Abstract
The ammonia monooxygenase (AMO)/particulate methane monooxygenase (pMMO) superfamily is a diverse group of membrane-bound enzymes of which only pMMO has been characterized on the molecular level. The pMMO active site is believed to reside in the soluble N-terminal region of the pmoB subunit. To understand the degree of structural conservation within this superfamily, the crystal structure of the corresponding domain of an archaeal amoB subunit from has been determined to 1.8 Å resolution. The structure reveals a remarkable conservation of overall fold and copper binding site location as well as several notable differences that may have implications for function and stability.
Keywords: AMO, amoB, copper, crystal structure, cupredoxin, hydrocarbon monooxygenase, methanotroph, pMMO
INTRODUCTION
The copper membrane-associated monooxygenases (CuMOs) are a diverse superfamily of multisubunit prokaryotic enzymes that oxidize a range of compounds, including ammonia and hydrocarbons.1 The founding members are ammonia monooxygenase (AMO)2 and particulate methane monooxygenase (pMMO),3 both produced by gram-negative bacteria. AMO converts ammonia to hydroxylamine in nitrifiers, and pMMO oxidizes methane to methanol in methanotrophs. Hydroxylamine and methanol are then further metabolized and serve as the primary energy source for the host organisms.4,5 A recent explosion in genomic data has led to the detection of numerous other homologs, including archaeal AMOs,6,7 putative butane monooxygenases (BMOs),8 hydrocarbon monooxygenases from gram-positive Mycobacteria9,10 and marine isolates (pHMOs),11,12 and pXMO systems, for which the substrate is unknown (Fig. 1A phylogenetic tree).1
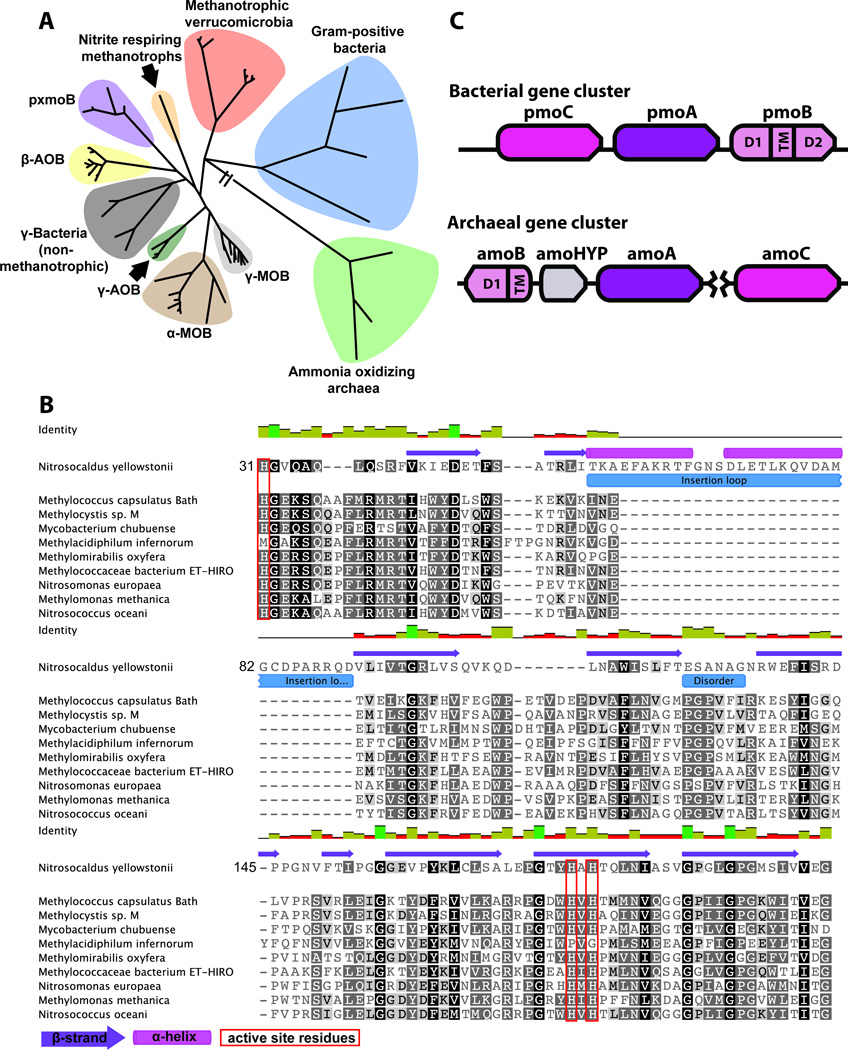
Figure 1.
