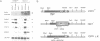Abstract
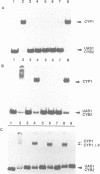
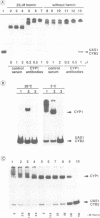
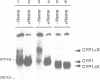
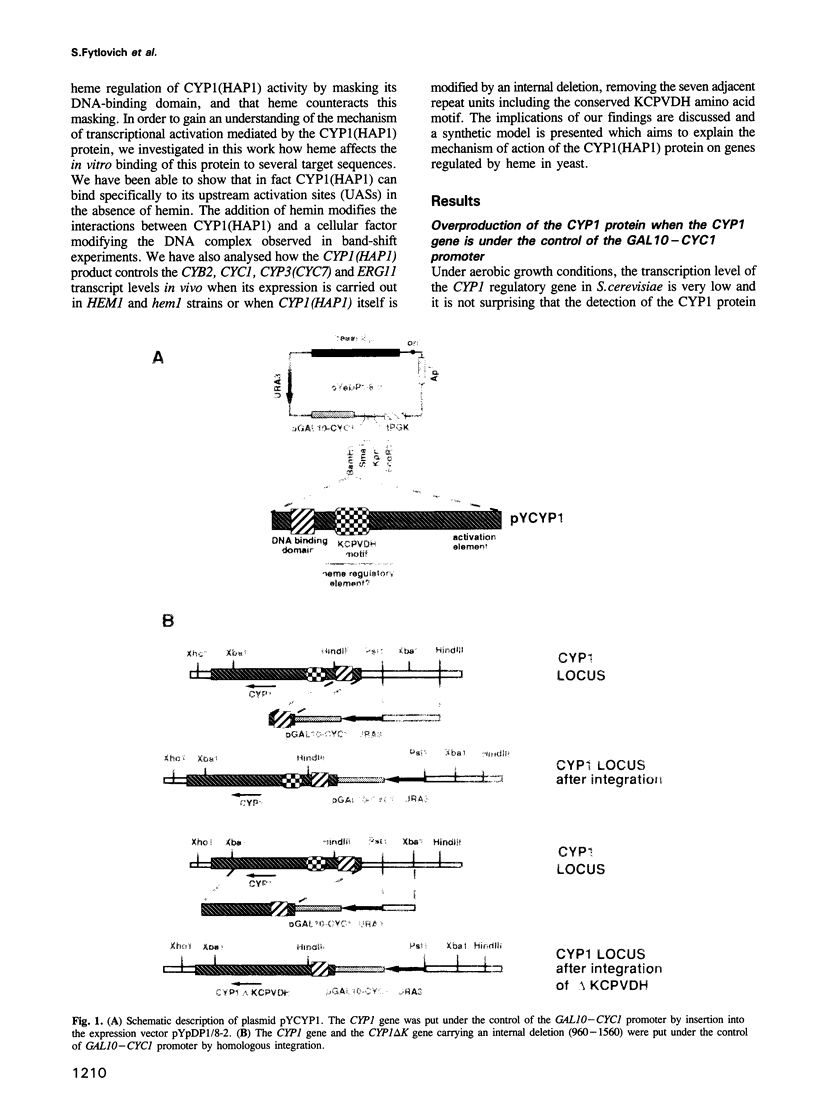
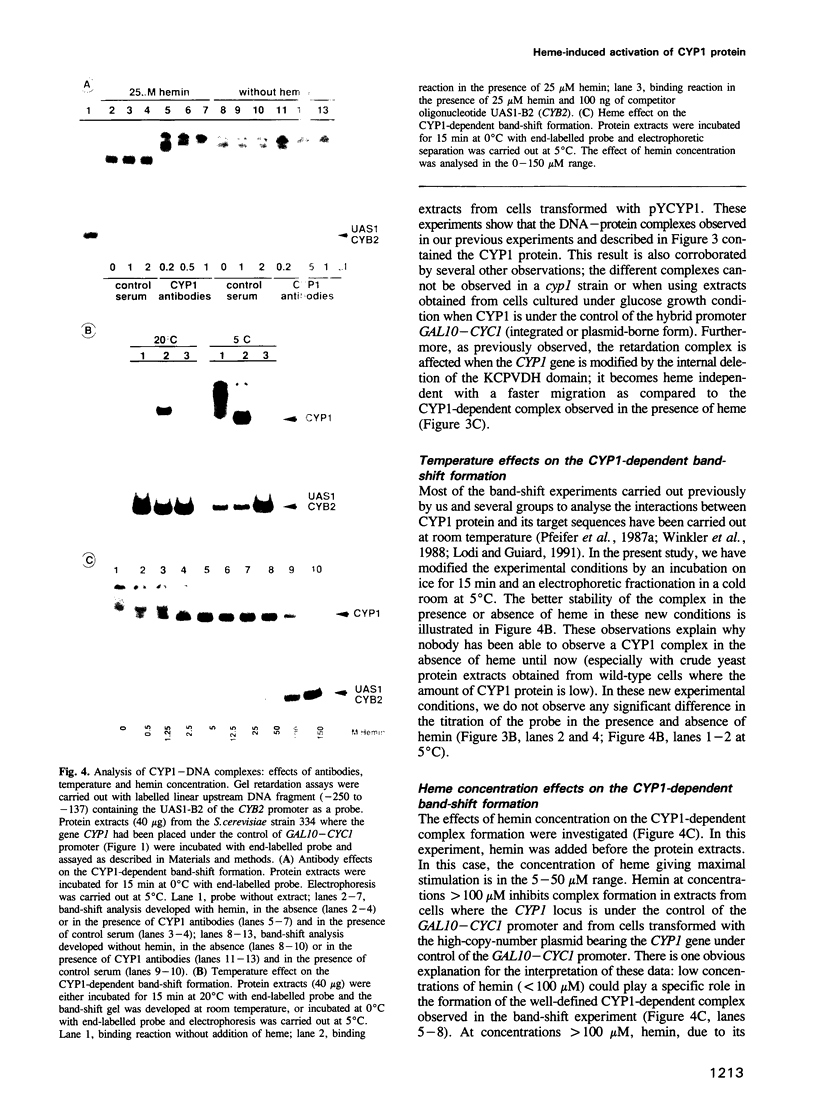
Previously, it was shown that the CYP1(HAP1) gene product mediates the transcription of several oxygen-regulated genes through a metabolic co-effector, heme, in the yeast Saccharomyces cerevisiae. This study investigates the overproduction of the CYP1 protein when the CYP1(HAP1) gene is placed under the control of the GAL10-CYC1 hybrid promoter (either at the locus of the CYP1(HAP1) gene or cloned in a high-copy-number plasmid). In these conditions, the CYP1 protein is detected by Western blot analysis and has a molecular mass in agreement with the open reading frame sequence. Band-shift experiments show that the CYP1(HAP1) protein is able to interact specifically with its target sequences in vitro without addition of hemin, and forms a large complex with one or several unidentified factors denoted as X. Addition of hemin allows the formation of a new complex which has a lower molecular mass. The internal deletion of the seven repeated amino acid sequences containing the KCPVDH motif in the CYP1(HAP1) protein modifies the heme responsiveness phenomenon observed in vitro in the band-shift experiments and in vivo in the transcription of the CYB2, CYC1, CYP3(CYC7) and ERG11 genes. On the basis of these data, we propose a new model for heme-induced activation of the CYP1 protein.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baleja J. D., Marmorstein R., Harrison S. C., Wagner G. Solution structure of the DNA-binding domain of Cd2-GAL4 from S. cerevisiae. Nature. 1992 Apr 2;356(6368):450–453. doi: 10.1038/356450a0. [DOI] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos O., Li G. Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991 Aug-Sep;7(6):609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chasman D. I., Kornberg R. D. GAL4 protein: purification, association with GAL80 protein, and conserved domain structure. Mol Cell Biol. 1990 Jun;10(6):2916–2923. doi: 10.1128/mcb.10.6.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot F., Verdière J., Gaisne M., Slonimski P. P. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J Mol Biol. 1988 Nov 20;204(2):263–276. doi: 10.1016/0022-2836(88)90574-8. [DOI] [PubMed] [Google Scholar]
- Cullin C., Pompon D. Synthesis of functional mouse cytochromes P-450 P1 and chimeric P-450 P3-1 in the yeast Saccharomyces cerevisiae. Gene. 1988 May 30;65(2):203–217. doi: 10.1016/0378-1119(88)90457-x. [DOI] [PubMed] [Google Scholar]
- Drygas M. E., Lambowitz A. M., Nargang F. E. Cloning and analysis of the Neurospora crassa gene for cytochrome c heme lyase. J Biol Chem. 1989 Oct 25;264(30):17897–17906. [PubMed] [Google Scholar]
