Abstract
In the model cyanobacterium Anabaena sp. PCC 7120, cells called heterocysts that are specialized in the fixation of atmospheric nitrogen differentiate from vegetative cells of the filament in the absence of combined nitrogen. Heterocysts follow a specific distribution pattern along the filament, and a number of regulators have been identified that influence the heterocyst pattern. PatS and HetN, expressed in the differentiating cells, inhibit the differentiation of neighboring cells. At least PatS appears to be processed and transferred from cell to cell. HetC is similar to ABC exporters and is required for differentiation. We present an epistasis analysis of these regulatory genes and of genes, hetP and asr2819, successively downstream from hetC, and we have studied the localization of HetC and HetP by use of GFP fusions. Inactivation of patS, but not of hetN, allowed differentiation to proceed in a hetC background, whereas inactivation of hetC in patS or patS hetN backgrounds decreased the frequency of contiguous proheterocysts. A HetC-GFP protein is localized to the heterocysts and especially near their cell poles, and a putative HetC peptidase domain was required for heterocyst differentiation but not for HetC-GFP localization. hetP is also required for heterocyst differentiation. A HetP-GFP protein localized mostly near the heterocyst poles. ORF asr2819, which we denote patC, encodes an 84-residue peptide and is induced upon nitrogen step-down. Inactivation of patC led to a late spreading of the heterocyst pattern. Whereas HetC and HetP appear to have linked functions that allow heterocyst differentiation to progress, PatC may have a role in selecting sites of differentiation, suggesting that these closely positioned genes may be functionally related.
Introduction
In response to deprivation of combined nitrogen, some filamentous cyanobacteria produce cells called heterocysts that are specialized in the fixation of N2 [1]. Heterocyst differentiation involves drastic changes in gene expression that are coordinated by two DNA-binding factors, the global regulator NtcA and the development-specific factor HetR [2]. The distribution of heterocysts in the diazotrophic filaments of cyanobacteria represents a simple and old example of developmental patterns in the living world. In strains of the genera Anabaena and Nostoc the pattern consists of long linear chains of cells with heterocysts separated by ca. 10 vegetative cells. Several gene products that influence the pattern of heterocyst distribution have been identified [2]. In Anabaena sp. strain PCC 7120 (hereafter Anabaena) the patS gene is expressed early in the differentiation process, specifically in the differentiating cells, and inhibits the differentiation of neighboring cells [3], [4]. Inactivation of patS produces a Mch (Multiple contiguous heterocysts) phenotype whereas overexpression of patS abolishes differentiation. The primary product of patS is a 17-amino acid peptide [5]. The 9-amino acid N-terminal stretch of PatS appears to be involved in processing the peptide, which is needed for immunity against PatS in the differentiating cells in which the peptide is produced. Processing of PatS would render a C-terminal peptide, likely of 8 amino acids, that acting as a morphogen is transferred to the neighboring vegetative cells [5]. PatS appears to interact with HetR and regulate its activity [6], [7], [8], but the pathway of intercellular transfer of PatS or a peptide derivative of PatS is unknown.
The hetN gene product, which exhibits similarity to short chain dehydrogenases, also affects the pattern of distribution of heterocysts in the filament [9], [10]. hetN is expressed as a monocistronic transcript starting ca. 6–12 h after N (nitrogen) step-down [10]. Contrasting results have been reported when hetN was inactivated by insertion of different constructs (or when hetN was over-expressed). A ΔhetN strain has recently been reported to yield increased heterocyst frequency and Mch 48 h after N step-down [11]. Finally, inactivation of patS together with under-expression of hetN produced massive heterocyst differentiation in the filaments of Anabaena [12]. Aside from the speculation that a RGSGR peptide resulting from hydrolysis of HetN in the cytoplasm would likely be transferred to neighboring cells directly through inter-cytoplasmic connections [11], neither the identity nor the mechanism of action of the actual HetN-derived signaling molecule is known.
The hetC gene product exhibits extensive similarity to ABC transporters, especially to those in the HlyB family of bacterial protein exporters [13], [14]. hetC is induced early during heterocyst differentiation and is regulated by NtcA and HetR [13], [15], and certain of its mutants do not form heterocysts [13]. However, after prolonged incubation in the absence of combined nitrogen, hetC mutants exhibit a pattern of weakly-fluorescent cells, a characteristic of heterocysts [13], but, in contrast to heterocysts, they can divide producing a pattern of spaced series of small cells [16]. Because heterocysts are terminal, non-dividing cells, this observation led to the proposal that HetC is involved in the transition to non-dividing cells during heterocyst differentiation [16].
Genes hetP and asr2819 are located downstream from hetC in the genome of Anabaena. The product of hetP bears no similarity to proteins of known function, and inactivation of hetP blocks heterocyst differentiation whereas its over-expression from a plasmid leads to over-differentiation [17], [18]. Because ectopic expression of hetP (from PpetE in a plasmid) in a hetR mutant leads to the formation of heterocyst-like cells, which however do not fix N2 aerobically, it has been proposed that HetP partially bypasses the requirement for HetR acting downstream from it during heterocyst differentiation [18]. Also, because in a ΔhetP mutant the pattern of expression of gfp transcriptional fusions to patS or hetR appeared similar to those in the wild type background, hetP was suggested to function downstream of pattern formation during heterocyst differentiation [18].
In this work, we have addressed the relationships of HetC with possible partners regulating heterocyst differentiation. We have investigated whether HetC may be involved in the export of portions of PatS and HetN from differentiating cells and whether the predicted peptidase domain of HetC may be involved in processing of PatS. Additionally, we have investigated possible relationships of HetC with the products of the heterocyst differentiation gene hetP and the previously unstudied gene asr2819, which are genomic neighbors of hetC. The study has also included analyses of the subcellular localization of some of these factors.
Materials and Methods
Strains and growth conditions
Anabaena sp. PCC 7120 and derivatives were grown photoautotrophically in a BG110-based medium supplemented with NH4Cl as described [19]. For bubbled cultures, the medium was supplemented with NaHCO3 and sparged with a mixture of air/CO2 (99∶1). Antibiotics, when required, were added to the media at final concentrations of: neomycin (Nm) 10 µg·mL−1, and each of streptomycin and spectinomycin 2 µg·mL−1 for liquid and 5 µg·mL−1 for solid cultures. For growth tests in solid medium, filaments grown in liquid BG110 medium supplemented with NH4 + were washed 3 times with BG110. Amounts of filaments corresponding to 5, 2.5, 1.25, 0.625 and 0.3125 ng Chl were spotted atop Petri dishes containing BG110 medium supplemented or not with NH4 +, without antibiotics, and the plates were incubated under standard conditions for 14 days.
Construction of mutants
To replace the hetC gene (alr2817) with the Smr Spr gene cassette C.S3, which includes transcriptional terminators at both ends [20], two DNA fragments, one encompassing sequences 5′ of the gene and the other including sequences 3′ of the gene, were amplified by PCR using DNA from Anabaena as template and the primer pairs alr2817-34/alr2817-35 and alr2817-36/alr2817-37, respectively, with the alr2817-35 primer complementing the alr2817-36 primer (all oligodeoxynucleotide primers are listed in Table 1). The upstream and downstream DNA fragments were cloned in plasmid pMBL-T (Dominion MBL, Spain; the plasmids used or constructed in this work are described in Table 2) and sequenced, and the C.S3 gene cassette was inserted in the EcoRV site generated at the junction of the two cloned DNA fragments. The resulting construct was inserted into plasmid pRL278 [21] (see Table 2). The resulting plasmid, pCSL20, bears Anabaena DNA from the hetC locus with 3051 bp of DNA within hetC substituted by gene cassette C.S3.
Table 1. Oligodeoxynucleotide primers used in this work.
| Name | Sequence 5′-3′ |
| alr2817-13 | GTTTCAAGAACGCAACG |
| alr2817-14 | GATATCAGCTAAGTGGTGATAAAG |
| alr2817-34 | TCTCTCTTGGGTGGGATTCT |
| alr2817-35 | ATCGCCATTAGTTCCGATATCTTTGCAGCCCTCCCT |
| alr2817-36 | AGGGAGGGCTGCAAAGATATCGGAACTAATGGCGAT |
| alr2817-37 | CTCGCCTACTGTGCATTTGAGACT |
| alr2817-38 | TGGTATCGGCTGAACAAA |
| alr2817-41 | TGTCGCCACCCCAGTCATAAT |
| alr2817-42 | CTTGAGGGCTTGAAAACTTTCTGGGTAGGAGTAGGACGA |
| alr2817-43 | TCGTCCTACTCCTACCCAGAAAGTTTTCAAGCCCTCAAG |
| alr2818-13 | CATGCTGAGCTCTGCTGGGTACAATCCTATGC |
| alr2818-8 | AAAGTAAAGCTTACAAGCCAAGCATGAAAGTGGT |
| alr2818-9 | CCGAAGGGTCTCACGCCATTATGAATAAAATC |
| alr2818-16 | TAAATACTCTTGGGCTTGTGTTCT |
| alr2818-17 | CTAGCTTCGGAGTTTTCTTTGAGT |
| alr2818-19 | AAAGTAGGTACCACAAGCCAAGCATGAAAGTGGT |
| alr2820-1 | CTTCTTGAGCTCTTACAGGAGAAGCCAGTCCAGGTT |
| alr5358-1 | TTATACACCTTGCGTCCCTTCCTC |
| alr5358-2 | CTCAACAGCTACATAGCGTGAAGCGCCGGT |
| alr5358-3 | ACCGGCGCTTCACGCTATGTAGCTGTTGAG |
| alr5358-4 | TGAAGTTCATCTCTGGCGCATTCC |
| asr2819-1 | CTAAATCCCCTTTACGATATCATTCGAGAAGTTGCA |
| asr2819-2 | TGCAACTTCTCGAATGATATCGTAAAGGGGATTTAG |
| asr2819-11 | TTATTCCCGTCACTTACCACCATC |
| asr2819-12 | TTCTAACTCAAGCACCACAAACTC |
| gfp-11 | TCTGGTACCTTATTTGTATAGTTC |
| gfp-13 | TTCTCCTTTGCTAGCACCTCCA |
| gfp-14 | TGGAGGTGCTAGCAAAGGAGAA |
| rnpB-4 | ACTCTTGGTAAGGGTGCAAAGGTG |
| rnpB-5 | AACCATAGTTCCTTCGGCCTTGCT |
Table 2. Plasmids used in this work.