Members of the AMO/pMMO superfamily. A: Phylogenetic tree. β-AOB, ammonia oxidizing β-proteobacteria; γ-AOB, ammonia oxidizing γ-proteobacteria; γ-MOB, methane oxidizing γ-proteobacteria; γ-bacteria, non methanotrophic γ-proteobacteria. B: Multiple sequence alignment of representative B subunits from each phylogenetic cluster. C: Gene cluster arrangement typically found in AOA and bacteria. The gene shown in grey is a conserved hypothetical protein of unknown function. The break between amoA and amoC indicates these genes are not always clustered in AOA.
Because of difficulties cultivating CuMO-producing organisms and isolating purified, active enzymes, little structural data are available for this superfamily. To date, the best-characterized CuMO is pMMO. Crystal structures of pMMO from three different methanotrophs show it is an α3β3γ3 heterotrimer of which each individual protomer comprises single copies of the pmoC, pmoA, and pmoB gene products.13–15 pMMO activity is dependent on copper, and the catalytic site is believed to be located at a crystallographically modeled dinuclear copper center in the pmoB subunit.16–18 The pmoB subunit consists of two cupredoxin-like domains connected by two transmembrane helices and a long linker region. The copper active site is housed in the N-terminal, periplasmic cupredoxin-like domain with three histidines and the N-terminal amino group of the polypeptide within coordinating distance. These three histidines are conserved in all pmoB, amoB (homologous subunit from AMO), and related sequences except those from the methanotrophic Verrucomicrobia (Fig. 1B sequence alignment).19
Interestingly, the degree of sequence conservation between methanotrophic pmoB subunits and some homologs is quite low with < 20% identity. The AMOs from ammonia oxidizing archaea (AOA) are the most phylogenetically distant. Not only is there a low degree of sequence conservation (Fig. 1B sequence alignment), but these amoB subunits lack the C-terminal cupredoxin domain; only an N-terminal cupredoxin domain followed by a single transmembrane helix is present (Fig. 1C operon).6,7 The impact of these sequence variations on the proposed location of the active site within this subunit is not clear. To address this question, we determined the 1.8 Å resolution crystal structure of the soluble region of an amoB subunit from the thermophilic AOA Nitrosocaldus yellowstonii.7
MATERIALS AND METHODS
The DNA encoding for amino acids 31 to 186 of Nitrosocaldus yellowstonii amoB (Ny_amoB) was amplified from genomic DNA and cloned into the vector pASK-IBA2 (IBA GmbH) using the BsaI restriction sites. Residue 31 corresponds to the predicted N-terminus of the mature polypeptide following signal sequence cleavage, and residue 186 marks the beginning of the predicted transmembrane region. The pASK-IBA2 vector was selected because it encodes a C-terminal Strep-tag for affinity chromatography and an N-terminal OmpA signal peptide that targets the protein for export to the periplasm where the signal peptide is cleaved, yielding the native N-terminus. Rosetta-2 (DE3) (EMD Millipore) cells transformed with the pASK-IBA2 vector containing the Ny_amoB gene were grown in Luria broth at 37 °C to an optical density at 600 nm of 0.6–1.0 and then induced with 200 μg L−1 anhydrotetracycline and 0.5 mM CuSO4 (final concentrations). Following induction, cells were grown overnight at 18 °C, harvested by centrifugation at 3,100 g for 10 min, and frozen as a solid pellet for later use.
