bisulfite sequencing (RRBS) data for HL-60 cells17 revealed that, compared with the whole genome, CpGs in epichromatin are very highly methylated (Fig. 5A; Fig. S3). Additional analyses, examining the proximity of epichromatin to various chromatin features (Fig. 5B; Table S6), indicated depletion of H3K4me3 (active promoter mark) until ~1.0 kb from epichromatin peaks. The overlap of epichromatin with Pol 2, DNase 1 hypersensitive, and CTCF binding sites appear lower than with random sequence DNA fragments. Similarly, epichromatin overlaps less with heterochromatin markers (H3K9me3 and H3K27me3) than do random sequences. In sum, comparisons with existing chromatin feature data sets illustrate that epichromatin is unlikely to correspond to transcriptionally active “open” chromatin and does not resemble heterochromatin. We suggest that epichromatin may represent a new form of chromatin, situated adjacent to the NE.
Figure 5. Epichromatin properties derived from available databases. (A) CpG Methylation: Epichromatin enriched regions have substantially more regions with higher levels of CpG methylation than the whole genome. The plots show the median (broad horizontal line), interquartile range (box) and range (whiskers) within each methylation level. The “Whole genome” plot is based upon a combination of all the samples for the two antibodies (PL2-6 and 1H6). Only epichromatin positions with at least 10 reads covering a CpG were used. (B) Chromatin Features: Overlap of epichromatin regions with databases that map insulator elements (CTCF), repressive histone modifications (H3K9me3, H3K27me3) and active “open” chromatin markers (DNase I hypersensitive sites [DHS], RNA polymerase 2 [Pol2] and active promoters [H3K4me3] (see Table S6). Epichromatin regions were extended up to 1 kb to investigate proximity relationships to chromatin features. Solid lines represent the average overlap combining all the samples for both antibodies. Dashed lines represent the average overlap of 100 random DNA sequences with the chromatin features.
Discussion
In the present study, we continued our characterization of interphase epichromatin regions (i.e., chromatin proximal to the NE)8,9 in undifferentiated and differentiated HL-60/S4 cells, using a modified chromatin immunoprecipitation (ChIP) protocol. Monoclonal antibodies PL2-6 and 1H6 were used to ChIP chromatin, followed by purification of epichromatin-associated DNA, “deep” sequencing, annotation of the sequences and mapping the enriched regions on to the human chromosomes. The most striking observations were the following: (1) Epichromatin is highly enriched in retrotransposon Alu; ~10-fold enrichment, compared with their average content in the human genome. (2) The epichromatin regions are distributed discontinuously along each chromosome, displaying chromosome-specific patterns (“epichromatin maps”). (3) The epichromatin maps are very similar for the two antibodies (PL2–6 and 1H6), despite their divergent origins and apparent specificities. (4) The epichromatin maps are very similar for each chromosome, whether derived from undifferentiated, granulocyte or macrophage cell states, implying that the enriched regions appear to be “constitutive.”
The modified chromatin immunoprecipitation protocol (denoted “xxChIP,” involving two formaldehyde fixations prior to sonication and IP) was developed because both antibodies bind to the surface of chromatin (epichromatin) in the intact interphase nucleus, but are capable of binding to all isolated mononucleosomes or to exposed chromatin from a disrupted nucleus. The double fixation protocol was designed to preserve and identify the “accessible” chromatin epitopes within the intact nucleus. It is important to emphasize that the two mouse monoclonal antibodies employed in this ChIP-Seq study are not specific for Alu-containing chromatin. Both antibodies can immunostain epichromatin in cells from various species that do not possess Alu repetitive elements (e.g., PL2-6 stains interphase epichromatin in Drosophila and tobacco cells8; 1H6, interphase epichromatin in mouse and Drosophila cells9). The conserved epitopes probably reflect a conserved nucleosome structure or conformation exposed at the surface of chromatin. Epichromatin may present a more “accessible” chromatin structure with “unaffiliated” histone basic tails that can interact with phosphatidylserine (presumed) embedded in the inner nuclear membrane.9 The exact structures of the epitopes recognized by these two antibodies remain to be elucidated. Current investigations are exploring the possibility that the chromatin conformational epitopes involve a complex of histone and phospholipid components.