| Name | Description | Strain generated | Reference |
| pCSAL39 | sf-gfp with N-terminal 4Gly linker-encoding sequence and BsaI site in a modified pMBL-T (Ap resistance substituted by C.K1 cassette) | - | A. López-Lozano and A. Herrero (unpublished) |
| pCSAM135 | Contains a translational fusion between the 5′-terminal region of sepJ and the gfp gene | Flores et al. 2007 | |
| pCSL18 | PCR product obtained with primers alr2817-34, alr2817-35, alr2817-36 and alr2817-37 (overlapping PCR), cloned in pMBL-T | - | This work |
| pCSL19 | C.S3 inserted in EcoRV between the two DNA fragments of pCSL18 | - | This work |
| pCSL20 | pCSL19 digested with BamHI and XhoI, and cloned in pRL278 | CSL3 | This work |
| pCSL22 | PCR product obtained with primers alr5358-1, alr5358-2, alr5358-3 and alr5358-4 (overlapping PCR), cloned in pMBL-T | - | This work |
| pCSL23 | pCSL22 digested with BamHI and XhoI and cloned in pRL278 | CSL7 | This work |
| pCSL24 | PCR product obtained with primers alr2817-38, alr2817-42, alr2817-43 and alr2817-41 (overlapping PCR), cloned in pMBL-T | - | This work |
| pCSL25 | pCSL24 digested with SpeI and XbaI and cloned in pCSRO (XbaI) | CSL16 | This work |
| pCSL68 | pCSV3-derived plasmid containing the mut2-gfp gene | - | This work |
| pCSL69 | PCR product obtained with primers alr2818-8 and alr2818-9, cloned in pCSAL39 digested with BsaI and HindIII | - | This work |
| pCSL70 | pCSL69 digested with KpnI and cloned in pCSV3 digested with the same enzyme | CSL67 | This work |
| pCSL111 | PCR product obtained with primers asr2818-13, asr2819-1, asr2819-2 and alr2820-1 (overlapping PCR), cloned in pCSRO digested with SacI | CSL97 | This work |
| pCSL123 | PCR product obtained with primers alr2818-19, GFP-13, GFP-14 and GFP-11 (overlapping PCR), cloned in pCSV3 digested with KpnI | CSL107 | This work |
| pCSM6 | hetC PCR product obtained with primers alr2817-13 and alr2817-14 fused to mut2-gfp through an EcoRV site, cloned in PstI-digested pCSV3 | CSM1 | This work |
| pCSRO | Plasmid derived from pRL278, with KmR gene substituted by C.S3 | - | Merino-Puerto et al. (2010) |
| pCSV3 | Plasmid derived from pRL500, with ApR gene substituted by C.S3 | - | Olmedo-Verd et al. (2006) |
| pMBL-T | Commercial vector for cloning purposes | - | Dominion MBL |
| pRL278 | Vector used for the positive selection of double recombinants in Anabaena | - | Black et al. (1993) |
Template Anabaena DNA and overlapping primer pairs alr2817-38/alr2817-42 and alr2817-43/alr2817-41 were used to delete, specifically, the predicted peptidase domain of hetC, and the PCR product was cloned in plasmid pMBL-T, sequenced and transferred to pCSRO [22]. The plasmid produced, pCSL25, bears DNA from the hetC locus with a deletion of 378 bp from inside hetC.
To delete the hetN gene, template Anabaena DNA and overlapping PCR primer pairs alr5358-1/alr5358-2 and alr5358-3/alr5358-4 were used for amplification. The resulting DNA fragments were cloned in pMBL-T and sequenced, and then transferred to pRL278. The resulting plasmid was named pCSL23.
For deletion of ORF asr2819, template Anabaena DNA and overlapping primer pairs alr2818-13/asr2819-1 and asr2819-2/alr2820-1 were used for amplification. The PCR product was cloned in pCSRO, and sequenced, producing plasmid pCSL111.
Plasmids pCSL20, pCSL23, pCSL25, and pCSL111 were transferred to Anabaena by conjugation, performed as described [23]. Exconjugants were selected by their resistance to Sm and Sp, or to Nm, and double recombinants were then selected by their resistance to sucrose (sucrose sensitivity is conferred by the sacB gene present in the vector portion of pRL278 or pCSRO), and the chromosome structure in the altered region was tested by PCR.
For construction of a fusion of hetC to the gfp-mut2 gene, a DNA fragment was amplified using Anabaena DNA as template and primer pair alr2817-13/alr2817-14. The PCR product was cloned in pMBL-T, and afterwards in pCSAM135, which bears the gfp-mut2 sequence [24], and the resulting hetC-gfp-mut2 construct was transferred to the mobilizable vector pCSV3 [25] to produce plasmid pCSM6. For construction of a hetP-sf-gfp fusion, the alr2818-8/alr2818-9 primer pair was used, and the resulting PCR product was cloned in plasmid pCSAL39 (which includes a sequence encoding a four-Gly linker preceded by a BsaI site just before the sf-gfp sequence). The resulting construct was cloned in pCSV3 producing plasmid pCSL70. For construction of a hetP-gfp-mut2 fusion, two DNA fragments were amplified by PCR. One contained the hetP gene and was amplified from plasmid pCSL70 with primer pair alr2818-19/gfp-13. The other contained the gfp-mut2 gene and was amplified from plasmid pCSL68 with the primer pair gfp-14/gfp-11. The fragments produced were joined by overlapping PCR, and the product was cloned in pCSV3 rendering plasmid pCSL123.
Plasmids pCSM6, pCSL70, and pCSL123 were transferred to Anabaena sp. by conjugation. Clones resistant to Sm and Sp, which had integrated the plasmid in the resident genomic locus by a single recombination event, were selected and their genomic structure was tested by PCR.
Northern blot analysis, qRT-PCR, Hgl determination and nitrogenase activity
Isolation of total RNA from different strains of Anabaena and northern blot analysis was performed as described [26]. Retrotranscription was performed with a QuantiTect Reverse Transcription kit (Qiagen) and the degenerated oligonucleotides “Random hexamer primers” (Bioline). Real-time PCR was performed in an iCycler iQ (BioRad) with the SensiFAST SYBR & Fluorescein kit using 1/10 volumes of the cDNA resulting from the retrotranscription step and primer pairs alr2818-16/alr2818-17, asr2819-11/asr2819-12, and rnpB-4/rnpB-5. Gene-expression ratios and statistical parameters were calculated with the REST 2009 software (http://rest-2009.gene-quantification.info).
Lipid analysis was performed as in [26]. Nitrogenase activity was determined under oxic and anoxic conditions in filaments incubated for 24 or 48 h in bubbled cultures without combined nitrogen and without antibiotics [26].
Microscopy
For light microscopy, filaments grown in BG110 + ammonium medium (in the presence of antibiotics when appropriate) were harvested, washed with nitrogen-free (BG110) medium and incubated for 24 h in bubbled cultures at 30°C in the light. At least 300 cells or 100 intervals were counted for each strain in each of two to six independent experiments. Dividing cells were counted as two cells. Cell suspensions were mixed (1∶10) with a 1% Alcian Blue (Sigma) solution [5]. Because Alcian Blue stains the polysaccharide (Hep) layer of the heterocyst envelope, the stained cells, to which we will refer collectively as (pro)heterocysts, comprise mature heterocysts and proheterocysts. In Anabaena, a proheterocyst is an intermediate between a vegetative cell and a heterocyst, which differs in shape and granularity from vegetative cells, and a mature heterocyst is a cell capable of aerobic fixation of N2 (see [27]).
Microscopy was performed with a confocal microscope as in [5], treating the images with the LAS AF Software (Leica), or with a Leica DM6000B fluorescence microscope and an ORCA-ER camera (Hamamatsu) using an FITC L5 filter (excitation, band-pass [BP] 480/40 filter; emission, BP 527/30 filter). BlindDeblur deconvolution of 3D images was made with the LAS AF Leica software.
