Abstract
Cajal bodies are important nuclear structures containing proteins that preferentially regulate RNA-related metabolism. We investigated the cell-type specific nuclear distribution of Cajal bodies and the level of coilin, a protein of Cajal bodies, in non-irradiated and irradiated human tumor cell lines and embryonic stem (ES) cells. Cajal bodies were localized in different nuclear compartments, including DAPI-poor regions, in the proximity of chromocenters, and adjacent to nucleoli. The number of Cajal bodies per nucleus was cell cycle-dependent, with higher numbers occurring during G2 phase. Human ES cells contained a high coilin level in the nucleoplasm, but coilin-positive Cajal bodies were also identified in nuclei of mouse and human ES cells. Coilin, but not SMN, recognized UVA-induced DNA lesions, which was cell cycle-independent. Treatment with γ-radiation reduced the localized movement of Cajal bodies in many cell types and GFP-coilin fluorescence recovery after photobleaching was very fast in nucleoplasm in comparison with GFP-coilin recovery in DNA lesions. By contrast, nucleolus-localized coilin displayed very slow fluorescence recovery after photobleaching, which indicates very slow rates of protein diffusion, especially in nucleoli of mouse ES cells.
Keywords: DNA repair, chromatin, nucleus, nucleolus, Cajal bodies, coilin
Introduction
Cajal bodies (CBs) were first reported by Santiago Ramón y Cajal in 1903. CBs are nuclear structures containing accumulated proteins with diverse functions. Most of these proteins play important roles in RNA processing.1,2 Small nuclear ribonucleoproteins (snRNPs) accumulate in Cajal bodies, associate with spliceosomes, and regulate splicing of pre-mRNA.3 These include five different snRNPs known as U1, U2, U3, U4, and U5. After transcription, snRNA is immediately exported to the cytoplasm, and each subunit is assembled with core Sm proteins to form SMN protein complexes. The snRNPs are relocated back into the cell nucleus and accumulate in CBs for final maturation. CBs then associate with transcription sites that mostly co-localize with nuclear speckles (summarized in ref. 2).
A main component of Cajal bodies is the p80 coilin protein. Coilin becomes increasingly phosphorylated during mitosis.4 During interphase, coilin is dispersed in the nucleoplasm or accumulates in CBs. These nuclear bodies (NBs) are non-membrane protein aggregates with diameters of 0.5−1.0 μm.5 Numerous studies characterized coilin and other CB-related proteins, and have begun to examine CB function.6,7 CBs also contain factors involved in pre-mRNA splicing, pre-rRNA processing, histone pre-mRNA 3ʹ maturation, and basal transcription. CBs are present in compartments containing polymerases I, II, and III, and telomerase RNA-positive compartments.5,8,9 CBs are highly mobile, kinetically independent structures.2,10 Coilin interacts with several components of CBs. For example, fluorescence resonance energy transfer (FRET) analysis revealed interactions between coilin and SMN protein, mutual coilin-coilin interactions, and SMN-SMN associations.10 These data unambiguously document the dynamic and functional properties of CBs.
CBs contain several nucleolar proteins including fibrillarin, NOPP140, and small nucleolar RNPs (snoRNPs).8 Transient expression of mutated p80 coilin (serine residues were replaced with aspartate) caused CB formation within nucleolar compartments. Expression of mutant coilin variants disrupted both CBs and nucleolar compartments.11,12 These experiments suggested that coilin, and potentially CBs, were important for functional properties of nucleolus.13 Because several nucleolar proteins respond to DNA injury, including UBFs, NPM, and fibrillarin,14 we postulated that coilin might respond to radiation-induced DNA damage. For example, Boulon et al.15 discussed UV-induced disruption of CBs into nucleoplasmic microfoci, and ionizing irradiation changed coilin-containing complexes.16 Thus, in the current study, we investigated not only morphology of Cajal bodies, but also biological properties of p80 coilin in response to DNA damage, which we induced by UVA- and γ-irradiation.
Inappropriate DNA repair can lead to mutations that severely injure the organism. A fundamental question concerns the responses of proteins and nuclear substructures to DNA injury, caused by genotoxic stress. Ionizing radiation can also induce local changes in chromatin conformation. DNA lesions are recognized by several proteins, which initiate different repair strategies based on the severity of DNA damage. DNA lesions include double-strand breaks (DSBs), which are recognized by specific protein complexes such as MRE11-RAD50-NBS1 that contribute to the repair DNA using homologous recombination (HR). This process is associated with activation of a DNA damage-related serine/threonine protein kinase, called ataxia telangiectasia mutated (ATM).17,18 ATM activation leads to phosphorylation of histone H2AX (γH2AX) and to MRE11-RAD50-NBS1 binding to chromatin lesions. This process also involves binding of the mediator protein MDC1 to damaged chromatin, and it leads to recruitment of the chromatin-remodeling factors, including 53BP1, SMC1, CHK2, or BRCA1. Another well-known DNA repair-related pathway represents non-homologous end joining (NHEJ), which is associated with binding of KU heterodimer to DSBs. Ku70/Ku80 attracts the catalytic sub-unit of DNA-dependent protein kinase and activates its kinase activity (summarized by ref. 19). This process initiates a cascade of events, which leads to recovery of damaged DNA. Hierarchical assembly of proteins, involved in DNA repair, occurs coordinately with the recruitment of additional chromatin-related factors, such as heterochromatin protein 1 (HP1) and the polycomb-group proteins BMI1 and Mel18.20-22
Another interesting DNA repair-related event (associated with nucleotide excision repair) is linked to inhibition of RNA pol II-driven transcription elongation, when ionizing radiation induces DNA lesions, and is called transcription-coupled repair (TCR).23 Moreover, double-strand breaks present a barrier for functioning of RNA polymerases, but inhibition of DNA protein kinase, for example, enables RNA pol II to bypass DNA breaks and continue in transcription elongation.24 Transcription of ribosomal genes could be also affected by exposure to ionizing radiation. Therefore, it is important to determine how the appearance of DNA lesions influences transcription and transcription factors in an irradiated genome. As a consequence of transcription-related events after DNA damage, changes in RNA processing could appear. Thus, proteins of Cajal bodies represent interesting candidates for such studies. Based on these observations, we studied coilin localization in UVA-induced DNA lesions. We determined that coilin, but not SMN (other CB protein), is recruited to DNA lesions induced by UVA-irradiation. Coilin recruitment to UVA-damaged chromatin was cell cycle-independent and occurred immediately after local micro-irradiation. Moreover, UVA-induced 53BP1-positive lesions did not mostly recruit coilin, γ-irradiation-induced foci (IRIF) did not co-localize with CBs, and exposure to γ-rays did not alter total coilin levels. However, localized CB movement was significantly reduced by cell exposure to γ-rays.