Fluorescent in situ hybridization (FISH) studies of human fibroblasts and lymphocytes using a consensus Alu probe18 have indicated that most of nuclear Alu is centrally located in the interphase nucleus. Although performed on different cell types than HL-60/S4, this conclusion is consistent with the present ChIP-Seq experiments. We find less than 30% of nuclear Alu within the epichromatin fraction. Indeed, the authors of the FISH study18 state “expansions from the more interior Alu-rich chromatin into the Alu-poor peripheral shell were seen on closer inspection, indicating complex spatial interactions of chromatin regions with high and low gene density.”
FISH studies with chromosome “paints” (probes) have indicated that the gene-dense, Alu-rich human chromosome 19 is located away from the NE in spherical lymphocytes and lymphoblasts or in flattened fibroblastic cells19,20, whereas the gene-sparse, Alu-poor chromosome 18 is found closer to the nuclear periphery. This conclusion contrasts markedly with our observation that in HL-60/S4 cells chromosome 19 is “epichromatin-rich” and chromosome 18 is “epichromatin-poor” (see Fig. S2). The disparity of interpretation could arise from a number of sources: (1) the difference in cell type (i.e., myeloid vs. lymphoid or fibroblast); (2) the intentional “suppression” of paint hybridization to human repetitive sequences by either co-hybridization with a vast excess of Cot1-DNA19 or the use of chromosome paints depleted of repetitive sequences20; (3) the existence of ill-defined territory boundaries, e.g., the extended territory in the interphase nucleus of chromosome 19 (described as “dispersed and irregular”19), compared with chromosome 18; and (4) the vastly different numbers of cell nuclei analyzed by FISH (e.g., ~10–50 per experiment) vs. ~2 × 107 cells for each ChIP-Seq experiment.22 It is conceivable that all chromosomes have regions that possess a finite probability of close association with the NE, but that large numbers of cells are required to map these regions and to estimate their probabilities.
In view of the “generic” nucleosome binding specificity of mAbs PL2-6 and 1H6, it was surprising to observe significant enrichment of retrotransposon Alu within the epichromatin region. It is possible that this enrichment is an adaptation specific for myeloid cells. Therefore, one should ask what attributes of Alu elements might relate to their concentration adjacent to the NE.
From a structural point-of-view, in vitro and in vivo evidence argue that two positioned nucleosomes are present within monomer Alu elements.23-25 The human genome contains ~7.8 × 105 Alu “monomers” (~280 bp), ~5.6 × 104 Alu “dimers,” ~7.5 × 103, Alu “trimers”, and ~1.4 × 103 “tetramers” (Table S2). Conceivably, Alu could position 2, 4, 6, or 8 nucleosomes and, it is suggested, constrain the mobility of adjacent nucleosomes.23-25 Stretches of positioned nucleosomes might facilitate ordering of chromatin fibers at the NE surface. Another structural attribute of Alu elements is their GC-richness and high CpG content (~1/3 of all genomic CpG sites are in Alu,14 with about 75% of Alu CpG methylated,26 accounting for ~25% of the total DNA methylation in the human genome21). These methylated DNA sites are clear candidates for the binding of MeCP2,27 which is associated with heterochromatin formation.
From a functional point of view, it is worth noting again that the major Alu subfamily enriched in epichromatin is AluS. It has been demonstrated that ~90% of retinoic acid receptor (RAR) response elements (DR2 motifs) are present in Alu repeats, with 95.5% of these Alu-DR2 elements distributed within AluS.28 Retinoic acid, via its interaction with RAR, is an important determinant of normal granulopoiesis.29-31 Evidence has been presented that subsets of Alu-DR2 elements in human embryonic stem cells respond to RA with Pol 3 transcription, supporting (neural) cell differentiation.32 Perhaps the enrichment of Alu-DR2 elements within HL-60/S4 epichromatin represents an evolved adaptation to present a “frontline” of RA response elements, facilitating subsequent myeloid differentiation. In a recent study of mouse chromatin megabase (topological) domains, evidence was presented that SINEs B1 and B2 (evolutionary relatives of Alu) are enriched at the boundaries of these domains.33 Combining this perspective with the present evidence for constitutive Alu-containing epichromatin regions suggests that a subset of megabase domains might “park” adjacent to the nuclear envelope.