- Dumont M. E., Cardillo T. S., Hayes M. K., Sherman F. Role of cytochrome c heme lyase in mitochondrial import and accumulation of cytochrome c in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Nov;11(11):5487–5496. doi: 10.1128/mcb.11.11.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M. E., Ernst J. F., Hampsey D. M., Sherman F. Identification and sequence of the gene encoding cytochrome c heme lyase in the yeast Saccharomyces cerevisiae. EMBO J. 1987 Jan;6(1):235–241. doi: 10.1002/j.1460-2075.1987.tb04744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989 Aug;3(8):1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Fraser R. S. Turnover of polyadenylated messenger RNA in fission yeast. Evidence for the control of protein synthesis at the translational level. Eur J Biochem. 1975 Dec 15;60(2):477–486. doi: 10.1111/j.1432-1033.1975.tb21026.x. [DOI] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Guiard B. Structure, expression and regulation of a nuclear gene encoding a mitochondrial protein: the yeast L(+)-lactate cytochrome c oxidoreductase (cytochrome b2). EMBO J. 1985 Dec 1;4(12):3265–3272. doi: 10.1002/j.1460-2075.1985.tb04076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Guarente L. Yeast HAP2 and HAP3: transcriptional activators in a heteromeric complex. Science. 1988 Apr 15;240(4850):317–321. doi: 10.1126/science.2832951. [DOI] [PubMed] [Google Scholar]
- Hennig B., Neupert W. Biogenesis of cytochrome c in Neurospora crassa. Methods Enzymol. 1983;97:261–274. doi: 10.1016/0076-6879(83)97138-0. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Pfeifer K., Powell L., Guarente L. Internal deletions in the yeast transcriptional activator HAP1 have opposite effects at two sequence elements. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4524–4528. doi: 10.1073/pnas.87.12.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P. J., Raine A. R., Gadhavi P. L., Laue E. D. Structure of the DNA-binding domain of zinc GAL4. Nature. 1992 Apr 2;356(6368):448–450. doi: 10.1038/356448a0. [DOI] [PubMed] [Google Scholar]
- Lodi T., Guiard B. Complex transcriptional regulation of the Saccharomyces cerevisiae CYB2 gene encoding cytochrome b2: CYP1(HAP1) activator binds to the CYB2 upstream activation site UAS1-B2. Mol Cell Biol. 1991 Jul;11(7):3762–3772. doi: 10.1128/mcb.11.7.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry C. V., Cerdán M. E., Zitomer R. S. A hypoxic consensus operator and a constitutive activation region regulate the ANB1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1990 Nov;10(11):5921–5926. doi: 10.1128/mcb.10.11.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue N. F., Chasman D. I., Buchman A. R., Kornberg R. D. Interaction of GAL4 and GAL80 gene regulatory proteins in vitro. Mol Cell Biol. 1987 Oct;7(10):3446–3451. doi: 10.1128/mcb.7.10.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccecchini M. L., Rudin Y., Blobel G., Schatz G. Import of proteins into mitochondria: precursor forms of the extramitochondrially made F1-ATPase subunits in yeast. Proc Natl Acad Sci U S A. 1979 Jan;76(1):343–347. doi: 10.1073/pnas.76.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Marmorstein R., Carey M., Ptashne M., Harrison S. C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Nicholson D. W., Hergersberg C., Neupert W. Role of cytochrome c heme lyase in the import of cytochrome c into mitochondria. J Biol Chem. 1988 Dec 15;263(35):19034–19042. [PubMed] [Google Scholar]