Results
Nuclear patterns and numbers of Cajal bodies in different cell types
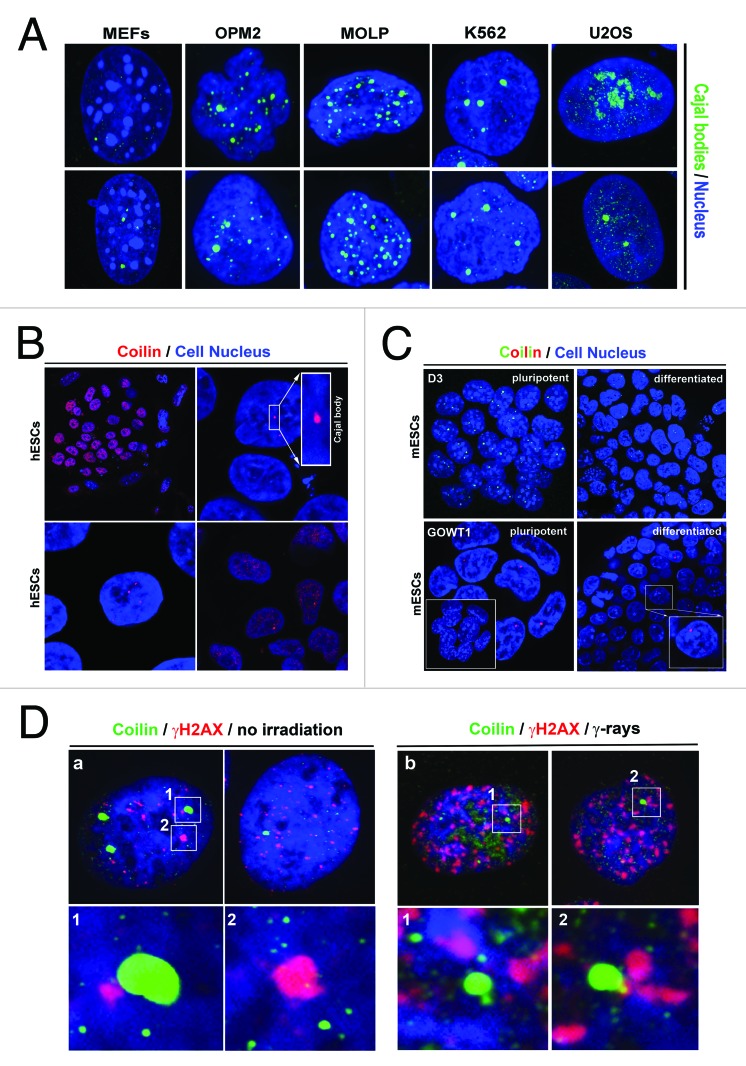
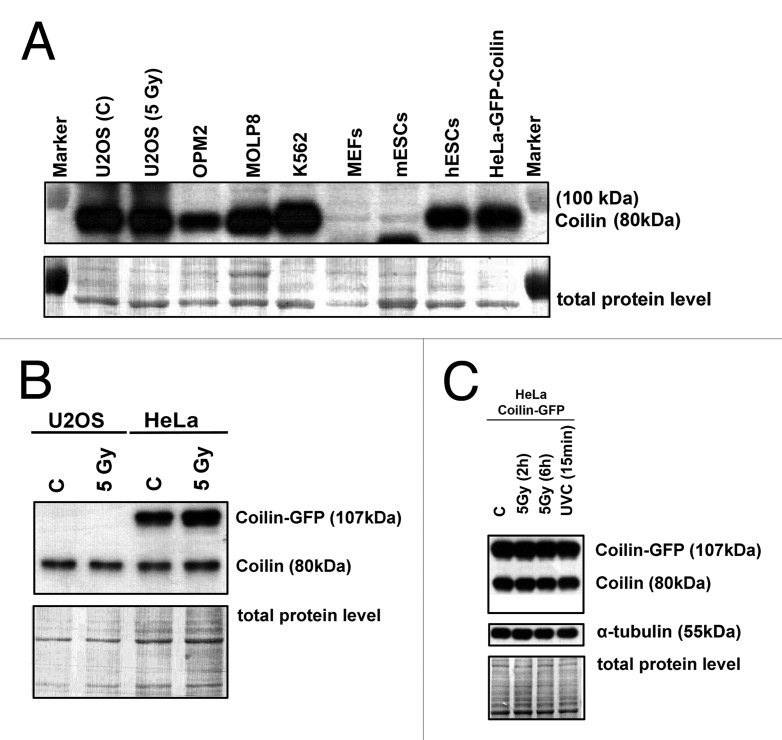
We studied the nuclear patterns of Cajal bodies in many cell types. The multiple myeloma cell lines OPM2 and MOLP8 contained multiple Cajal bodies, MEFs contained 1–4 subtle Cajal bodies (diameter 0.5–0.8 µm), and leukemia K562 cells contained 3–4 robust Cajal bodies (diameter > 1 µm) (Fig. 1A). In 50% of U2OS cells, we observed high coilin levels inside nucleoli (Fig. 1A). In hESCs, we found high coilin levels in nucleoplasm, and approximately 50% of the cell population contained visible Cajal bodies (Fig. 1B). Moreover, we observed 1–4 Cajal bodies in mouse ESCs (D3 line). Localization pattern of CBs was not changed by retinoic acid-induced differentiation (Fig. 1C). Treatment with γ-radiation did not affect the degree of association between coilin and γ-H2AX foci that appear spontaneously in non-irradiated cells or as irradiation-induced foci (IRIF) after γ-irradiation (Fig. 1D). In non-irradiated and irradiated cells, we observed either Cajal bodies associated with γ-H2AX foci or Cajal bodies located away from these foci (Fig. 1D, number 1 or 2). Western blot analyses of coilin levels supported the fact that Cajal bodies did not change after γ-radiation, because after irradiation the levels of endogenous and ectopically expressed coilin were not changed (Fig. 2A-C). Additionally, no changes in coilin levels were observed after UVC-irradiation (Fig. 2C).The level of coilin was cell type-specific, low in OPM2 cells, and subtle 80-kDa coilin fragments were found in MEFs and mESCs (Fig. 2A).
Figure 1. Nuclear pattern of Cajal bodies in several cell types. (A) Cajal body (green) morphology was studied in mouse embryonic fibroblasts (MEFs), multiple myeloma cells (OPM2 and MOLP8), K562 leukemia cells, and U2OS human osteosarcoma cells. (B) Nuclear distribution of coilin in embryonic stem cells. Coilin was homogeneously distributed in interphase nuclei of human ESCs, or accumulated into body-like structures (Cajal bodies) in hESCs. (C) Cajal bodies were observed in pluripotent mouse ESCs (line D3), and in GOWT1 mESCs and in their differentiated counterparts. (D) Morphological association between Cajal bodies (green) and γ-H2AX-positive foci (red) in non-irradiated and γ-irradiated U2OS cells.
Figure 2. western blot analysis of coilin levels in selected cell types. (A) Levels of coilin (80 kDa) in non-irradiated and γ-irradiated U2OS, OPM2, MOLP8, K562, MEFs, mESCs, hESCs, and HeLa cells stably expressing GFP-coilin. (B) Levels of coilin in non-treated and γ-irradiated U2OS and HeLa cells stably expressing GFP-coilin (irradiation by 5 Gy of γ-rays). (C) Comparison of endogenous and ectopically expressed coilin in non-irradiated HeLa cells stably expressing GFP-coilin. Coilin levels were assessed in HeLa cells irradiated with 5 Gy of γ-rays at 2 and 6 h after irradiation, and after irradiation with UVC. Coilin levels were normalized to the total protein levels or α-tubulin.