Results
Deletion of hetC in patS and hetN backgrounds
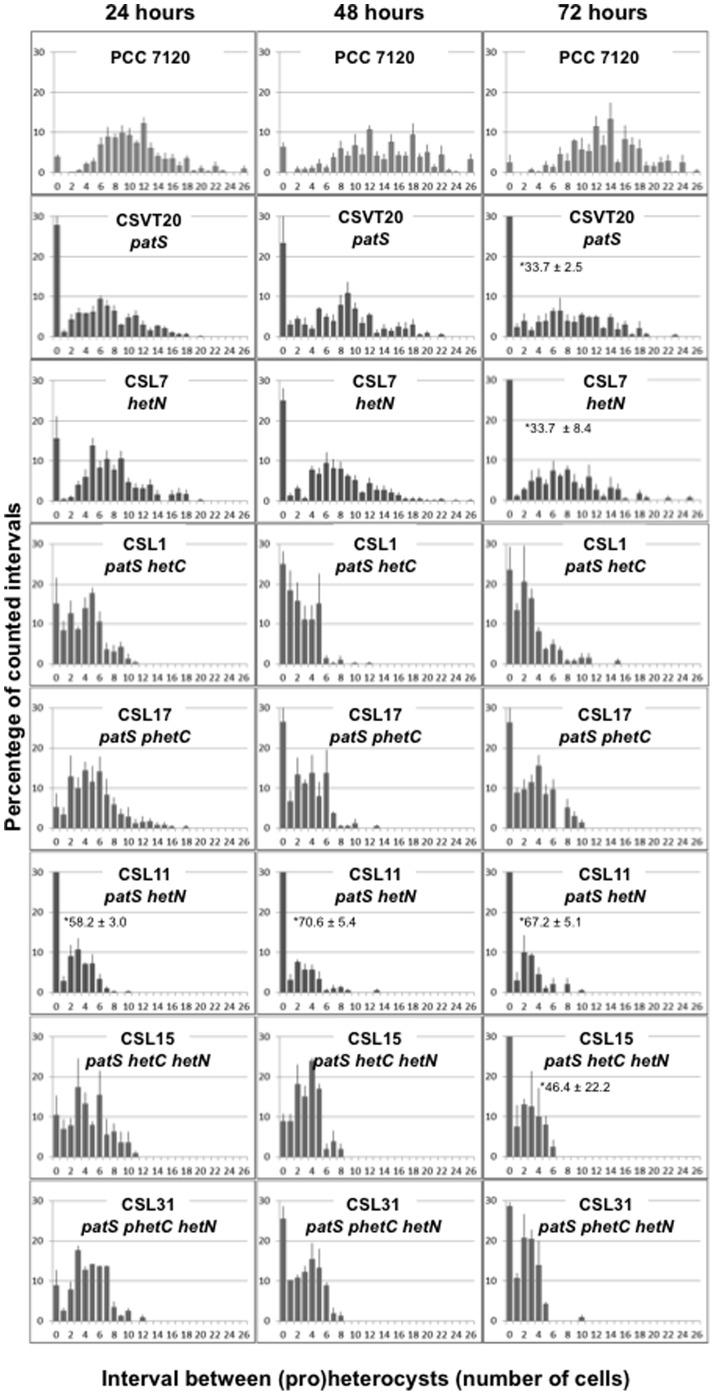
To test a possible functional relationship between the HetC protein required for heterocyst differentiation and the differentiation negative regulatory factors PatS and HetN, we generated mutant strains of Anabaena lacking one, two or the three of these factors (see schemes of the genomic regions in Fig. 1). Strain CSVT20 (ΔpatS) has been described previously [5]. The mutants were analyzed in terms of frequency and distribution of cells stained with Alcian Blue and the presence or absence of heterocyst-specific glycolipids (Hgl). The data are presented in Table 3 and Figures 2 and 3.
Figure 1. Schematics of the hetC, hetN and patS genomic regions in Anabaena sp. strain PCC 7120 and mutant derivatives.
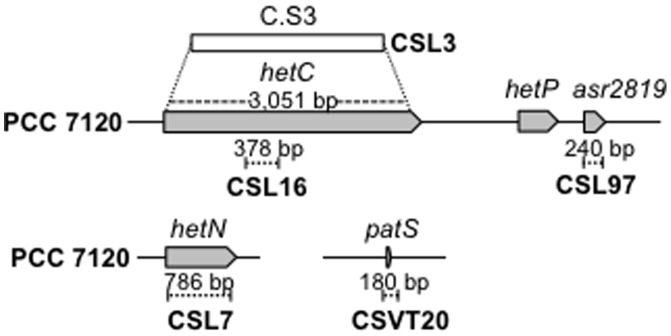
The gene map is from [38]. The Anabaena genes are represented with grey arrows, the deleted portions (of the specified sizes) with dashed segments, and C.S3 gene-cassette insertion with a white bar. The names of the resulting mutant strains in the wild-type genetic background are indicated.
Table 3. Spatial pattern of heterocysts in Anabaena sp. PCC 7120 mutant strains.
| Strain | Genotype | Time (h) | Percentage heterocysts | Contiguous heterocysts | Mean interval |
| PCC 7120 | 24 | 10.4±0.4 | 3.9±0.6 | 10.5±0.7 | |
| 48 | 8.6±0.4 | 6.4±1.0 | 13.1±0.8 | ||
| 72 | 7.5±0.7 | 2.5±1.9 | 13.3±0.8 | ||
| CSL3 | hetC | 24 | 0.03±0.02 | - | - |
| 48 | 0.00±0.0 | - | - | ||
| 72 | 0.00±0.0 | - | - | ||
| CSL16 | hetC-p | 24 | 1.1±0.7 | - | - |
| 48 | 2.8±1.6 | - | - | ||
| 72 | 1.4±1.0 | - | - | ||
| CSVT20 | patS | 24 | 18.6±2.2 | 27.8±8.0 | 5.5±0.7 |
| 48 | 17.2±1.0 | 23.4±6.7 | 6.8±0.4 | ||
| 72 | 17.6±1.0 | 33.7±2.5 | 6.0±0.6 | ||
| CSL7 | hetN | 24 | 14.3±1.0 | 15.7±5.5 | 6.8±0.1 |
| 48 | 13.9±0.6 | 25.0±3.1 | 6.2±0.2 | ||
| 72 | 17.7±1.0 | 33.7±8.4 | 5.6±1.2 | ||
| CSL1 | patS hetC | 24 | 16.4±1.5 | 15.1±6.4 | 3.8±0.4 |
| 48 | 29.4±1.8 | 25.0±3.2 | 2.3±0.2 | ||
| 72 | 33.0±2.7 | 23.7±5.7 | 2.7±0.2 | ||
| CSL17 | patS hetC-p | 24 | 18.5±3.4 | 5.4±3.4 | 5.2±0.7 |
| 48 | 25.2±2.6 | 26.6±6.0 | 3.0±0.5 | ||
| 72 | 24.7±3.1 | 26.5±4.0 | 3.1±0.2 | ||
| CSL11 | patS hetN | 24 | 42.7±4.8 | 58.2±3.0 | 1.5±0.2 |
| 48 | 62.5±10.0 | 70.6±5.4 | 1.1±0.3 | ||
| 72 | 65.0±6.7 | 67.2±5.1 | 1.1±0.02 | ||
| CSL12 | hetC hetN | 24 | 2.5±0.8 | - | - |
| 48 | 2.1±0.8 | - | - | ||
| 72 | 1.8±0.8 | - | - | ||
| CSL30 | hetC-p hetN | 24 | 2.1±1.3 | - | - |
| 48 | 1.2±0.6 | - | - | ||
| 72 | 0.8±0.6 | - | - | ||
| CSL15 | patS hetC hetN | 24 | 28.8±2.0 | 10.5±4.8 | 4.3±1.0 |
| 48 | 36.5±0.4 | 8.9±1.9 | 3.3±0.1 | ||
| 72 | 57.3±12.7 | 46.4±22.2 | 1.7±0.6 | ||
| CSL31 | patS hetC-p hetN | 24 | 20.2±1.1 | 8.9±3.8 | 4.5±0.3 |
| 48 | 34.8±7.8 | 25.5±3.2 | 2.8±0.1 | ||
| 72 | 43.5±6.2 | 28.6±1.0 | 2.0±0.03 | ||
| CSL101 | patS hetC-p (patS8) | 24 | 1.6±0.7 | - | - |
| CSL102 | patS hetC-p (patS17) | 24 | 1.8±0.3 | - | - |
| CSL67 | hetP-sfgfp | 24 | 11.5 ±0.8 | 9.1±2.5 | 8.6±0.6 |
| 48 | 11.4±0.6 | 20.5±0.4 | 9.8±0.4 | ||
| 72 | 11.0±0.6 | 14.7±1.2 | 11.1±0.9 | ||
| CSL68 | hetP-sfgfp hetC-p | 24 | 12.1±1.8 | 13.4±2.4 | 7.7±0.4 |
| 48 | 11.5±1.2 | 13.7±5.6 | 9.7±0.3 | ||
| 72 | 14.7±1.8 | 19.4±3.9 | 10.4±0.6 | ||
| CSL69 | hetP-sfgfp patS | 24 | 25.5 ±6.3 | 26.3±6.9 | 5.7±1.2 |
| 48 | 27.0±9.6 | 19.8±0.8 | 7.5±0.2 | ||
| 72 | 14.9±2.7 | 21.2±3.1 | 8.6±1.1 | ||
| CSL70 | hetP-sfgfp hetN | 24 | 17.6±1.3 | 29.2±1.0 | 4.8±0.3 |
| 4872 | 22.9±3.116.1±0.2 | 47.3±0.0144.4±4.8 | 4.4±0.15.2±0.1 | ||
| CSL97 | asr2819 | 24 | 13.8±2.6 | 1.8±0.9 | 8.2±0.6 |
| 48 | 12.5±1.1 | 4.9±0.9 | 8.2±0.7 | ||
| 72 | 15.8±0.4 | 5.5±1.3 | 10.5±1.1 | ||
| CSL98 | asr2819 hetC-p | 24 | 1.2±0.1 | - | - |
| 48 | 3.0±1.1 | - | - | ||
| 72 | 1.1±0.4 | - | - |
Filaments were grown with ammonium and incubated for the indicated times in the absence of combined nitrogen. Heterocyst frequency (as percentage of total cells), the mean size of vegetative cell intervals between heterocysts, and the percentage of contiguous heterocysts (interval size = 0, percentage of total intervals) were calculated (for the strains included in Figs. 2, 6 and 7 values are from the data in the figures).
Figure 2. Heterocyst distribution in Anabaena mutant strains.
Filaments from bubbled, ammonium-supplemented cultures of the indicated strains were washed three times with BG110 medium, resuspended in BG110 and incubated under the same culture conditions for 24, 48 or 72 h (as indicated). Cells were counted after staining with Alcian Blue. Data are the mean and standard deviation of the mean of two to six independent experiments (see Materials and Methods).