Our present view of epichromatin parallels the earlier conception of “lamina-associated domains” (LADs34). However, there are differences, which must be underscored. It was suggested that “constitutive LADs” are enriched in LINES, rather than SINES35; whereas our “constitutive” epichromatin is enriched in SINE Alu, rather than LINE L1. The microarrays employed to identify the LAD sequences “cover the entire non-repetitive genome”35, whereas our ChIP-Seq analyses directly map Alu sequences into the epichromatin regions. In the HL-60/S4 cell system, we estimate that epichromatin constitutes ~4% (range, 2.5–6.0) of the total chromatin; the recovery of LADs is ~30–40% of total chromatin.35 This high yield of LADs is consistent with the prolonged incubation time needed to mark the LADs DNA by methylation (“...in a typical DamID experiment the methylation patterns represent the average over a time period of ~24 h or more36). In our ChIP experiments, cells are fixed for 10 min in 1% HCHO. Thus, our study is a “snapshot,” compared with the prolonged DamID procedure. Furthermore, different cell types have been analyzed by the different methods. Collectively, these differences make any correlation between LADs and epichromatin premature. In a more recent study,37 evidence was presented that LADs are “stochastically reshuffled” during mitosis, with H3K9me2 being involved in the LADs association to the NE. This is particularly interesting, in view of our earlier prediction that the N-terminal Tudor Domain of Lamin B Receptor, one of the first inner nuclear membrane proteins to engage with decondensing telophase chromosomes, has a binding preference for H3K9me2.38
What is the relationship of epichromatin to heterochromatin and euchromatin? The paucity of epichromatin overlap with heterochromatin histone modifications (i.e., H3K9me3 and H3K27me3, see Fig. 5B and Table S6) argues against a strict correspondence between epichromatin and heterochromatin; likewise, the unimpressive overlap of epichromatin with DNase I hypersensitive chromatin, RNA polymerase 2 binding sites and H3K4me3 suggests that epichromatin does not resemble “highly expressing” genes.39 It is of interest to point out that LADs exhibit enrichments of H3K9me3 (internally, within LADs) and of H3K27me3 (at the “boundaries” of LADs),34,40 further underscoring the apparent differences between epichromatin and LADs. Currently, there is very little information on which to base a hypothesis about the functional significance of epichromatin. However, it is interesting to speculate that epichromatin might correspond to regions containing “housekeeping genes”41, which appear to be enriched in Alu and depleted in L1.42 Furthermore, Alu-rich regions are negatively correlated with H3K27me3.43 Identification of epichromatin with housekeeping genes would be consistent with their “constitutive” distribution within the HL-60/S4 cell system. Future studies will explore the genetic content of epichromatin and its comparison to RNA-Seq data for the three different cell states of HL-60/S4.
Materials and Methods
Cell Culture and differentiation
HL-60/S4 cells were maintained in RPMI-1640 medium supplemented with 10% fetal calf serum plus 1% penicillin/streptomycin, as previously described.10 Cells were differentiated into granulocytes by addition of all-trans retinoic acid (RA) to a final concentration of 1 µM for 4 d and into macrophage by addition of phorbol ester (TPA) to 16 nM for 4 d, as previously described.10,11 For the ChIP experiments, undifferentiated cells (O) and granulocytes (RA) were maintained in T-75 flasks as suspensions yielding ~2 × 107 total cells per experiment. Macrophage differentiation was also performed in T-75 flasks with the cells adhering and frequently clumping by day 1. Cells were scraped off the flasks after fixation, but prior to antibody treatment. Because of clumping, it was difficult to obtain an accurate cell titer; yields were estimated to be ~1 × 107 total cells.
Immunostaining and microscopy
The 3D SIM microscope images of U2OS cells stained with PL2-6 and 1H6 are from data collected as previously described.9 The computed tangential images presented in this paper (Fig. 1) are from different cells and have not been previously published. Immunostaining of the Xenopus erythrocyte nuclei stained with PL2–6 (Fig. 3A and B) was performed using conventional epifluorescence. Immunostaining of the various cell states of HL-60/S4 with PL2–6 (Fig. 3C–E) and imaging with the DeltaVision deconvolution microscope has been described earlier.8,9
Electrophoretic mobility shift assay (EMSA)
For the titration experiments, stock solutions included: Hela mononucleosomes (1.2 µM) in 0.1 M NaCl plus TE buffer (pH 7.5); PL2–6 (8.38 µM, based upon A280 measurements) in PBS; normal mouse IgG (6.67 µM) dissolved in PBS. Mixtures at various molar ratios (indicated in Fig. 2) were made in 0.1 M NaCl plus TE, incubated at 37 °C for 30 min, followed by addition of sample buffer, electrophoresis on 1% agarose horizontal gels (150 V, 1.8 h) and subsequent staining with ethidium bromide and imaging.