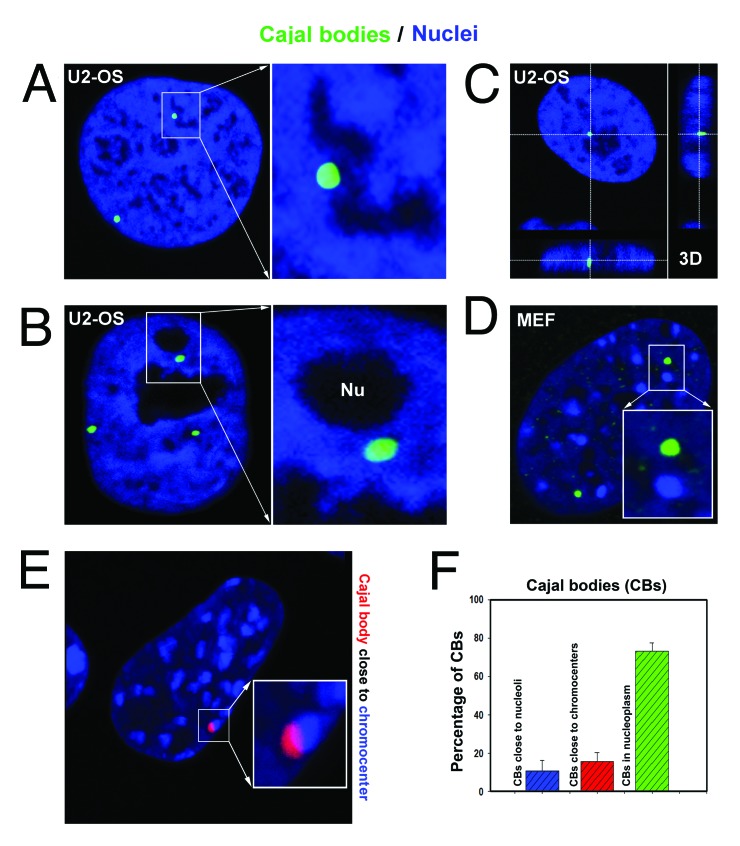
In terms of morphology, Cajal bodies were observed in DAPI-poor nuclear regions (Fig. 3A), in proximity to nucleoli (Fig. 3B and C), distant from clusters of centromeric heterochromatin called chromocenters, and in proximity to chromocenters (Fig. 3D-F).
Figure 3. Morphology of Cajal bodies. Cajal bodies (green in panels A−D; red in panel E) were localized in (A) DAPI-poor genomic regions; (B, C) in proximity to nucleoli; (D) away from chromocenters; and (E) very close to chromocenters in MEFs. Studies were performed in U2OS and MEF cells. (F) Analysis of morphology of CBs in MEFs containing well visible chromocenters after DAPI staining. Data represent mean percentage of events with respect to the total number of CBs analyzed ± standard error (S.E.).
Cell cycle-dependent nuclear distribution and pattern of Cajal bodies
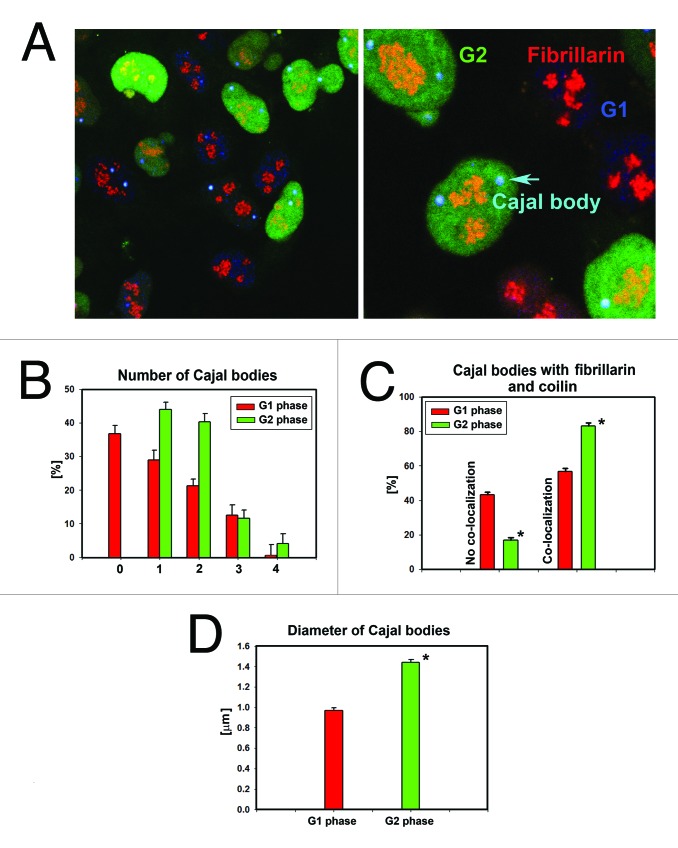
We analyzed the nuclear pattern of Cajal body localization during the cell cycle (Fig. 4A-D), with a focus on G1 and G2 phases. We used HeLa-Fucci cells expressing RFP-cdt1 for analysis of the G1 phase, and GFP-geminin for analysis of the G2 phase. In HeLa-Fucci cells, we observed different numbers of Cajal bodies in G1 and G2 phases. We found an increased number of Cajal bodies in G2 compared with those in G1 cell cycle phase (Fig. 4B; Fig. S1). We also studied the association of Cajal bodies with fibrillarin (Fig. 4A and C). The number of Cajal bodies containing both coilin and fibrillarin and the positivity of both proteins in Cajal bodies increased in G2 phase (Fig. 4C). The G2 phase of the cell cycle also was characterized by the appearance of Cajal bodies with larger diameters (Fig. 4D).
Figure 4. Cell-cycle-dependent nuclear pattern of Cajal bodies. (A) Cajal bodies in G1 phase (red) and G2 phase (green) of the cell cycle. Studies were performed in HeLa-Fucci cells. Fibrillarin (red) and CBs (blue) were visualized by primary antibodies and stained by secondary antibodies [conjugated with Alexa 594 (red) and Alexa 405 (blue)]. (B) Number of Cajal bodies in G1 and G2 phases of the cell cycle. (C) Cajal bodies positive for both coilin and fibrillarin in G1 and G2 phases. (D) Diameter of Cajal bodies in G1 and G2 phases. The following numbers of cell were analyzed: for number of Cajal bodies in G1 = 193; number of Cajal bodies in G2 = 268; Cajal bodies associated with fibrillarin in G1 = 118; Cajal bodies associated with fibrillarin in G2 = 189; diameter of Cajal bodies in G1 = 70; diameter of Cajal bodies in G2 = 94. Data in panels B and C represent mean percentage of events with respect to the total number (set as 100%) of CBs analyzed ± standard error (S.E.). Asterisks in panels C and D show statistical significance at P ≤ 0.05. Analysis was performed by Student’s t test.