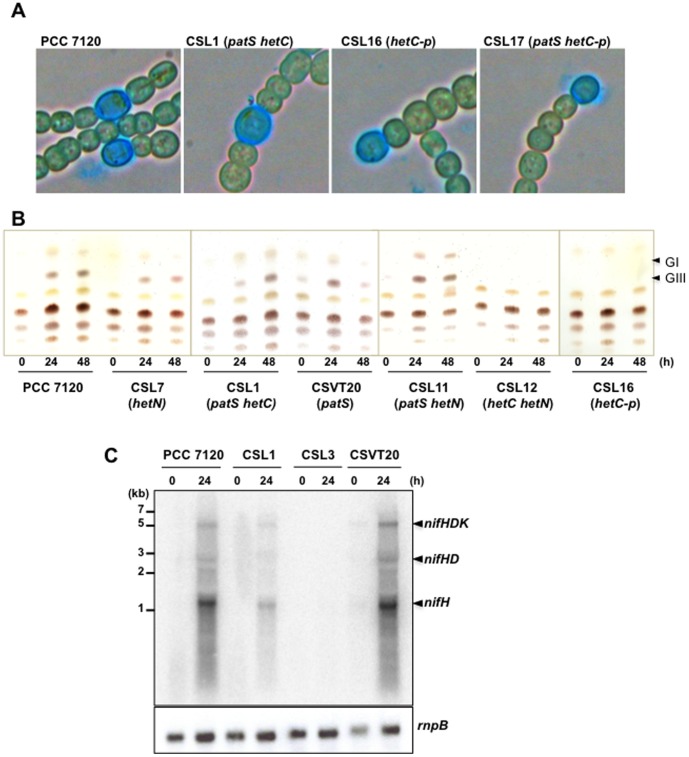
Figure 3. Microscopy, lipids and nifHDK expression in Anabaena mutant strains.
Filaments from bubbled, ammonium-supplemented cultures of the indicated strains were washed three times with BG110 medium, resuspended in BG110 medium and incubated under the same culture conditions for the times indicated in h. (A) Samples taken at 24 h were stained with Alcian Blue prior to being photographed under a light microscope. (B) Lipids (GI and GIII are heterocyst envelope glycolipids) were isolated and separated by TLC. (C) Total RNA was isolated and used in northern blot analysis with a probe of the nifH gene or, as a loading and transfer control, the rnpB gene. A size standard is indicated at the left and the observed transcripts at the right.
As is the case with other previously described mutants of hetC, strain CSL3 (a hetC single mutant) did not produce (pro)heterocysts. As previously described, the patS single mutant, strain CSVT20, shows a Mch phenotype with more heterocysts and shorter vegetative-cell intervals between heterocysts than in the wild-type strain [5]. However, in contrast to previous reports describing that the Mch phenotype of a patS mutant was alleviated at 72 h after N step-down [4], strain CSVT20 maintained the increased frequency of single and contiguous heterocysts at this time (Table 3, Fig. 2). In the patS hetC double mutant, strain CSL1 (see Fig. 3A), (pro)heterocyst frequency was similar to that of the patS single mutant at 24 h, and somewhat higher at 48 (1.67-fold) and 72 (1.87-fold) h after N step-down. In strain CSL1, the mean interval size of vegetative cells between (pro)heterocysts was smaller than in the patS single mutant, and the percentage of contiguous (pro)heterocysts (interval size = 0) was lower (at 24 and 72 h) than in CSVT20. Indeed, aside from the contiguous (pro)heterocysts, the distribution of interval sizes is notably different in strains CSL1 and CSVT20, with more tendency to short intervals different from zero in the former (Fig. 2). Finally, Hgls, which are not detectable in the hetC mutant (not shown), were present in strain CSL1 as well as in CSVT20 (Fig. 3B). Therefore, the requirement for a functional HetC product can be overridden by mutation of patS at least up to the stage of the formation of proheterocysts with Hep and Hgl layers.
Strain CSL7 (hetN) showed more heterocysts, shorter intervals and more contiguous heterocysts than the wild type at all time-points, with the percentage of contiguous heterocysts increasing during incubation in the absence of combined nitrogen. These observations differ from those in a previous report of the effects of down-regulation of hetN, expressed from the heterologous PpetE gene promoter, which led to Mch at 48 h but not earlier [28]. The differences between our results and those reported for other patS and hetN mutants could be due to differences in the genetic structure of the compared strains. Strain CSL12 (hetN hetC) was also able to form (pro)heterocysts, although at frequencies considerably lower than in the hetN single mutant or the wild type (Table 3), and, like the hetC single mutant, lacked Hgl (Fig. 3B). Thus, deletion of hetN does not compensate for the lack of HetC, although it provides a capacity to proceed slightly with differentiation that is not observed in the hetC single mutant.
Finally, we generated strain CSL11, in which patS and hetN were deleted (in the patS hetN double mutant described previously [12], hetN was conditionally down-regulated), and strain CSL15, a triple hetC patS hetN deletion mutant. In strain CSL11, the frequencies of (pro)heterocysts and contiguous heterocysts were considerably higher, and the mean interval shorter, than in the patS or hetN single mutants at all time points. As might be expected, like the patS and hetN single mutants, strain CSL11 produced Hgl (Fig. 3B). In strain CSL15, the percentage of (pro)heterocysts was considerably higher than in the hetC patS double mutant (strain CSL1), reaching ca. 57% of the total cells after 72 h in the absence of combined nitrogen. In CSL15 the mean vegetative cell interval between heterocysts was shorter than in CSL1 (0.6 times) at 72 h, and the percentage of contiguous heterocysts was lower at 24 and 48 h but about twice at 72 h. Thus, inactivation of hetN led to a further increase in the differentiation capacity relative to the hetC patS mutant. In comparison to CSL11 (patS hetN), CSL15 (hetC patS hetN) had fewer heterocysts (0.67-fold at 24 h, 0.58-fold at 48 h and 0.88-fold at 72 h), many fewer contiguous heterocysts (0.18-fold, 0.13-fold and 0.69-fold, respectively) and more short intervals different from zero, especially at 24 and 48 h (Table 3, Fig. 2). Thus, in the absence of both PatS and HetN, the lack of HetC appears again to shift the Mch pattern to more individual heterocysts.
An altered hetC gene version
The hetC gene product includes a putative peptidase domain of the C39 family (InterPro, IPR005074; residues 339-465) (see Fig. 4B), which according to topological predictions would be located in the cytoplasm (Toppred). To test the functionality of this domain of HetC, mutant strains were generated that bore a hetC gene (hetC-p) that encodes a HetC version that lacks residues 338–463 that comprises the putative peptidase domain (see Fig. 1). The strain bearing this construct in the wild-type background (CSL16) was severely impaired in heterocyst differentiation although, in contrast to the mutant lacking the whole hetC gene (CSL3), produced a low percentage of (pro)heterocysts (Table 3) that could be stained with Alcian blue (Fig. 3A) but bore no Hgl (Fig. 3B). On the other hand, the percentage and distribution of (pro)heterocysts in strains bearing the hetC-p version in a patS background (CSL17), in a hetN background (CSL30) or in a patS hetN background (CSL31) followed a trend similar to the strains bearing the full hetC deletion in the same backgrounds (strains CSL1, CSL12 and CSL15, respectively) (Table 3, Fig. 2, see also Fig. 3A). Thus, the region that contains the putative peptidase domain appears necessary for proper HetC function. However, the percentage of contiguous (pro)heterocysts at 24 h was ca. 2.8 times higher in CSL1 than in CSL17, and at 48 and 72 h ca. 2.9 and 1.6 times higher, respectively, in CSL15 than in CSL31.
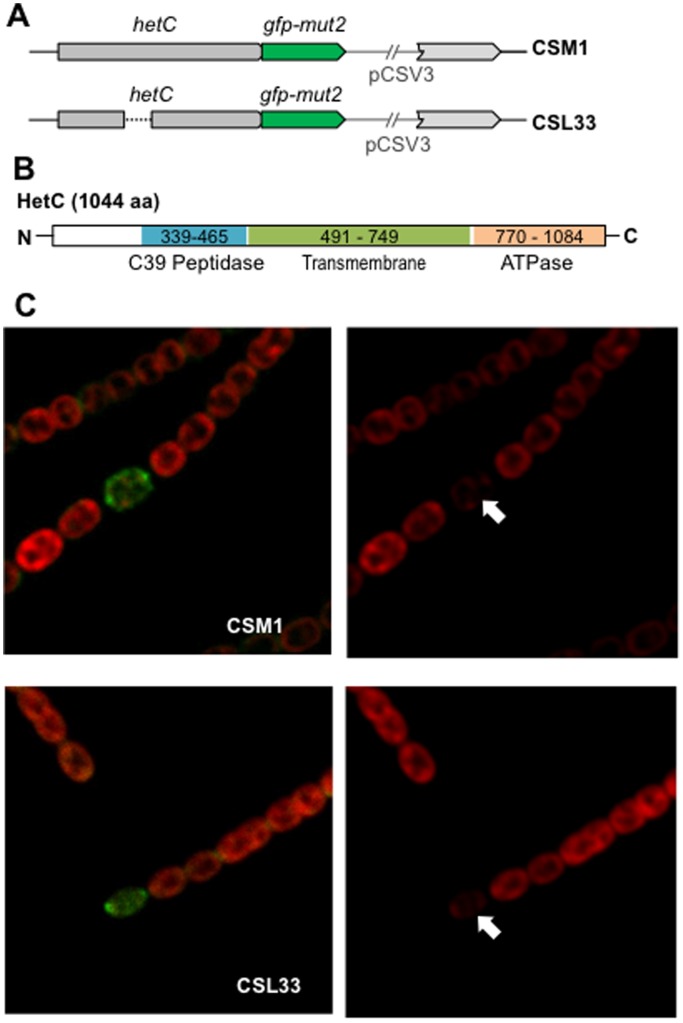
Figure 4. Localization of HetC-GFP.