xxChIP-Seq protocol
This method is based, in part, on information supplied by Covaris (Covaris Inc., Woburn, MA), maker of the focused acoustic sonicator E210, used in these studies. Suspension cells (~2 × 107) were centrifuged (2000 rpm, 5 min) and gently resuspended in Fixation Buffer (50 mM Hepes pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) to which was added a volume of 37% Formaldehyde to a final concentration of 1%. Cells were oscillated for 10 min at RT. Fixation was stopped by the addition of 2.5 M glycine to a concentration of 0.125 M for 5 min. Cells were washed in PBS and lysed in LB1 (Lysis Buffer 1: 50 mM Hepes pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.25% Triton X-100) on ice, with intermittent inversion for 10 min. Protease inhibitors (Sigma-Aldrich P8340) were added to LB1 just prior to use. Cells were washed with LB2 (Lysis Buffer 2: 10 mM Hepes pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) and then incubated in LB2 containing 5% bovine serum albumin (BSA) for 30 min with a rocking motion. We next added 5 µg of primary antibody (PL2-6 or 1H6) and incubated at least 1 h at RT with rocking. Cells were washed with LB2 and fixed again in 1% formaldehyde for 10 min at RT. The reaction was stopped with glycine, as before. Cells were washed in LB2, suspended in 90 µl LB3 (Lysis Buffer 3: 10 mM Tris pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, 0.5% N-lauryl sarcosine), transferred into Covaris micro-chambers for sonication and brought to 130µl with LB3. Sonication for 30 min was performed at 7 °C, Duty Cycle 20%, Intensity 5, cycles per burst 200. The sonicate was transfered to an eppendorf tube, followed by 2 sequential washes of the micro-chamber with 75 µl of LB3 to yield a final volume of 280 µl. The sonicated chromatin/antibody solution was centrifuged at 14 000 RPM for 10 min and the supernatant harvested for incubation with the uncoupled agarose column.
Uncoupled agarose was prepared by adding 40 µl of agarose to each micro-column, washing twice with 200 µl of LB3 and rocking with 200 µl LB3 for at least 30 min before use. At the same time, Protein A/G High-Capacity Agarose (Thermo Scientific Pierce CL-6B) columns were prepared for the ChIP fractionation by adding 27 µl of Protein A/G agarose to a micro-column, washing twice with 200 µl LB3 and rocking with 200 µl 5% BSA/LB3 for at least 30 min.
The entire sonicate was equilibrated with uncoupled agarose on a rocker plate for at least 30 min. After brief centrifugation, 180 µl was incubated with Protein A/G agarose for 1 h at RT or overnight at 4 °C, to bind the epichromatin-antibody complex. The remainder of the chromatin sonicate (“input”) represents the total nuclear chromatin. The Protein A/G agarose, with bound epichromatin, was washed 7 times with 200 µl of LB3. The final wash was preceded by 30 min of rocking at RT. Epichromatin was eluted with two pooled washes of 50 µl fresh 0.05 M NaHCO3 containing 1% SDS, rocking for 15 min and centrifuged. These epichromatin preps were brought to 0.2 M NaCl, treated with RNase for 30 min at 37 °C, followed by Proteinase K for at least 4 h at 65 °C with shaking. DNA purification was with a QIAquick PCR Purification Kit (Qiagen # 28106). For each experiment, both input and ChIP fractions were measured for DNA amount and sequenced. PCR and High Throughput DNA sequencing was performed in the Genomics and Proteomics Core Facility of the German Cancer Research Center.
Short read mapping
ChIP-Seq and input control reads were trimmed to remove low quality bases and adaptor contamination using TrimGalore (http:www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Reads were trimmed for a consensus adaptor sequence (-a AGATCGGAAG AGCG) with at least 6 bp overlap, 10% error rate (–stringency 6 –e 0.1) and bases with quality lower than 20 (on phred33 scale, -q 20). If the resultant read was less than 20 bp, it was discarded (-length 20). The trimmed reads were mapped to the hg19 build 37.1 genome assembly using Bowtie212 version 2.0.0.7beta7, fragment length range of 80 to 500 bp (-I 80 -X 500), trimming a single 3′ nucleotide of each read (-3 1), surpressing reads not aligning as pairs (–no-mixed). Unique alignments were extracted using samtools44 v0.1.18 r982:295 by setting a mapping quality filter of 20, which equates to a 1% probability that the alignment does not correspond to the correct position. The subsequent uniquely mapping reads had read duplicates removed and were converted to BED format using a custom script.