Coilin recruitment to UVA-induced DNA lesions and coilin recovery after photobleaching
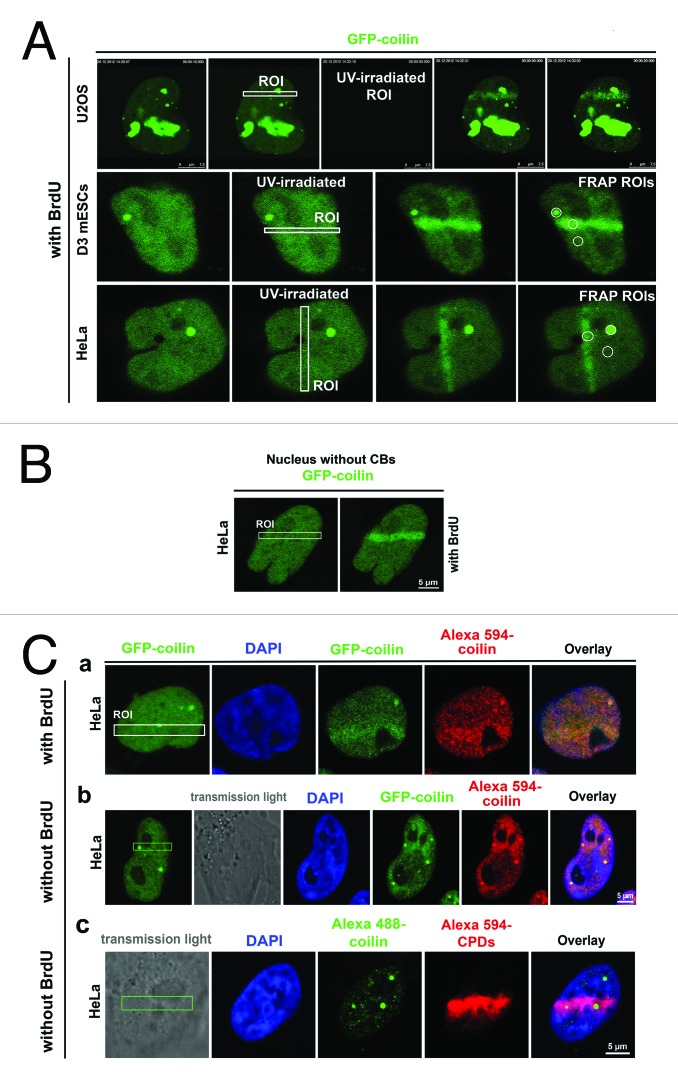
We observed high levels of coilin in nucleolar compartments of U2OS cells, which could be a consequence of ectopically expressed GFP-coilin. This interpretation is consistent with the previous report of reference 25. However, HeLa cells stably expressing GFP-coilin were not characterized by high coilin levels in nucleoli (Fig. 5A). Therefore, we used these cells for additional analyses. In both U2OS and HeLa cells, genomic regions that were locally micro-irradiated using a UVA-laser displayed pronounced recruitment of GFP-coilin. Coilin was recruited to DNA lesions within 15–20 s after micro-irradiation (Fig. 5A). Increased coilin levels in DNA lesions were maintained up to 5 min after irradiation. This was observed in experiments with and without BrdU pre-sensitization, and in cells lacking CBs (Fig. 5A-C). We also observed that endogenous coilin is recruited to DNA lesions (Fig. 5C part a, b, and c). These results contribute to those of reference 16, which reports focal patterns of coilin accumulation after UVC-irradiation of entire cell population. Here we showed that UVA-induced DNA lesions that recruited coilin contained cyclobutane pyrimidine dimers (CPDs) (Fig. 5C part c), 53BP1 (Fig. 6A part a), and phosphorylated histone H2AX (Fig. 6A part b). This was observed even without BrdU pre-sensitization. However, γ-irradiation did not potentiate co-localization of CBs and 53BP1, or CBs and γH2AX (Fig. 1D and 6B part a, b). This result was confirmed at different time points after γ-irradiation (0.5, 2, 6, and 24 h after irradiation). Simultaneous irradiation with UVA and γ-rays caused both locally induced DNA lesions and IRIF (Fig. 6C).
Figure 5. Recruitment of coilin to UVA-induced DNA lesions. (A) In live U2OS cells transiently expressing GFP-coilin and HeLa cells stably expressing GFP-coilin. Coilin was recruited to micro-irradiated genomic regions. This was also observed in transiently transfected D3 mESCs. These experiments were performed with BrdU pre-sensitization. (B) Recruitment of coilin to UVA-induced DNA lesions in HeLa cells stably expressing GFP-coilin, but lacking CBs. (C) Cells were locally irradiated using a UVA-laser in defined regions of interest (ROI) and endogenous coilin was analyzed under conditions of (a) BrdU pre-sensitization or (b) without BrdU pre-sensitization. (c) CPDs were analyzed without BrdU pre-sensitization. CPDs were visualized by red fluorescence and CBs as green. Experiments were performed in HeLa cells stably expressing GFP-coilin (green). The cell nuclei were clearly visible before local micro-irradiation, and GFP fluorescence was maintained after formaldehyde fixation and immunostaining. To assess endogenous coilin levels, HeLa cells were micro-irradiated and endogenous coilin and CPDs were visualized using antibodies (panel Cc).

Figure 6. Recruitment of coilin, 53BP1 and γH2AX to UVA-induced DNA lesions. (A) (a) Recruitment of coilin (green) and 53BP1 (red) to locally induced DNA lesions without BrdU pre-sensitization. (b) Increased levels of coilin (green) and γH2AX (red) were observed in UVA-induced DNA lesions without BrdU pre-sensitization. (B) Nuclear localization patterns of coilin (green) and 53BP1 (red), or coilin (green) and γH2AX (red) in HeLa cells irradiated with 5 Gy of γ-rays and in control non-irradiated cells; cells were analyzed 0.5, 2, 6, and 24 h after γ-irradiation. (C) HeLa cells were irradiated with 5 Gy of γ-rays and then with UVA in selected regions of interest denoted by the white box. Coilin (green) and 53BP1 (red) were recruited to DNA lesions and γ-irradiation-induced foci appeared (white arrow).
Locally induced DNA lesions accumulated higher levels of coilin, as also evidenced by the observed localization of endogenous coilin in irradiated HeLa cells expressing GFP-H2B (Fig. 7A part a), but did not accumulate higher levels of Cajal body-associated SMN protein (Fig. 7A part b). In addition, in BrdU pre-sensitized cells, UVA-induced DNA lesions that were positive for γH2AX (Fig. 7A part c) contained high levels of phosphorylated HP1α protein (phosphor-Ser92) (Fig. 7A part d). However, after γ-irradiation, no changes in nuclear localization patterns of CBs and HP1α or CBs and SMN were observed (Fig. 7B parts a, b).
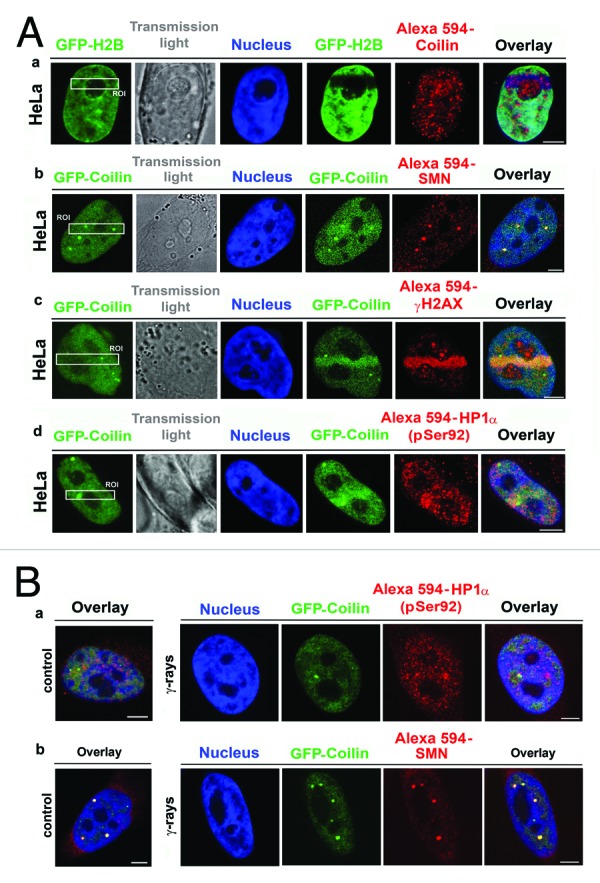
Figure 7. Visualization of selected proteins localized to DNA lesions. (A) (a) Recruitment of endogenous coilin (red) to DNA lesions was studied in HeLa cells stably expressing histone H2B (green). (b) GFP-coilin (green), but not other CB-related protein, called SMN, was recruited to UVA-damaged chromatin. (c) High levels of GFP-coilin (green) and γ-H2AX (red) co-localized with DNA lesions in BrdU pre-sensitized cells. (d) GFP-coilin (green) and phosphorylated HP1α at serine 92 (red) were recruited to DNA lesions in BrdU pre-sensitized cells. (B) Nuclear localization patterns of (a) phosphorylated HP1α at serine 92 and (b) SMN were identical in non-irradiated cells and cells irradiated with 5 Gy of γ-rays.