(A) Scheme of the genomic hetC region of strains CSM1 (expressing a HetC-GFP-mut2 fusion protein) and CSL33 (expressing a HetC-p-GFP-mut2 fusion protein). The pCSV3 vector portion integrated in the hetC locus is represented as a thin line. (B) Scheme of the putative domains of the HetC protein. (C) Confocal microscopy of filaments of strains CSM1 and CSL33 grown in bubbled, ammonium-supplemented medium and incubated for 24 h in medium containing no combined nitrogen. Cyanobacterial autofluorescence (red) is shown in the right-hand images, and merged autofluorescence and GFP fluorescence (green) in the left-hand images. Heterocysts (indicated with white arrows) are identified by their greatly diminished autofluorescence.
To test whether the function of the peptidase domain of HetC is related to processing of PatS, we expressed from the patS promoter in the patS locus, in the hetC-p patS background, patS minigenes that encode the full 17-residue peptide (strain CSL102) or a peptide consisting of a Met residue followed by the PatS eight C-terminal residues (CSL101) [5] as the only patS version. In both strains, the (pro)heterocyst frequency was comparable, ca. 1.8% and 1.6% for CSL102 and CSL101, respectively, at 24 h (Table 3), and similar to that of strain CSL16 (hetC-p), whereas the parental strain CSL17 (patS hetC-p) had 18.5% of heterocysts at 24 h. Thus, both patS minigenes complemented the lack of patS in a hetC-p background.
Nitrogenase activity and diazotrophic growth in hetC, patS and hetN mutants
Nitrogenase activity, determined under both oxic and anoxic assay conditions, and diazotrophic growth were investigated in some of the mutants described above (Table 4; see Materials and Methods for details). The two hetC single-mutant strains, CSL3 (hetC) and CSL16 (hetC-p), exhibited negligible levels of nitrogenase activity under oxic or anoxic conditions. Strains CSL1 (hetC patS) and CSL17 (hetC-p patS) developed appreciable levels of nitrogenase activity under anoxic conditions, although those levels were much lower than in the wild type. Notably, in contrast to CSL3, significant expression of nifHDK took place in CSL1, although at 24 h after N step-down the transcript levels were lower than those observed in the wild type or the patS single mutant (Fig. 3C). However, as is the case for CSL3, strain CSL1 did not grow diazotrophically (Table 4). Thus, the lack of patS does not allow heterocyst function in the absence of a functional hetC gene. Strains CSL12 (hetC hetN) and CSL30 (hetC-p hetN) exhibited negligible nitrogenase activity under any tested condition and they were incapable of diazotrophic growth (Table 4). Thus, deletion of hetN appears unable to compensate for inactivation of hetC. In the hetC patS hetN (strain CSL15) or hetC-p patS hetN (CSL31) triple mutants, appreciable nitrogenase activity developed under anoxic conditions, as was the case in the patS hetN double mutant (CSL11). None of these strains was capable of effective diazotrophic growth (Table 4).
Table 4. Nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120 mutant strains.
| Strain | Genotype | Time (h) | Nitrogenase activityOxic Anoxic | Diazotrophic growth | |
| PCC 7120 | 24 | 16.0±3.6 (4) | 17.7±1.1 (12) | YES | |
| 48 | 8.3±2.4 (6) | 11.6±0.7 (7) | |||
| CSL3 | hetC | 24 | 0.0±0.0 (3) | 0.1±0.03 (7) | NO |
| 48 | 0.1±0.1 (4) | 0.1±0.1 (3) | |||
| CSL16 | hetC-p | 24 | 0.0±0.0 (2) | 0.0 (1) | NO |
| 48 | 0.1±0.1 (3) | 0.0 (1) | |||
| CSVT20 | patS | 24 | 3.2 (1) | 12.7±1.0 (6) | YES |
| 48 | 1.1±0.2 (2) | 7.8±0.7 (2) | |||
| CSL7 | hetN | 24 | 3.6 (1) | 10.5±3.1 (3) | YES |
| 48 | 2.1±0.7 (2) | 6.1±0.9 (3) | |||
| CSL1 | patS hetC | 24 | 0.0±0.02 (2) | 0.8±0.5 (6) | NO |
| 48 | 0.0±0.01 (2) | 0.4±0.3 (4) | |||
| CSL17 | patS hetC-p | 24 | 0.1±0.04 (3) | 0.2±0.1 (3) | NO |
| 48 | 0.7±0.4 (4) | 1.3±0.8 (4) | |||
| CSL11 | patS hetN | 24 | 0.0 (1) | 1.7±1.4 (3) | NO |
| 48 | 0.03±0.04 (2) | 0.5±0.4 (4) | |||
| CSL12 | hetC hetN | 24 | 0.0 (1) | 0.0 (1) | NO |
| 48 | 0.01±0.1 (2) | 0.1±0.03 (2) | |||
| CSL30 | hetC-p hetN | 24 | 0.0 (1) | 0.0±0.0 (2) | NO |
| 48 | 0.02±0.02 (2) | 0.1±0.1 (2) | |||
| CSL15 | patS hetC hetN | 24 | 0.1±0.1 (2) | 0.2±0.1 (3) | NO |
| 48 | 0.1±0.1 (2) | 2.2±1.8 (3) | |||
| CSL31 | patS hetC-p hetN | 24 | 0.1±0.03 (2) | 0.3±0.1 (3) | NO |
| 48 | 0.8±0.4 (3) | 2.4±0.7 (4) | |||
| CSL67 | hetP-sfgfp | 24 | 10.2±3.3 (2) | 22.1 (1) | YES |
| 48 | 8.2±3.0 (2) | 14.9 (1) | |||
| CSL68 | hetP-sfgfp hetC-p | 24 | 12.5 (1) | 14.0 (1) | YES |
| 48 | 4.3±0.02 (2) | 7.9±1.8 (2) | |||
| CSL69 | hetP-sfgfp patS | 24 | 1.4 (1) | 7.6 (1) | YES |
| 48 | 1.6 (1) | 7.4 (1) | |||
| CSL70 | hetP-sfgfp hetN | 24 | 6.6 (1) | 9.8 (1) | YES |
| 48 | 3.8 (1) | 9.2 (1) | |||
Nitrogenase activity was determined in ammonium-grown filaments incubated in the absence of combined nitrogen for 24 or 48 h and expressed in µmol ethylene produced (mg Chl)−1 h−1. Data are the mean and standard deviation of the mean (number of independent experiments indicated in parenthesis). Diazotrophic growth was tested in solid BG110 medium.
HetC localization
It has been described that a hetC-gfp transcriptional fusion is expressed most strongly in proheterocysts and heterocysts [16]. Predictions of the topology of the putative HetC protein (SOSUI) identified a membrane domain with six transmembrane helices (residues 491–749) between the putative peptidase and ATPase stretches (Fig. 4B). To study the subcellular localization of this protein, Anabaena derivatives producing a HetC-GFP (strain CSM1) or a HetC-p-GFP (Δpeptidase HetC-GFP) (strain CSL33) fusion protein as the only HetC version were generated (Fig. 4A). In these constructs, the GFP is added to the C-terminus of the corresponding HetC protein. Upon N step-down, fluorescence above the background was detected after ca. 6 h, located throughout the (pro)heterocyst surface but especially concentrated within the polar region of the heterocyst, apparently in the narrow area corresponding to the heterocyst neck (Fig. 4C). Because strain CSM1 expresses HetC fused to the gfp-mut2-encoded GFP, which only folds efficiently in the cytoplasm, our results support that the C terminus of HetC is located in the cytoplasm. No difference in GFP fluorescence was apparent between strains CSM1 and CSL33 (Fig. 4C), indicating that the putative peptidase stretch of HetC does not appreciably influence the production and sub-cellular localization of HetC.
HetP localization and effects of over-expression of hetP
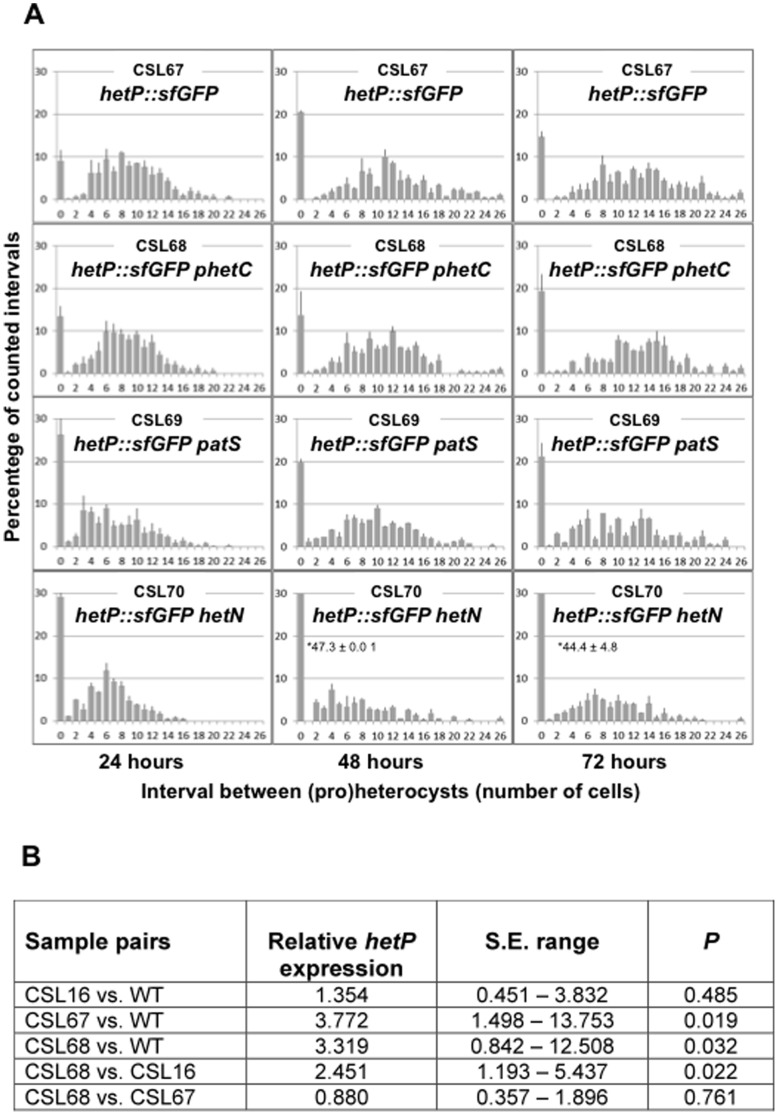
Previously, a transcriptional fusion of hetP to luxAB was reported to increase 2.5-fold in expression 6 h after N step-down [17], and a PhetP-gfp fusion was shown to produce higher fluorescence in heterocysts than in vegetative cells at 24 h [18]. We have constructed strains bearing a hetP-gfp-mut2 or hetP-sf-gfp gene fusion that encode, respectively, a HetP protein C-terminally fused to conventional GFP (strain CSL107) or to sf-GFP [29] (strain CSL67). These strains bear the fusion gene preceding the inserted pCSV3 plasmid vector and a native hetP copy in the chromosome (Fig. 5A; see Materials and Methods). The hetP-sf-gfp construct was also introduced into the hetC-p, the patS and the hetN mutant backgrounds producing strains CSL68, CSL69 and CSL70, respectively. (Pro)heterocyst frequency and distribution was studied in each of those strains (Table 3, Fig. 6A; for comparison, see also Fig. 2). In general terms, strains CSL67, CSL69 and CSL70 exhibited (pro)heterocyst patterns similar to those of their respective parental strains (strains PCC 7120, CSVT20 and CSL7), although with a tendency to increase heterocyst frequency and the frequency of doublets (considerably higher in strain CSL67 than in PCC 7120), and to decrease the mean interval size between heterocysts. However, in contrast to strain CSL16 (hetC-p), which presented a very low (pro)heterocyst frequency (Table 3), strain CSL68 showed a frequency of heterocysts and mean interval size similar to those in the wild type. The frequency of contiguous heterocysts in strain CSL68 was much higher (7.8 times at 72 h) than in the wild type.