- Nogi Y., Shimada H., Matsuzaki Y., Hashimoto H., Fukasawa T. Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae. II. The isolation and dosage effect of the regulatory gene GAL80. Mol Gen Genet. 1984;195(1-2):29–34. doi: 10.1007/BF00332719. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Arcangioli B., Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987 Apr 10;49(1):9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Prezant T., Pfeifer K., Guarente L. Organization of the regulatory region of the yeast CYC7 gene: multiple factors are involved in regulation. Mol Cell Biol. 1987 Sep;7(9):3252–3259. doi: 10.1128/mcb.7.9.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. C., Guarente L. Regulation of the yeast CYT1 gene encoding cytochrome c1 by HAP1 and HAP2/3/4. Mol Cell Biol. 1991 Oct;11(10):4934–4942. doi: 10.1128/mcb.11.10.4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness M., Schafer W., D'Ari L., Rine J. Positive and negative transcriptional control by heme of genes encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5702–5712. doi: 10.1128/mcb.9.12.5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turi T. G., Loper J. C. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14 alpha-demethylase (ERG11). J Biol Chem. 1992 Jan 25;267(3):2046–2056. [PubMed] [Google Scholar]
- Verdière J., Creusot F., Guérineau M. Regulation of the expression of iso 2-cytochrome c gene in S. cerevisiae: cloning of the positive regulatory gene CYP1 and identification of the region of its target sequence on the structural gene CYP3. Mol Gen Genet. 1985;199(3):524–533. doi: 10.1007/BF00330769. [DOI] [PubMed] [Google Scholar]
- Verdière J., Gaisne M., Guiard B., Defranoux N., Slonimski P. P. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. II. Missense mutation suggests alternative Zn fingers as discriminating agents of gene control. J Mol Biol. 1988 Nov 20;204(2):277–282. doi: 10.1016/0022-2836(88)90575-x. [DOI] [PubMed] [Google Scholar]
- Verdière J., Gaisne M., Labbe-Bois R. CYP1 (HAP1) is a determinant effector of alternative expression of heme-dependent transcribed genes in yeast [corrected]. Mol Gen Genet. 1991 Aug;228(1-2):300–306. doi: 10.1007/BF00282480. [DOI] [PubMed] [Google Scholar]
- Winkler H., Adam G., Mattes E., Schanz M., Hartig A., Ruis H. Co-ordinate control of synthesis of mitochondrial and non-mitochondrial hemoproteins: a binding site for the HAP1 (CYP1) protein in the UAS region of the yeast catalase T gene (CTT1). EMBO J. 1988 Jun;7(6):1799–1804. doi: 10.1002/j.1460-2075.1988.tb03011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zitomer R. S., Lowry C. V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992 Mar;56(1):1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]