Peak identification, quality control, and annotation
Epichromatin regions were identified using SICER v1.113 to call peak region by comparing the ChIP-Seq alignment BED file to the corresponding input control alignment BED file. SICER is especially suitable for peak calling in epichromatin, since it was developed to analyze diffuse broad histone markers. The effective genome size was set to 85% of hg19. The fragment length parameter was set to the Bioanalyzer reported fragment size for each ChIP-Seq sample. Read enrichment was scanned over 200 bp windows, allowing for merging of enriched regions that were up to 200 bp apart, and were filtered for an FDR of 0.01, after calculating significance of read enrichment compared with the input control. The resultant peaks were converted to broadPeak format using a custom script. Quality control statistics were calculated, as outlined by Landt et al.45 including the fraction of read in peaks (FRiP), PCR bottleneck coefficient (PBC) using custom scripts, and the normalized and relative strand correlations (NSC/RSC) using SPP v1.046 in R v2.15.0 (http://www.R-project.org). The resultant peaks were annotated for feature overlap using the Genomatix Genome Analyzer suite version v3.00801 (Genomatix Software GmbH).
Normalized read enrichment signal tracks
Read enrichment signal tracks were generated using SPP v1.0. The SPP script was modified so that it was able to work with the previous produced alignments in BED format. The protocol used was adapted from the SPP tutorial (http://compbio.med.harvard.edu/Supplements/ChIP-seq/tutorial.html). Read enrichment/depletion signals were generated in wig format. Normally SPP generates the read density profiles from wherever the first read in each chromosome is observed. This makes comparison of signals between experiments difficult, as the first read of each chromosome does not always match. For this reason, the ranges for the signal were predefined for the entire length of each chromosome starting from the first base. For file size optimisation, the read density wig files were converted from a bed-like format to the wig-fixed step format using a custom script. Window sizes of 100 bp, 1 kb, 5 kb, 10 kb, 25 kb, and 50 kb and bandwidths of one and two times the window size were used to generate profiles. We found that at a fine scale both 5 kb and 10 kb signals without bandwidth smoothing were informative, while the 50 kb signal with bandwidth smoothing was most interpretable for visualization of larger genomic regions. Resultant wig files were visualized using IGV.47
Analysis of repeats
Repeat annotations of the hg19 genome were downloaded from the UCSC genome browser48 RepeatMasker track of SINE Alu repeat elements (excluding free alu monomers) on the 4th of July 2013. We examined the read density of ChIP-Seq experiments with PL2-6 and 1H6 antibodies over all extracted Alu elements on Chromosome 1, and found enrichment of some but not all Alu elements (data not shown). We performed genome wide enrichment analysis of the Alu subfamilies (AluJ, AluS, and AluY) and their subfamily members (Table S1). To calculate the location of higher order Alu multimers, we attempted to plot the head-to-head distance of Alus, as previously performed by Bettecken et al.,25 but were unable to observe distinct Alu multimers. Instead, we decided to merge all Alu elements that were within 30 bp of each other to better represent Alu multimers by virtue of size of the merged Alu elements. Histogram analysis showed good separation of Alu multimers (Fig. S1). Manual histogram segmentation separated Alu multimers (from 0.5-mers to tetramers) at 200 bp, 381 pb, 525 bp, 695 bp, 839 bp, 991 bp, 1144 bp, and 1296 bp. This analysis also revealed the average Alu monomer to be ~300 bp, which is the known average size of an Alu element. Alu and L1 density tracks (Fig. 4; Fig. S2) were calculated as the occupancy of the repeat elements per 10 kb window.
Analysis of chromatin features
We downloaded chromatin feature annotation from the ENCODE databases: wgEncodeAwgDnaseUwHl60UniPk.narrowPeak, wgEncodeAwgTfbsUwHl60CtcfUniPk.narrowPeak, wgEncodeHaibTfbsHl60Pol24h8V0422111PkRep2.broadPeak and wgEncodeUwHistoneHl60H3k4me3StdPkRep1.narrowPeak.