Figure 5. Localization of HetP-GFP.
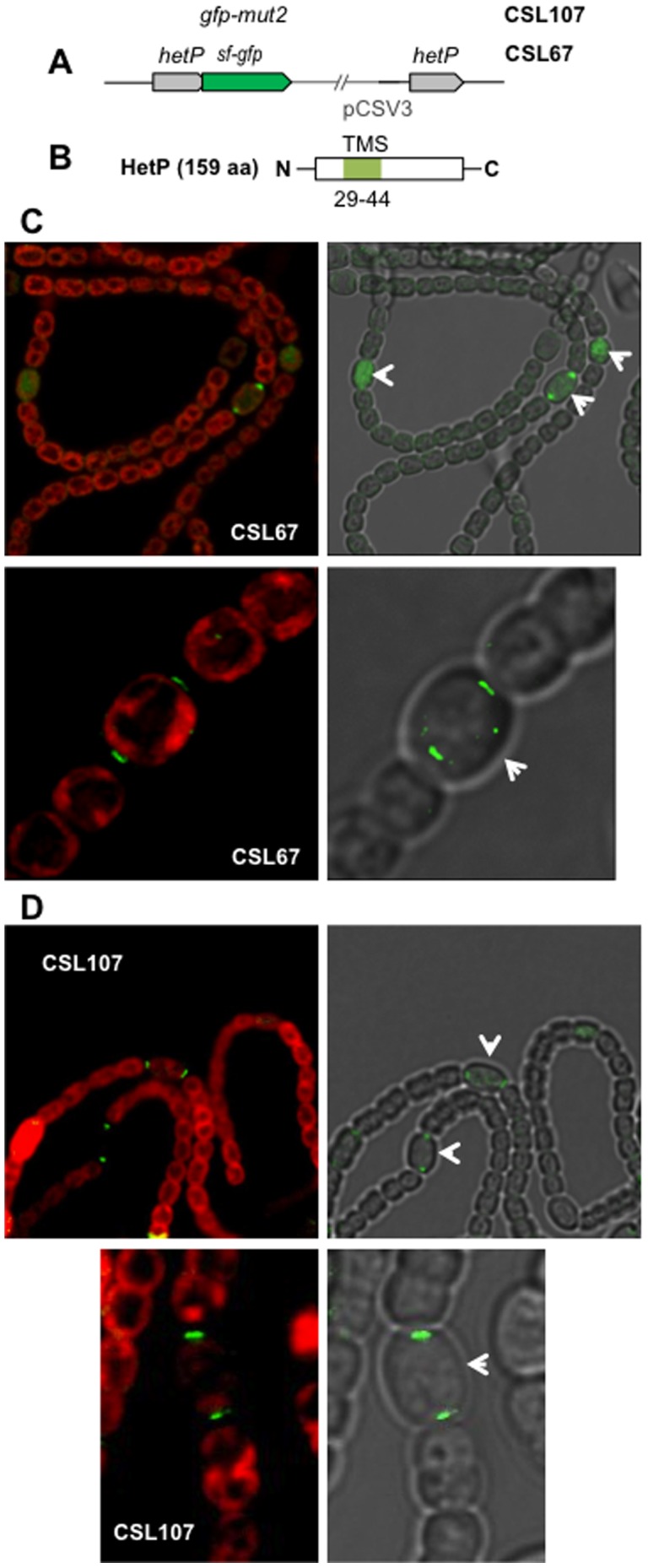
(A) Scheme of the hetP genomic region of strains CSL67 (HetP-sf-GFP fusion protein) and CSL107 (HetP-GFP-mut2 fusion protein). The pCSV3 vector portion integrated in the hetP locus is represented as a thin line. (B) Scheme of the HetP protein showing a predicted transmembrane segment (TMS). (C) Confocal microscopy of filaments of strain CSL67 grown in BG110 solid medium (upper part) or deconvoluted fluorescence microscopy image of filaments of strain CSL67 grown in bubbled ammonium-supplemented medium and incubated for 40 h in medium containing no combined nitrogen (lower part). (D) Fluorescence microscopy of filaments of strain CSL107 grown in BG110 solid medium. Merged images of autofluorescence and GFP fluorescence are shown at the left side, and of bright field and GFP fluorescence at the right side. Heterocysts and some proheterocysts are indicated with white arrows.
Figure 6. Heterocyst distribution and hetP expression levels in Anabaena mutant strains altered in hetP.
(A) Heterocyst distribution in the indicated strains grown in bubbled cultures with ammonium and incubated for the indicated times in the absence of combined nitrogen under culture conditions (see legend to Fig. 2 for details). (B) Ratios of the expression levels of hetP of the indicated strains 18 h after N step-down, measured by qRT-PCR normalized to the rnpB gene. S.E. range indicates the “standard error change” and P (the hypothesis test P) represents the probability that the difference between the sample and control groups is due only to chance [39]. Data are the mean of two independent experiments.
Nitrogenase activity under oxic and anoxic conditions was comparable in strains CSL67, CSL69 and CSL70 and their respective parental strains, and those three mutant strains were capable of diazotrophic growth (Table 4). In contrast to its parental strain, CSL16, which showed negligible activity levels, strain CSL68 showed high levels of oxic and anoxic nitrogenase activity, and indeed it was capable of diazotrophic growth (Table 4). Because the hetP gene influences heterocyst differentiation positively [17], [18], the above results suggest that the HetP-sf-GFP fusion protein is functional. The relative expression levels of hetP (or hetP-sf-gfp) were studied by qRT-PCR in strains PCC 7120, CSL16 (hetC-p), CSL67 (hetP-sf-gfp in wild-type background) and CSL68 (hetP-sf-gfp in hetC-p background). Results included in Figure 6B indicate that 18 h after N step-down the hetP transcript levels were ca. 3- to 4-fold higher in strains CSL67 and CSL68 than in their respective parental strains. Thus, the increased differentiation in strain CSL67 with respect to the wild type could result from the increased expression of a functional hetP gene. Moreover, the comparison of (pro)heterocyst pattern and hetP expression in strains CSL16 and CSL68 indicates that over-expression of hetP compensates for the lack of a functional hetC gene.
Fluorescence from sf-GFP was tracked in strains CSL67, CSL68, CSL69 and CSL70. In CSL67 (hetP-sf-gfp in a wild-type background), fluorescence was located throughout the cell area in proheterocysts and focalized near the cell poles in mature heterocysts (Fig. 5C). Deconvolution analysis of the images from mature heterocysts showed the GFP fluorescence in the cytoplasm (Fig. 5C, see merged GFP/bright-field image) but external to the chlorophyll fluorescence (Fig. 5C, see merged GFP/autofluorescence image). Localization of fluorescence was similar in strains CSL68, CSL69 and CSL70 (not shown). Similar sf-gfp expression levels were detectable by qRT-PCR in strains CSL67 and CSL68 (not shown). In strain CSL107 bearing a hetP-gfp-mut2 gene version, GFP fluorescence was similar to that in CSL67 (Fig. 5D), suggesting that HetP-GFP is folded in the cytoplasm.