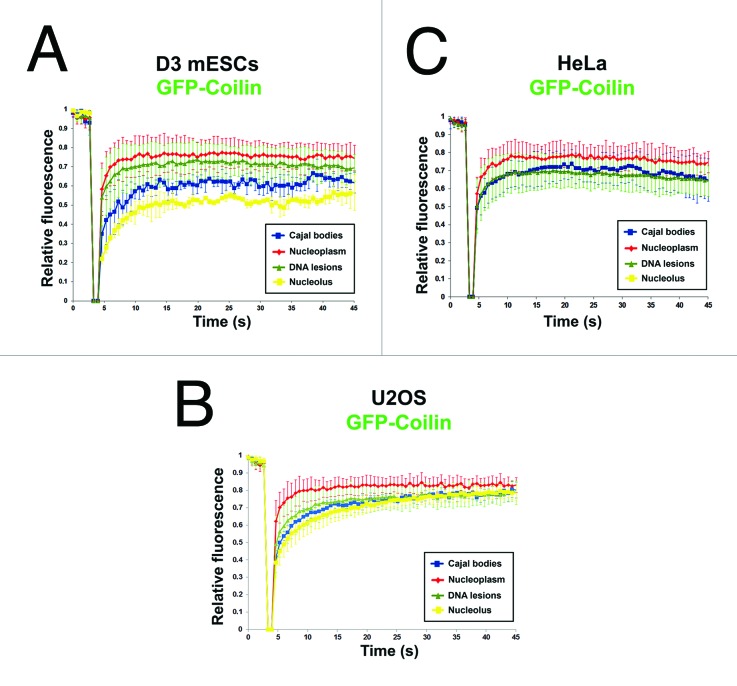
We also performed FRAP analysis to investigate coilin diffusion properties. The slowest recovery of GFP-coilin after photobleaching occurred in nucleoli in D3 mESCs, whereas the most rapid recovery of fluorescence occurred in nucleoplasm of D3 mESCs and U2OS cells (Fig. 8A and B). The kinetics of coilin recovery in Cajal bodies after photobleaching was approximately the same as the kinetics of coilin recruitment to UVA-induced DNA lesions (Fig. 8A-C). In HeLa cells, the observed differences in coilin recovery after photobleaching in different nuclear regions were not statistically significant (Fig. 8C).
Figure 8. FRAP analysis of coilin diffusion in various nuclear regions. Kinetics of GFP-coilin were studied in nucleoli, DNA lesions, nucleoplasm, and non-irradiated Cajal bodies in (A) D3 mESCs; (B) U2OS cells; and (C) HeLa cells. FRAP data were normalized to 1, and data represent means ± standard error (S.E.).
Localized movement of Cajal bodies and cell-cycle dependent coilin status in DNA lesions
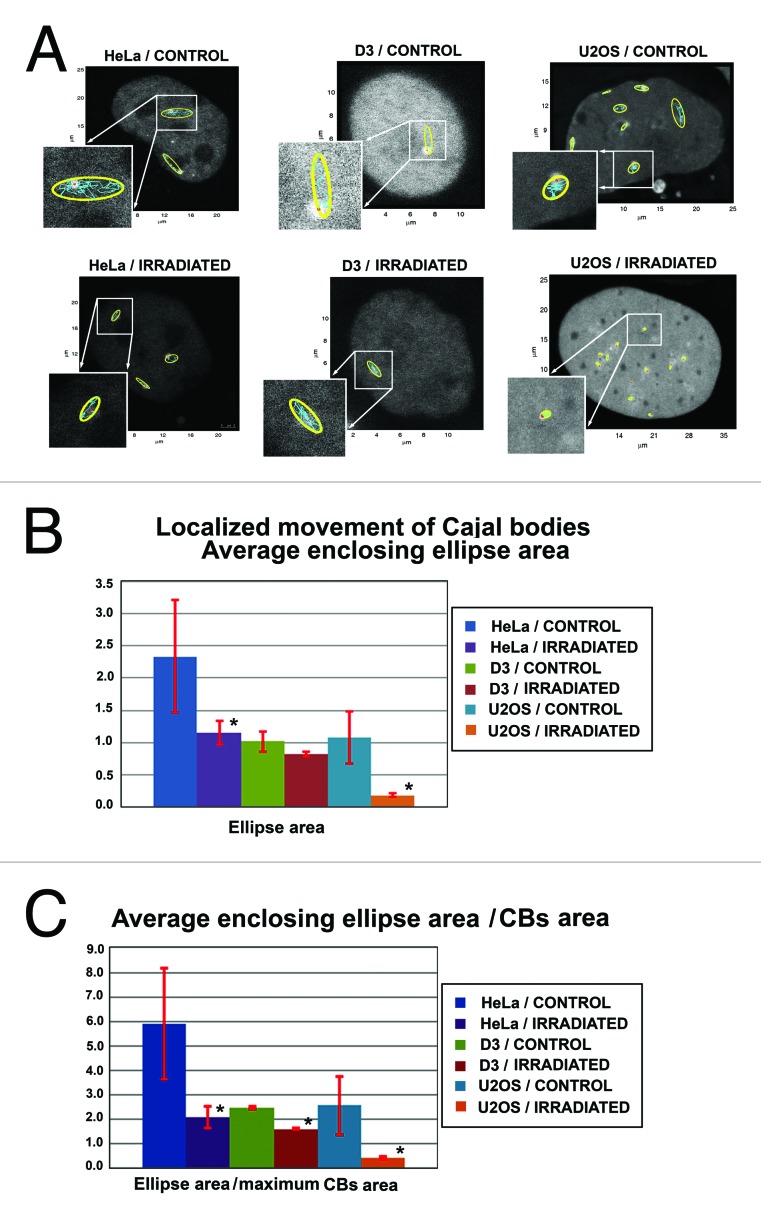
We used HeLa cells, mouse ESCs (line D3), and U2OS cells to study the localized movement of Cajal bodies in non-irradiated and irradiated cells by 5 Gy of γ-rays. In all cases tested, we observed that irradiation influenced the movement of Cajal bodies. In all cell types studied, γ-irradiation reduced the average size of the enclosing ellipse around Cajal body trajectories (Fig. 9A and B). This conclusion was supported by normalization of the average enclosing ellipse area with respect to the area of CBs. This parameter also was reduced after cell irradiation with 5 Gy of γ-rays (Fig. 9C). These results indicate that γ-radiation substantially restricts Cajal body localized movement.
Figure 9.