We downloaded additional H3K27me3, H3K9me3, and input control bigWig signal files from the Canadian Epigenetics, Environment and Health Research Platform (CEEHRC, http://www.epigenomes.ca/downloads.html). Information about CEEHRC and the participating investigators and institutions can be found at (http://www.cihr-irsc.gc.ca/e/43734.html). We converted the bigWig signal files into a simulated alignment file based on a fragment size of 202 bp and equal distribution of reads originating from the positive and negative strand. We identified regions of enrichment using SICER, using similar parameters used for epichromatin peak identification, except using a gap size of 400 bp and an FDR of 0.0001.
To examine the random overlap with chromatin features, we performed 100 permutations of shuffling each of the epichromatin peak files using bedtools, excluding gaps in the chromosomes. This way, we maintained the size distribution of the epichromatin peaks in random regions of the genome. We performed similar overlap analysis and calculated the average overlap of all 100 permuted shuffles of each epichromatin peak file.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
A.L.O. and D.E.O. express their gratitude to the German Cancer Research Center (DKFZ, Heidelberg) for the awards of Visiting Scientist fellowships, allowing collaborations within the Langowski laboratory. We also thank Peter Lichter, Roland Eils, and Bendikt Brors for helpful comments, Harald Herrmann and Michaela Hergt for use of the DeltaVision microscope, reagents and advice, and Stephan Wolf, Nadine Wehran, Ute Ernst, and Sabine Schmidt of the DKFZ Genomics and Proteomics Core Facility. We are grateful to Yolanda Markaki (LMU Munich) and Lothar Schermelleh (University of Oxford) for helping to acquire the 3D-SIM images. The data reported in this paper have been submitted to ArrayExpress Experiments Archive, which can be found at www.ebi.ac.uk with accession numbers: E-MTAB-2360.
Glossary
Abbreviations:
- NE
nuclear envelope
- PL2-6
mouse mAb anti-H2A/H2B/DNA
- 1H6
mouse mAb anti-phosphatidylserine
- ChIP
chromatin immunoprecipitation
- xxChIP
ChIP employing two sequential formaldehyde fixations
- ChIP-Seq
ChIP followed by DNA sequencing
- EMSA
electrophoretic mobility shift assay
- SINE
short interspersed nuclear element
- LINE
long interspersed nuclear element
- TPA
phobol ester
- RA
retinoic acid
- FISH
fluorescent in-situ hybridization
- RAR
retinoic acid receptor
- LADs
lamina-associated domains
References
- 1.Olins DE, Olins AL. Chromatin history: our view from the bridge. Nat Rev Mol Cell Biol. 2003;4:809–14. doi: 10.1038/nrm1225. [DOI] [PubMed] [Google Scholar]
- 2.Grigoryev SA, Woodcock CL. Chromatin organization - the 30 nm fiber. Exp Cell Res. 2012;318:1448–55. doi: 10.1016/j.yexcr.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–91. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 6.Wandke C, Kutay U. Enclosing chromatin: reassembly of the nucleus after open mitosis. Cell. 2013;152:1222–5. doi: 10.1016/j.cell.2013.02.046. [DOI] [PubMed] [Google Scholar]
- 7.Schooley A, Vollmer B, Antonin W. Building a nuclear envelope at the end of mitosis: coordinating membrane reorganization, nuclear pore complex assembly, and chromatin de-condensation. Chromosoma. 2012;121:539–54. doi: 10.1007/s00412-012-0388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olins AL, Langhans M, Monestier M, Schlotterer A, Robinson DG, Viotti C, Zentgraf H, Zwerger M, Olins DE. An epichromatin epitope: persistence in the cell cycle and conservation in evolution. Nucleus. 2011;2:47–60. doi: 10.4161/nucl.2.1.13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prudovsky I, Vary CP, Markaki Y, Olins AL, Olins DE. Phosphatidylserine colocalizes with epichromatin in interphase nuclei and mitotic chromosomes. Nucleus. 2012;3:200–10. doi: 10.4161/nucl.19662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olins AL, Buendia B, Herrmann H, Lichter P, Olins DE. Retinoic acid induction of nuclear envelope-limited chromatin sheets in HL-60. Exp Cell Res. 1998;245:91–104. doi: 10.1006/excr.1998.4210. [DOI] [PubMed] [Google Scholar]
- 11.Olins AL, Hoang TV, Zwerger M, Herrmann H, Zentgraf H, Noegel AA, Karakesisoglou I, Hodzic D, Olins DE. The LINC-less granulocyte nucleus. Eur J Cell Biol. 2009;88:203–14. doi: 10.1016/j.ejcb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–8. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grover D, Mukerji M, Bhatnagar P, Kannan K, Brahmachari SK. Alu repeat analysis in the complete human genome: trends and variations with respect to genomic composition. Bioinformatics. 2004;20:813–7. doi: 10.1093/bioinformatics/bth005. [DOI] [PubMed] [Google Scholar]