Mutation of asr2819
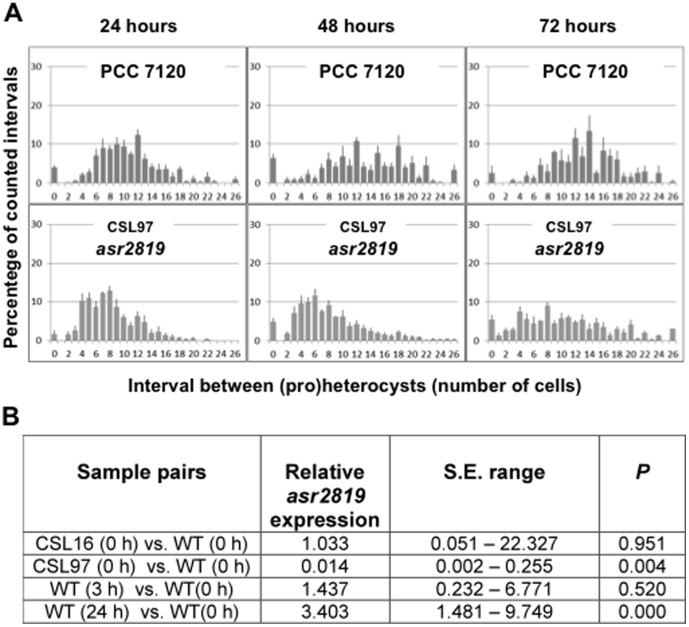
The Anabaena ORF asr2819, which presumptively encodes an 84-amino-acid protein, was induced upon N step-down (ca. 3- to 4-fold increase after 24 h [Fig. 7B]; see also [30]). To study the role of Asr2819, we sought to generate mutant strains that lack this peptide. Two hundred and forty bp, including all of the asr2819 ORF but its final 15 bp (see Fig. 1), were removed from the genome of the wild type, the hetC-p mutant (CSL16), and the patS hetN (CSL11) mutant, generating, respectively, strains CSL97, CSL98, and CSL106. In strain CSL97 the percentage of (pro)heterocysts was somewhat higher, and the mean interval size shorter, than in the wild type (Table 3). Notably, at 72 h this strain showed a very relaxed pattern of (pro)heterocyst spacing, reflected in a greater dispersion of size intervals than in the wild type (Fig. 7A). In strain CSL98, (pro)heterocyst frequency was similar to that of its parental strain (CSL16) (Table 3). Thus, the hetC-p mutation is epistatic over the asr2819 mutation. Like its parental strain (CSL11), strain CSL106 produced a very high proportion of heterocysts (in fact, given the normal stickiness of heterocysts, and thus of filaments, it was problematic to distinguish vegetative cell intervals). Because the asr2819 gene product contributes to the formation of the spatial pattern of heterocyst distribution along the Anabaena filament (Fig. 7A), we name it the patC gene.
Figure 7. Heterocyst distribution and asr2819 expression levels in Anabaena sp. strains PCC 7120 and CSL97.
(A) Heterocyst distribution in the indicated strains grown in bubbled cultures with ammonium and incubated for the indicated times in the absence of combined nitrogen under culture conditions (see legend to Fig. 2 for details). (B) Ratios of the expression levels of asr2819 in the indicated strains at the indicated times after N step-down, measured by qRT-PCR normalizing with the rnpB gene. Data are the mean of two to three independent experiments.
Discussion
There has been a long-standing speculation that the HetC protein, which is similar to ABC exporters and required for differentiation, might act by transferring the negative regulators PatS and/or HetN from the heterocysts to the neighboring cells. To address this issue, we have undertaken a study of epistatic effects between the genes hetC, patS and hetN. In a hetC patS double mutant (strain CSL1), the frequency of (pro)heterocysts is high, even higher than in the single patS mutant after a long incubation in the absence of combined nitrogen. Moreover, strain CSL1 produces heterocyst-specific glycolipids and shows detectable expression of the nifHDK genes encoding nitrogenase, although both are found at lower levels than in the wild type. Thus, the lack of the negative factor PatS mitigates the effect of the lack of HetC. In contrast, the lack of hetN alone does not alleviate the lack of HetC, although it further increases differentiation in the absence of HetC and PatS. These results would be compatible with the idea of HetC-dependent export of a PatS-derived morphogen. The results would not eliminate the possibility that HetC exports a morphogen that is derived from HetN if one assumes that in the absence of HetC a previous accumulation of PatS in the proheterocyst suffices to inhibit differentiation.
However, two sets of data speak against HetC function being solely PatS (or PatS and HetN) export from the (pro)heterocyst. First, the cells stained with Alcian Blue in the double patS hetC (or patS hetC-p) and triple patS hetN hetC (or patS hetN hetC-p) mutants never look like mature heterocysts (see Fig. 3A). Indeed, nitrogenase activity was not effectively complemented, albeit in the double and triple mutants lack of nitrogenase activity could be related to an excessive frequency of differentiating cells, which may result in an unsustainably high ratio of heterocysts to vegetative cells in the filament. Indeed, this appears to be the case already for the patS hetN double mutant (strain CSL11), which lacks oxic nitrogenase activity (Table 4). (In this regard, our results with strain CSL11 contrast with the observations, reported in [12], that nitrogenase activity was highly expressed in a patS mutant in which hetN was diminished in expression.) None of those mutants grew diazotrophically (Table 4). Second, the distribution of heterocysts differs in the patS hetC (or patS hetC-p) double mutant and in the single hetC or patS mutants, and differs also between the patS hetN hetC (or patS hetN hetC-p) triple mutant and the hetC or patS hetN mutants (Fig. 2, Table 3).
Deletion of the putative peptidase motif of HetC results in a greatly reduced frequency of (pro)heterocysts, albeit not as low as does deletion of hetC (Table 3). Because lack of processing of the primary product of patS to produce an active peptide results in lack of differentiation [5], the phenotype of strains that bear a hetC-p version might be consistent with an involvement of the peptidase domain of HetC in processing of PatS. However, although the HetC-p-GFP fusion protein appears to be correctly localized (Fig. 4C), one cannot be sure that the HetC-p protein retains other possible activities such as any putative transport activity. In a wild-type background, expression of PatS-17 or PatS-8 (a putative product of processing of PatS-17 in the proheterocysts) as the only PatS version recreates a heterocyst pattern similar to that of the wild-type strain, whereas lack of processing of PatS-17 provokes inhibition of differentiation [5]. It might be expected that if the peptidase domain of HetC were involved in processing of PatS-17, expression of PatS-17, but not of PatS-8, in a hetC-p background would reduce the frequency of heterocysts. However, both strains CSL101 and CSL102 exhibited low heterocyst frequencies (Table 3), indicating that expression of PatS-8 does not compensate for the lack of the peptidase domain of HetC. In the scenario that HetC were not directly involved in PatS (or PatS and HetN) processing or transfer, the partial compensation of the lack of hetC by deletion of patS (or patS and hetN) could be explained by the assumption that the elimination by mutation of the negative factors makes the requirement for HetC, a positive element, less strict during heterocyst differentiation.
To get clues for the function of HetC and HetP, we have studied the subcellular localization of those two proteins in (pro)heterocysts by making use of C-terminal GFP fusion domains expressed from gene constructs present in the cells with copy numbers similar to those of the native genes. In mature heterocysts, HetC-GFP is localized through the heterocyst periphery, and appears especially concentrated near the heterocyst poles. Because different bioinformatics programs predict that HetC has a number of transmembrane segments, it is evident that this protein is targeted to a membrane, and according to Fig. 4C may be targeted to the plasma membrane. In both CSM1 and CSL33, there is more fluorescence near the cell poles. However, it is unclear whether that localization results from the presence of two close membrane units in the heterocyst neck (see e.g. Fig. 2d in [31]), or whether there is a preferential targeting near the cell poles. In Gram-negative bacteria, genes that encode exporters of toxic peptides are frequently linked to genes that encode a protein that belongs to the membrane-fusion protein family that spans the periplasmic space linking the ABC exporter to an outer membrane channel [14]. Because no homolog of membrane-fusion proteins has been detected in the hetC genomic region, it has been speculated that the HetC substrate could be released to the Anabaena periplasm (see [14]). It is also possible that HetC acts in connection with a different type of protein to promote intercellular molecular transfer. As yet another alternative, HetC might not mediate transport but have a different function that may involve interactions with another membrane factor. Given that HetC has been involved in the inhibition of cell division in the differentiating cells leading to commitment to differentiation [16], HetC might interact with components of the cell-division complex. Such a possibility would be reminiscent of the interaction of the ABC-transporter-like FtsEX complex with FtsZ in E. coli [32].
It is notable that inactivation of genes that encode negative factors of heterocyst differentiation, patS, hetN, or patU3 [33], produce a Mch phenotype, which is also the tendency with over-expression of the positive elements hetP (Fig. 6) or hetF [34], [35]. In contrast, inactivation of hetC tends to alleviate the Mch distribution in patS or patS hetN backgrounds, shifting the heterocyst distribution pattern to a higher frequency of short intervals different from zero (Fig. 2, Table 3). The tendency to differentiation in clusters of cells when the negative regulators are eliminated could involve the action of a putative PatN factor if its role in Anabaena were similar to that described in Nostoc punctiforme at establishing clusters of cells permissive for differentiation (those lacking PatN) by biased inheritance during cell division [36]. PatN is a membrane factor and its inactivation results in an Msh (increased frequency of single heterocyst) phenotype. It is tempting to speculate that the roles of HetC and PatN could be related, e.g., HetC might act only in cells devoid of PatN.
HetP-GFP is present throghout the cell area in proheterocysts, and later is almost restricted to the polar regions, where it remains in mature heterocysts (Fig. 5C). HetP-GFP proteins appear to be present in soluble fractions from whole Anabaena filaments, which could correspond to the protein present throughout proheterocysts, and in membrane fractions, which could correspond to the protein in cells more advanced in differentiation, 12 h after N step-down (our unpublished observations). Several bioinformatics programs (Psipred, TMHMM) predict that HetP bears a single putative membrane segment comprising residues ca. 29-44 (Fig. 5B). This segment could maintain HetP anchored to membranes at the cell poles. Moreover, the HetP-GFP fluorescence near the heterocyst poles appears peripheral to the chlorophyll fluorescence in those cells. It would be of much interest to study the mechanism that sorts this protein to membranes specifically at the heterocyst poles. As a possibility, membrane regions with a specific curvature could be selected in the differentiated and complex heterocyst pole, as is the case for the localization of SpoVM during spore formation in Bacillus subtilis [37]. Also, HetP could be anchored to the membrane and maintained in the heterocyst poles by interaction with septal proteins. Our results on epistasis of hetC and hetP show that a moderate over-expression of hetP (in strain CSL68) fully complements the lack of a functional hetC gene (hetC-p; Fig. 6). It is worth to stress that in strain CSL68 nitrogenase activity and diazotrophic growth are similar to those in the wild type (Table 4), in contrast to a partial complementation of the hetC mutation by PatS deletion (in strain CSL1) or of hetR inactivation by hetP over-expression [18]. These results are consistent with a functional link between HetC and HetP, whose roles may overlap. This idea is also supported by the similar effects of the mutation of each gene that results in strains capable of initiation of differentiation, which however arrests at an early stage.