Localized movement of Cajal bodies. (A) Average enclosing ellipses (yellow) calculated for Cajal bodies in HeLa cells, D3 mESCs, and U2OS cells. Non-treated cells were used as control cell population, and data were compared with average enclosing ellipse of Cajal bodies in cells irradiated by 5 Gy of γ-rays. Tracking of ellipse centroids is shown in blue. Selected ellipses are enlarged in individual frames. Small red dots represent the starting points of tracking analysis. (B) Data represent average enclosing ellipse areas in non-irradiated and γ-irradiated HeLa cells, D3 mESCs, and U2OS cells. (C) Average enclosing ellipse area per Cajal body area in HeLa cells, D3 mESCs, and U2OS cells. Data in panels B and C represent mean ± standard error (S.E.). Asterisks in panels (B) and (C) show statistically significant differences from control values at P ≤ 0.05. Analysis was performed by Student’s t test.
To further clarify coilin function during the DNA damage response (DDR), we studied the possible coilin-coilin dimerization in DNA lesions using FRET analysis. We observed very low FRET efficiencies, with 5.4% ± 3.3% in DNA lesions and 7.8% ± 4.9% in non-irradiated nucleoplasm. For closely interacting partners such as p53 and 53BP1, the FRET efficiency in our microscope system is usually ~30–40%. So, by this experimental approach we can exclude the potential significance of coilin dimerization in DNA lesions. FRET analysis of coilin/53BP1 (2.8 ± 2.1%), coilin/γH2AX (3.0 ± 2.6%), and coilin/CPDs (0.0 ± 0.0) also showed no interaction.
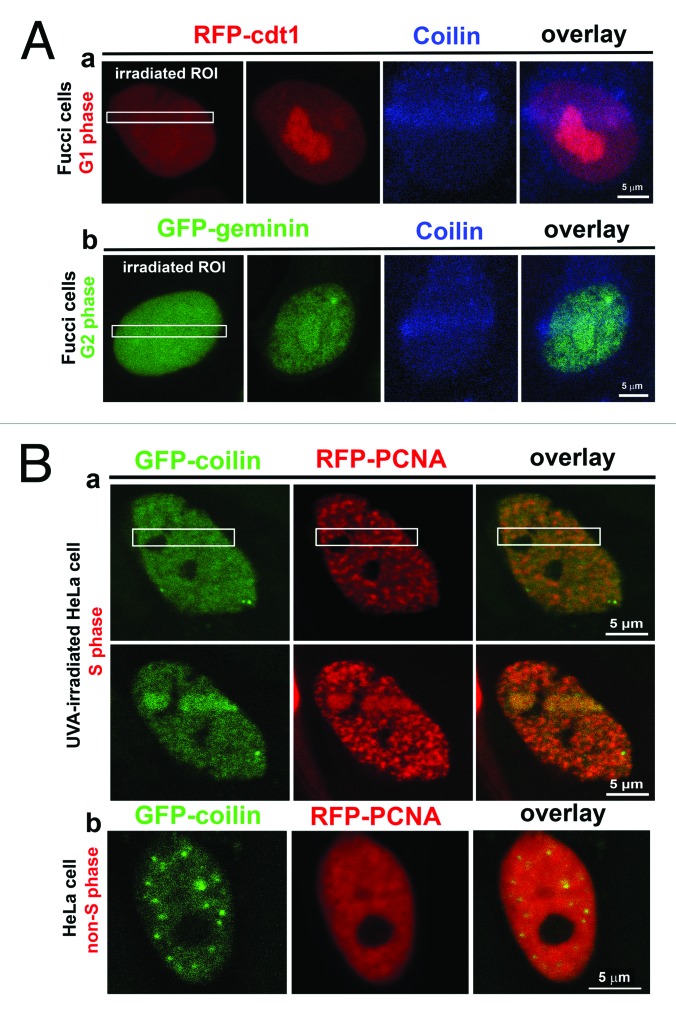
We assessed whether the recruitment of coilin to DNA lesions is dependent on the cell cycle. In HeLa cells expressing RFP-cdt1 in G1 phase and GFP-geminin in G2 phase, we analyzed endogenous coilin in DNA lesions. We observed coilin recruitment to UVA-induced lesions in both G1 and G2 phase (Fig. 10A parts a, b). Simultaneous detection of GFP-coilin and RFP-PCNA in S-phase cells indicated that both proteins localized to UVA-induced DNA lesions (Fig. 10B part a showing S phase; Fig. 10B part b shows non-irradiated control non-S phase cell). This can be explained by the fact that UVA-irradiation by 355-nm UVA-laser induces both γH2AX-positive DSBs and CPDs. Interestingly, simultaneous detection of coilin and CPDs indicated that coilin was not present in irradiated regions containing CPDs (Fig. 5C). Moreover, many DNA lesions contained 53BP1 but lacked coilin, and vice versa (Fig. 6A part a). Only γH2AX co-localized with GFP-coilin in UVA-induced DNA lesions (Fig. 6A part b). Therefore, at this time any conclusions drawn from these results are speculative because we cannot rule out competitive effects in the visualization of proteins by GFP technology and antibodies.
Figure 10.
Coilin localization in DNA lesions in individual cell cycle phases. (A) (a) G1 phase was characterized by expression of RFP-ctd1 (red); (b) G2 phase was characterized by expression of GFP-tagged geminin in HeLa-Fucci cells. Cell-cycle-independent accumulation of coilin (blue) at DNA lesions was observed. (B) (a) Recruitment of GFP-coilin (green) and RFP-PCNA (red) to UVA-induced DNA lesions at S phase. (b) Nuclear localization patterns of GFP-coilin (green) and RFP-PCNA (red) in non-irradiated non-S phase cells.
Discussion
Proteomic studies are useful to identify precise functions of proteins in DDR. The majority of studies have investigated DDR in non-nucleolar regions of the genome (summarized by ref. 17, 18). However, the DDR-related function of proteins such as coilin, which can appear in nucleoli of tumor cells, is less understood.14,25-27 Recently, it was observed that nucleolar accumulation of coilin was influenced by both cisplatin and γ-radiation, which induced co-localization of coilin and the RNA Pol I subunit RPA194. Radiation also enhances the interaction between coilin and UBF factors that modulate the association of RNA Pol I with rDNA.25 Association between coilin and fibrillarin-positive regions of nucleoli appeared after cisplatin treatment and irradiation with 4 Gy of γ-rays.25 These studies unambiguously demonstrate nuclear re-arrangement of CBs after exposure to genotoxic stress. In the current study, we report that local micro-irradiation with UVA induces pronounced accumulation of coilin in non-nucleolar genomic regions (Fig. 5A and 7A).
These data suggest a novel function for coilin, which generally regulates snRNP biogenesis and histone mRNA processing. It is likely that the rapid recruitment of coilin to DNA lesions reflects chromatin conformational changes caused by genotoxic stress (ref. 25; Fig. 5A and B and 7A). The function of coilin in DDR might also be ascribed to its asymmetric arginine methylation and arginine methyltransferase functions. For example, PRMT1 methyltransferase is known to be responsible for methylation of other DDR-related proteins such as 53BP1.28,29 These results suggest the importance of post-translational modification of proteins involved in DNA repair and imply that coilin could be involved in DNA repair-related processes. This is supported by the fact that the morphology of Cajal bodies is also affected by UVC-irradiation,16 and a reduced level of coilin correlated with an increased level of γH2AX. This phenomenon was observed after etoposide treatment, which is a powerful pro-apoptosis agent that induces apoptotic DNA fragmentation.1
It is well known that Cajal bodies undergo pronounced localized movement.10,13 We observe that γ-irradiation affects CB motility (Fig. 9). In non-irradiated cells, other authors showed that Cajal bodies move toward and away from the periphery of nucleoli. Thus, two classes of Cajal bodies were found. FRAP analysis shows that coilin is dynamically exchanged with Cajal bodies, but this depends on coilin interactions with other proteins.10,13 Our study showed that γ-radiation reduced the average area of enclosing ellipses; these parameters were used to measure Cajal body movement in three different cell lines, including mouse ESCs (Fig. 9). These experiments documented that the morphology and movement of Cajal bodies were affected by γ-radiation-induced cell stress. Thus, our results suggest that the coilin recruitment to DNA lesions is relevant, because other CB-related proteins such as SMN were not recruited to UVA-damaged chromatin (Fig. 7A).