- 16.Grover D, Kannan K, Brahmachari SK, Mukerji M. ALU-ring elements in the primate genomes. Genetica. 2005;124:273–89. doi: 10.1007/s10709-005-3086-8. [DOI] [PubMed] [Google Scholar]
- 17.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–31. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–67. doi: 10.1023/A:1012495201697. [DOI] [PubMed] [Google Scholar]
- 21.Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Huang S. Nuclear function of Alus. Nucleus. 2014;5:5. doi: 10.4161/nucl.28005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Englander EW, Howard BH. Nucleosome positioning by human Alu elements in chromatin. J Biol Chem. 1995;270:10091–6. doi: 10.1074/jbc.270.17.10091. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka Y, Yamashita R, Suzuki Y, Nakai K. Effects of Alu elements on global nucleosome positioning in the human genome. BMC Genomics. 2010;11:309. doi: 10.1186/1471-2164-11-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bettecken T, Frenkel ZM, Trifonov EN. Human nucleosomes: special role of CG dinucleotides and Alu-nucleosomes. BMC Genomics. 2011;12:273. doi: 10.1186/1471-2164-12-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie H, Wang M, Bonaldo MdeF, Smith C, Rajaram V, Goldman S, Tomita T, Soares MB. High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum. Nucleic Acids Res. 2009;37:4331–40. doi: 10.1093/nar/gkp393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertulat B, De Bonis ML, Della Ragione F, Lehmkuhl A, Milden M, Storm C, Jost KL, Scala S, Hendrich B, D’Esposito M, et al. MeCP2 dependent heterochromatin reorganization during neural differentiation of a novel Mecp2-deficient embryonic stem cell reporter line. PLoS One. 2012;7:e47848. doi: 10.1371/journal.pone.0047848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laperriere D, Wang TT, White JH, Mader S. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics. 2007;8:23. doi: 10.1186/1471-2164-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kastner P, Lawrence HJ, Waltzinger C, Ghyselinck NB, Chambon P, Chan S. Positive and negative regulation of granulopoiesis by endogenous RARalpha. Blood. 2001;97:1314–20. doi: 10.1182/blood.V97.5.1314. [DOI] [PubMed] [Google Scholar]
- 30.Gaines P, Berliner N. Retinoids in myelopoiesis. J Biol Regul Homeost Agents. 2003;17:46–65. [PubMed] [Google Scholar]
- 31.Drumea K, Yang ZF, Rosmarin A. Retinoic acid signaling in myelopoiesis. Curr Opin Hematol. 2008;15:37–41. doi: 10.1097/MOH.0b013e3282f20a9c. [DOI] [PubMed] [Google Scholar]
- 32.Hu Q, Tanasa B, Trabucchi M, Li W, Zhang J, Ohgi KA, Rose DW, Glass CK, Rosenfeld MG. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nat Struct Mol Biol. 2012;19:1168–75. doi: 10.1038/nsmb.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 35.Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–80. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–78. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 37.Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–92. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 38.Olins AL, Rhodes G, Welch DB, Zwerger M, Olins DE. Lamin B receptor: multi-tasking at the nuclear envelope. Nucleus. 2010;1:53–70. doi: 10.4161/nucl.1.1.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Gräf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–67. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luperchio TR, Wong X, Reddy KL. Genome regulation at the peripheral zone: lamina associated domains in development and disease. Curr Opin Genet Dev. 2014;25C:50–61. doi: 10.1016/j.gde.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 41.Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends Genet. 2013;29:569–74. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 42.Eller CD, Regelson M, Merriman B, Nelson S, Horvath S, Marahrens Y. Repetitive sequence environment distinguishes housekeeping genes. Gene. 2007;390:153–65. doi: 10.1016/j.gene.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, Willard HF. Evidence for sequence biases associated with patterns of histone methylation. BMC Genomics. 2012;13:367. doi: 10.1186/1471-2164-13-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–31. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–9. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–9. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.