In summary, our results are consistent with a double role of HetC at regulation of heterocyst differentiation. HetC might be involved in processing or transfer of the negative regulators (PatS and HetN) from (pro)heterocysts to the neighboring cells. Besides that, HetC (and HetP) might interact with membrane factors leading to inhibition of cell division, perhaps in cells devoid of PatN. In this model, PatN might not influence the selection of cells that initiate differentiation but those in which, though the action of HetC, differentiation could proceed to completion. On the other hand PatC, which is not necessary for differentiation, might participate in differentiation site selection especially during established diazotrophic growth. Finally, the commitment to differentiation could involve still more elements. It is noteworthy that the inactivation of sepJ, which encodes a septal protein that is important for cell-to-cell anchoring and intercellular transfer in the filament, interferes, as is the case of hetC, with inhibition of cell division during heterocyst differentiation [24]. Deciphering the structure of the multiprotein complexes at the vegetative cell-heterocyst septa represents an outstanding research challenge for the future.
Acknowledgments
We thank Marta León and Alicia M. Muro-Pastor for construction of strain CSM1, and Vicente Mariscal for help with microscopy and useful discussions.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
Work was supported by grant BFU2010-17980 from the Spanish Government, co-financed by FEDER. L.C.-G. was the recipient of a JAE-predoc fellowship from CSIC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Flores E, Herrero A (2010) Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol 8: 39–50. [DOI] [PubMed] [Google Scholar]
- 2. Herrero A, Picossi S, Flores E (2013) Gene expression during heterocyst differentiation. Adv. Bot. Res 65: 281–329. [Google Scholar]
- 3. Yoon HS, Golden JW (1998) Heterocyst pattern formation controlled by a diffusible peptide. Science 282: 935–938. [DOI] [PubMed] [Google Scholar]
- 4. Yoon HS, Golden JW (2001) PatS and products of nitrogen fixation control heterocyst pattern. J. Bacteriol 183: 2605–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corrales-Guerrero L, Mariscal V, Flores E, Herrero A (2013) Functional dissection and evidence for intercellular transfer of the heterocyst-differentiation PatS morphogen. Mol. Microbiol 88: 1093–1105. [DOI] [PubMed] [Google Scholar]
- 6. Huang X, Dong Y, Zhao J (2004) HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci.USA 101: 4848–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khudyakov I, Golden JW (2004) Different functions of HetR, a master regulator of heterocyst differentiation in Anabaena sp. PCC 7120, can be separated by mutation. Proc. Natl. Acad. Sci. USA 101: 16040–16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Risser DD, Callahan SM (2009) Genetic and cytological evidence that heterocyst patterning is regulated by inhibitor gradients that promote activator decay. Proc. Natl. Acad. Sci. USA 106: 19884–19888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Black TA, Wolk CP (1994) Analysis of a Het− mutation in Anabaena sp. PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J. Bacteriol 176: 2282–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bauer CC, Ramaswamy KS, Endley S, Scappino LA, Golden JW, et al. (1997) Suppression of heterocyst differentiation in Anabaena PCC 7120 by a cosmid carrying wild-type genes encoding enzymes for fatty acid biosynthesis. FEMS Microbiol. Lett 151: 23–30. [DOI] [PubMed] [Google Scholar]
- 11. Higa KC, Rajagopalan R, Risser DD, Rivers OS, Tom SK, et al. (2012) The RGSGR amino acid motif of the intercellular signaling protein, HetN, is required for patterning of heterocysts in Anabaena sp. strain PCC 7120. Mol. Microbiol 83: 682–693. [DOI] [PubMed] [Google Scholar]
- 12. Borthakur PB, Orozco CC, Young-Robbins SS, Haselkorn R, Callahan SM (2005) Inactivation of patS and hetN causes lethal levels of heterocyst differentiation in the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol 57: 111–123. [DOI] [PubMed] [Google Scholar]
- 13. Khudyakov I, Wolk CP (1997) hetC, a gene coding for a protein similar to bacterial ABC protein exporters, is involved in early regulation of heterocyst differentiation in Anabaena sp. strain PCC 7120. J. Bacteriol 179: 6971–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michiels J, Dirix G, Vanderleyden J, Xi C (2001) Processing and export of peptide pheromones and bacteriocins in Gram-negative bacteria. Trends Microbiol 9: 164–168. [DOI] [PubMed] [Google Scholar]
- 15. Muro-Pastor AM, Flores E, Herrero A (2009) NtcA-regulated heterocyst differentiation genes hetC and devB from Anabaena sp. PCC 7120 exhibit a similar tandem promoter arrangement. J. Bacteriol 191: 5765–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu X, Wolk CP (2001) Role for hetC in the transition to a nondividing state during heterocyst differentiation in Anabaena sp. J. Bacteriol 183: 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fernández-Piñas F, Leganés F, Wolk CP (1994) A third genetic locus required for the formation of heterocysts in Anabaena sp. strain PCC 7120. J. Bacteriol 176: 5277–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Higa KC, Callahan SM (2010) Ectopic expression of hetP can partially bypass the need for hetR in heterocyst differentiation by Anabaena sp. strain PCC 7120. Mol. Microbiol 77: 562–574. [DOI] [PubMed] [Google Scholar]
- 19. López-Igual R, Picossi S, López-Garrido J, Flores E, Herrero A (2012) N and C control of ABC-type bicarbonate transporter Cmp and its LysR-type transcriptional regulator CmpR in a heterocyst-forming cyanobacterium Anabaena sp. Environ. Microbiol 14: 1035–1048. [DOI] [PubMed] [Google Scholar]
- 20. Elhai J, Wolk CP (1988) A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68: 119–138. [DOI] [PubMed] [Google Scholar]
- 21. Black TA, Cai Y, Wolk CP (1993) Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena . Mol. Microbiol 9: 77–84. [DOI] [PubMed] [Google Scholar]
- 22. Merino-Puerto V, Herrero A, Flores E (2013) Cluster of genes that encode positive and negative elements influencing filament length in a heterocyst-forming cyanobacterium. J. Bacteriol 195: 3957–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP (1997) Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol 179: 1998–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flores E, Pernil R, Muro-Pastor AM, Mariscal V, Maldener I, et al. (2007) Septum-localized protein required for filament integrity and diazotrophy in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol 189: 3884–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valladares A, Rodríguez V, Camargo S, Martínez-Nöel GMA, Herrero A, et al. (2011) Specific role of the cyanobacterial PipX factor in the heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol 193: 1172–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Merino-Puerto V, Mariscal V, Mullineaux CW, Herrero A, Flores E (2010) Fra proteins influencing filament integrity, diazotrophy and localization of septal protein SepJ in the heterocyst-forming cyanobacterium Anabaena sp. Mol. Microbiol 75: 1159–1170. [DOI] [PubMed] [Google Scholar]
- 27. Wolk CP, Ernst A, Elhai J (1994) Heterocyst metabolism and development. In: The Molecular Biology of Cyanobacteria. Bryant, DA (ed.) Dordrecht: Kluwer Academic Publishers pp. 769–823. [Google Scholar]
- 28. Callahan SM, Buikema WJ (2001) The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol. Microbiol 40: 941–950. [DOI] [PubMed] [Google Scholar]
- 29. Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol 24: 79–88. [DOI] [PubMed] [Google Scholar]
- 30. Flaherty BL, Van Nieuwerburgh F, Head SR, Golden JW (2011) Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics 12: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flores E, Herrero A, Wolk CP, Maldener I (2006) Is the periplasm continuous in filamentous multicellular cyanobacteria? Trends Microbiol. 14: 439–443. [DOI] [PubMed] [Google Scholar]
- 32. Corbin BD, Wang Y, Beuria TK, Margolin W (2007) Interaction between cell division proteins FtsE and FtsZ. J. Bacteriol 189: 3026–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang W, Du Y, Khudyakov I, Fan Q, Gao H, et al. (2007) A cluster that regulates both heterocyst differentiation and pattern formation in Anabaena sp. strain PCC 7120. Mol. Microbiol 66: 1429–1443. [DOI] [PubMed] [Google Scholar]
- 34. Wong FC, Meeks JC (2001) The hetF gene product is essential to heterocyst differentiation and affects HetR function in the cyanobacterium Nostoc punctiforme . J. Bacteriol 183: 2654–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Risser DD, Callahan SM (2008) HetF and PatA control levels of HetR in Anabaena sp. strain PCC 7120. J. Bacteriol 190: 7645–7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Risser DD, Wong FC, Meeks JC (2012) Biased inheritance of the protein PatN frees vegetative cells to initiate patterned heterocyst differentiation. Proc. Natl. Acad. Sci. USA 109: 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramamurthi KS, Lecuyer S, Stone HA, Losick R (2009) Geometric cue for protein localization in a bacterium. Science 323: 1354–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, et al. (2001) Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Research 8: 205–213. [DOI] [PubMed] [Google Scholar]
- 39. Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software Tool (REST©) for group-wise comparison and statistical analysis of relative expression results in Real-Time PCR. Nucleic Acid. Res 30(9): e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.