Conclusions
This study showed cell-type-specific CB morphology and determined that coilin recognized UVA-induced DNA lesions in human tumor cells and mESCs. The morphological changes of CBs after DNA damage were described previously.15,16 We additionally observed that coilin, but not SMN, was recruited to and accumulated at UVA-induced DNA lesions immediately after irradiation. These observations raise questions about coilin dimerization in locally induced DNA lesions, and suggest that other proteins could potentially interact with coilin in UVA-damaged chromatin. In summary, coilin is rapidly recruited to UVA-induced DNA lesions within 15–20 s after irradiation. This event could be a primary stress response, which might attract other proteins involved in DDR-related pathways. Radiation can induce multiple DNA-damage-response pathways,30 which could explain the fact that we observed cell cycle-independent coilin recruitment at DNA lesions.
Materials and Methods
Cell lines and cell cultures
We used the following cell lines for these experiments: MOLP8 and OPM-2 multiple myeloma cells (DSMZ); K562 leukemia cells (ATCC, USA); U2OS human osteosarcoma cells (ATCC); and immortalized mouse embryonic wild-type fibroblasts (iMEFs; a generous gift from the laboratory of Prof Thomas Jenuwein [Max Planck Institute of Immunobiology and Epigenetics]). We analyzed the nuclear pattern of Cajal bodies in mouse embryonic stem cells (mESCs) (line D3, ATCC), in GOWT1 cells (gift from Dr Hitoshi Niwa), and in human ESCs (hESCs).31 Differentiation of mouse ESCs was induced by all-trans retinoic acid according to previously published protocols.32
We also used HeLa cells stably expressing GFP-colin (a generous gift from the laboratory of Prof Angus Lamond, University of Dundee16). For detection of endogenous proteins in DNA lesions, we used HeLa cells stably expressing GFP-H2B to visualize nuclei. This was essential in order to see cell nuclei before local micro-irradiation by UVA-laser in selected regions of interest (ROIs) (see below). HeLa cells stably expressing GFP-H2B were a generous gift from Dr Marion Cremer (Ludwig-Maximilians-University).
For cell cycle studies, we used commercially available Fucci system-related cells expressing RFP-cdt1 in G1 phase and GFP-geminin in S/G2/M phases, which have previously been described in detail (ref. 33) (Life Technologies; http://www.lifetechnologies.com).
Adherent cells were cultivated in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum at 37 °C in a humidified atmosphere containing 5% CO2. Cells growing in suspension were cultivated in RPMI-1640 medium supplemented with 10% fetal calf serum. HeLa cells stably expressing GFP-coilin were cultivated in DMEM containing 0.4 g/l geneticin G418 (Gibco Life Technologies). Human and mouse ESCs were cultivated according to references 32 and 34.
Western blot analysis
Western blots were performed according to reference 32. For analysis of coilin levels by western blots, we used antibody against coilin (#sc-32860; Santa Cruz) at a dilution of 1:1,000. Colin levels were analyzed in the following cell types: MOLP8, OPM-2, K562, U2OS, MEFs, mESCs, hESCs, and HeLa. We tested the effect of 5 Gy of γ-radiation on coilin levels. For secondary antibody, we used the peroxidase-conjugated anti-rabbit IgG (#A-4914; Sigma Aldrich) at a dilution of 1:2000. Equal protein was loaded in each gel lane. Protein levels were normalized to the total protein levels, measured by µQuant spectrophotometer and KCjunior software (Bio-Tek Instruments, Inc), or α-tubulin (#LF-PA0146, Fisher Scientific)
Immunofluorescence analysis
Immunofluorescence methods were published previously in references 31 and 34. For the current experiments, we used the following antibodies: fibrillarin (#ab5821 and #ab4566; Abcam, UK); coilin (#sc-32860; Santa Cruz); phospho-HP1α (serin-92) (#MBS 852359, My Biosource); SMN (#ab124438, Abcam); γ-H2AX (#ab2893, Abcam); 53BP1 (#ab21083, Abcam); and CPDs (#NMDND001, Cosmo Bio Co., Ltd).
Confocal microscopy live-cell studies and analysis of Cajal body movement
Cells were seeded on uncoated, γ-irradiated, 50-mm glass-bottomed dishes used for inverted microscopy (No. 0; MatTek Corporation, #P50G-0-30-F). Cells were transfected with plasmid DNA encoding GFP-colin (generous gift from Dr Petr Kaláb, National Cancer Institute, NIH, Bethesda, USA). Cell transfection was performed using METAFECTAN transfection reagent (Biontex Laboratories GmbH). D3 mESCs were transfected using Effectene transfection reagent (Qiagen, Bio-Consult). For confocal microscopy analyses, we used a Leica SP5 X confocal microscope and a white-light laser (wavelengths 460−670 nm). For image acquisition, we used the following settings: resolution 1024 × 1024 pixels, 400 Hz, bidirectional mode, zoom 8–12. Analysis of CB localized movement was performed after compensation of cell global motion. The detailed methodological approach was described previously.32 For Cajal body analysis, we analyzed the average areas enclosed by ellipses determined by the area occupied by CBs. The average enclosing ellipse is the ellipse with minimal area that enclosed all the positions of the CB centroids that appeared in time. A detailed description of this approach is given by reference 32. The current analyses were performed in non-irradiated and γ-irradiated HeLa, D3 mESCs, and U2OS cells.
Induction of double-strand breaks by UVA-irradiation in live cells
For local micro-irradiation using a UVA-laser (wavelength 355 nm), cells were seeded on uncoated, γ-irradiated, 50-mm glass-bottomed dishes used for inverted microscopy (No. 0; MatTek Corporation, #P50G-0-30-F). At 70% confluence, the cells were sensitized with BrdU according to reference 22, and irradiated using a UVA-laser connected to the Leica SP5 X confocal microscope. Defined regions of interest (ROIs) were irradiated. We selected individual CBs and also regions distant from CBs (for comparison). ROIs were irradiated by 100% laser output for 4–5 s. Laser intensity was not reduced at the acousto-optic tunable filter. UVA-laser irradiation was performed according to reference 34. After fixation of irradiated cells in 4% formaldehyde, phosphorylated histone H2A.X (γH2AX) and endogenous coilin were detected with rabbit polyclonal antibody against γH2AX (phospho S139; #ab2893; Abcam); 53BP1 (#ab21083, Abcam) and coilin (#sc-32860, Santa Cruz). Also, we analyzed endogenous phospho-HP1α (serine-92) (#MBS 852359, My Biosource) and SMN (#ab124438, Abcam). After immunodetection, locally irradiated cells were found according to known coordinates marked on the microscope dishes, or by the use of tissue culture dishes with microscope slide grids. For experiments on irradiation of the whole cell population, cells were irradiated by 5 Gy of γ-rays delivered by cobalt-60. Immunofluorescence analyses were performed 2 h after γ-irradiation.
Fluorescence recovery after photobleaching (FRAP)
FRAP was performed according to reference 35. We used HeLa, D3 mESCs, and HeLa cells stably expressing GFP-coilin. FRAP was performed in the following regions: (1) in a genomic region containing dispersed coilin (but without UVA-irradiation); (2) in non-irradiated Cajal bodies with accumulated coilin; and (3) in genomic regions without Cajal bodies, but containing accumulated coilin after UVA-irradiation. FRAP was performed using an argon laser (wavelength 488 nm). FRAP data were normalized to 1, and average values ± standard error (S.E.) were calculated by the use of Excel software. Statistical analysis was performed in Sigma Plot software 8.0, and Student’s t test was used for statistical analysis.
Fluorescence resonance energy transfer (FRET)
For FRET, we used a Leica SP5 X confocal microscope and the FRET mode of LEICA LAS AF software (version 2.1.2). To determine interactions between proteins, we used the FRET acceptor photobleaching technique.36 Proteins were labeled by mCherry and GFP, or by Alexa 594 and Alexa 488. In this study, we investigated potential coilin-coilin dimerization (coilin-GFP and coilin-Alexa 594) in DNA lesions induced by irradiation with a UVA-laser.
Supplementary Material
Disclosure of Potential Conflict of Interest
The authors indicate no potential conflict of interest.
Acknowledgments
This work was supported by grants P302/12/G157 and 13-07822S from the Grant Agency of the Czech Republic. D.V.S., E.B., and S.K. are members of a project supported by the European Social Fund in Czech Republic, the Education for Competitiveness Operational Programme (ECOP) (No. CZ.1.07/2.3.00/30.0030). Linguistic revision of the manuscript was performed by BioScience Writers (Houston, TX, USA). We would like to thank Dr Miroslav Dundr from Rosalind Franklin University in Chicago for discussing our results.
Author Contributions
The authors declare the following contributions. E.B. designed experiments, coordinated the experimental efforts, analyzed data, analyzed images, and wrote the paper. V.F. cultivated U2OS and HeLa cell lines and performed local micro-irradiation by UVA-laser, immunodetection, and confocal microscopy. S.L. was responsible for ES and HeLa-Fucci cell cultivation, western blot data, and immunodetection after local micro-irradiation. P.S. performed FRET analysis, UVA-irradiation, and confocal microscopy. D.V.S. analyzed localized of movement CBs. J.S. analyzed CB levels in individual cell-cycle phases, and S.K. performed statistical analyses.
References
- 1.Velma V, Carrero ZI, Allen CB, Hebert MD. Coilin levels modulate cell cycle progression and γH2AX levels in etoposide treated U2OS cells. FEBS Lett. 2012;586:3404–9. doi: 10.1016/j.febslet.2012.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol. 2012;24:415–22. doi: 10.1016/j.ceb.2012.03.010. http://dx.doi.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis--evidence that the coiled body is a kinetic nuclear structure. J Cell Biol. 1993;120:841–52. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- 6.Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–19. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raska I, Andrade LE, Ochs RL, Chan EK, Chang CM, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-H. [DOI] [PubMed] [Google Scholar]
- 8.Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–9. doi: 10.1016/S0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. In vivo kinetics of Cajal body components. J Cell Biol. 2004;164:831–42. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohmann K, Ferreira JA, Lamond AI. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;131:817–31. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sleeman J, Lyon CE, Platani M, Kreivi JP, Lamond AI. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp Cell Res. 1998;243:290–304. doi: 10.1006/excr.1998.4135. [DOI] [PubMed] [Google Scholar]
- 13.Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol. 2000;151:1561–74. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore HM, Bai B, Boisvert FM, Latonen L, Rantanen V, Simpson JC, Pepperkok R, Lamond AI, Laiho M. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics. 2011;10:009241. doi: 10.1074/mcp.M111.009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–27. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cioce M, Boulon S, Matera AG, Lamond AI. UV-induced fragmentation of Cajal bodies. J Cell Biol. 2006;175:401–13. doi: 10.1083/jcb.200604099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–33. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eppink B, Essers J, Kanaar R. (2012). Interplay and Quality Control of DNA Damage Repair Mechanisms. In Rippe K (Edited by), Genome Organization and Function in the Cell Nucleus (pp. 395-410), 1st edition, Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA. [Google Scholar]
- 20.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiácovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–80. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sustáčková G, Kozubek S, Stixová L, Legartová S, Matula P, Orlova D, Bártová E. Acetylation-dependent nuclear arrangement and recruitment of BMI1 protein to UV-damaged chromatin. J Cell Physiol. 2012;227:1838–50. doi: 10.1002/jcp.22912. [DOI] [PubMed] [Google Scholar]
- 23.Moses RE, O’Malley BW. DNA transcription and repair: a confluence. J Biol Chem. 2012;287:23266–70. doi: 10.1074/jbc.R112.377135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19:276–82. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 25.Gilder AS, Do PM, Carrero ZI, Cosman AM, Broome HJ, Velma V, Martinez LA, Hebert MD. Coilin participates in the suppression of RNA polymerase I in response to cisplatin-induced DNA damage. Mol Biol Cell. 2011;22:1070–9. doi: 10.1091/mbc.E10-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16:2395–413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foltánková V, Legartová S, Kozubek S, Hofer M, Bártová E. DNA-damage response in chromatin of ribosomal genes and the surrounding genome. Gene. 2013;522:156–67. doi: 10.1016/j.gene.2013.03.108. [DOI] [PubMed] [Google Scholar]
- 28.Boisvert FM, Cote J, Boulanger MC, Cleroux P, Bachand F, Autexier C, Richard S. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J Cell Biol. 2002;159:957–69. doi: 10.1083/jcb.200207028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boisvert FM, Déry U, Masson JY, Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005;19:671–6. doi: 10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Revet I, Feeney L, Bruguera S, Wilson W, Dong TK, Oh DH, Dankort D, Cleaver JE. Functional relevance of the histone gammaH2Ax in the response to DNA damaging agents. Proc Natl Acad Sci USA. 2011; 108:8663-8667. PMID:21555580; http://dx. doi: 10.1073/pnas.1105866108 [DOI] [PMC free article] [PubMed]
- 31.Bártová E, Galiová G, Krejcí J, Harnicarová A, Strasák L, Kozubek S. Epigenome and chromatin structure in human embryonic stem cells undergoing differentiation. Dev Dyn. 2008;237:3690–702. doi: 10.1002/dvdy.21773. [DOI] [PubMed] [Google Scholar]
- 32.Stixová L, Matula P, Kozubek S, Gombitová A, Cmarko D, Raška I, Bártová E. Trajectories and nuclear arrangement of PML bodies are influenced by A-type lamin deficiency. Biol Cell. 2012;104:418–32. doi: 10.1111/boc.201100053. [DOI] [PubMed] [Google Scholar]
- 33.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008; 132:487-98; PMID: 19101468, http://dx. doi: 10.1016/j.chembiol.2008.10.015 [DOI] [PubMed]
- 34.Bártová E, Šustáčková G, Stixová L, Kozubek S, Legartová S, Foltánková V. Recruitment of Oct4 protein to UV-damaged chromatin in embryonic stem cells. PLoS One 2011; 12:e27281, PMID:22164208; 10.1371/ journal.pone. 0027281. [DOI] [PMC free article] [PubMed]
- 35.Stixová L, Bártová E, Matula P, Daněk O, Legartová S, Kozubek S. Heterogeneity in the kinetics of nuclear proteins and trajectories of substructures associated with heterochromatin. Epigenetics Chromatin. 2011;4:5. doi: 10.1186/1756-8935-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piston DW, Kremers GJ. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–14. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.