For protein purification, cells were resuspended in buffer containing 100 mM Tris, pH 8.0, 500 mM NaCl, and 10% (v/v) glycerol, and lysed either by multiple passes through a 15,000 PSI microfluidizer or by sonication for a total of 8 min in 1 s pulses with 3 s resting time. To remove insoluble material, the cell extract was centrifuged for 30 min to 1 hr at 125,000 g. The supernatant was then applied to a 20 mL streptactin affinity column (Qiagen) equilibrated with buffer containing 500 mM NaCl, 100 mM Tris, pH 8.0, and 10% glycerol. Following extensive washing, Ny_amoB was eluted with buffer containing 100 mM Tris, pH 8.0, 500 mM NaCl, 2.5 mM d-desthiobiotin, and 10% glycerol. The eluted protein was then concentrated in an Amicon concentrator with 10 kDa nominal molecular weight cut off and applied to a 120 mL Superdex 75 (GE Healthcare) size exclusion column equilibrated with 20 mM Pipes, pH 7.2, 20 mM NaCl, and 10% glycerol. Notably, Ny_amoB is extremely stable and can be subjected to temperatures of > 65 °C for extended periods of time without precipitating.
Crystals of Ny_amoB were obtained using the sitting drop vapor diffusion method by mixing 1 μL of solution containing 5 mg mL−1 protein and 3.5 μL of well solution containing 1 M NH4SO4, 100 mM sodium formate pH 4.0, and 2% polyethylene glycol 8000. Football shaped crystals appeared within 2–3 days. Data collection was performed at sector 21 (Life Sciences Collaborative Access Team, LS-CAT) of the Advanced Photon Source at Argonne National Laboratory. HKL2000 was used to process and integrate all data sets.20 Copper single wavelength anomalous dispersion (SAD) was used to determine phases with a high redundancy data set (Table 1), and an initial model was built using the program COOT.21 This model was then used as a molecular replacement search model for a 1.8 Å resolution data set (Table 1). Phaser was used to obtain molecular replacement solutions, and the structure was refined with REFMAC.22,23 The final coordinates were deposited in the Protein Data Bank (PDB accession code 4O65). Structural comparisons to pMMO were performed using the secondary structure matching (SSM) algorithm at http://www.ebi.ac.uk/msd-srv/ssm.24 The pMMO crystal structures were inspected manually to determine which portions of the pmoB sequences correspond to the N-terminal cupredoxin domain, and sequence comparisons to Ny_amoB were carried out using the program EMBOSS Needle.25
Table I.
Data Collection and Refinement Statistics
| Cu SAD | High resolution | |
|---|---|---|
| Space group | P43212 | P43212 |
| Resolution (Å) | 2.5 (2.5–25.0) | 1.8 (1.80–22.43) |
| Rsym | 0.058 (0.084) | 0.056 (0.384) |
| I/σI | 120.0 (41.0) | 33.5 (3.5) |
| Completeness (%) | 99.8 (100) | 99.5 (98.4) |
| Redundancy | 27 (28) | 7(7.7) |
| Rwork‡/Rfree§ (%) | 18.1/22.9 | |
| Average B-factor | 20.9 | |
| r.m.s deviations | ||
| Bond lengths (Å) | 0.019 | |
| Bond angles (°) | 2.200 | |
The phylogenetic tree found in Figure 1A was constructed using software available at www.phylogeny.fr. In sum 61 CuMO sequences were curated to contain domain 1 only and to remove the putative signal peptides, Muscle was used for sequence alignment, Gblocks was used for additional sequence curation, PhyML using the approximate likelihood-ratio test was used to calculate phylogenetic relationships, and TreeDyn was used for sequence visualization. The sequence alignment in Figure 1B was constructed in Geneious using Muscle to perform the alignment. The region around the insertion loop was manually aligned to reflect structural knowledge.
RESULTS AND DISCUSSION
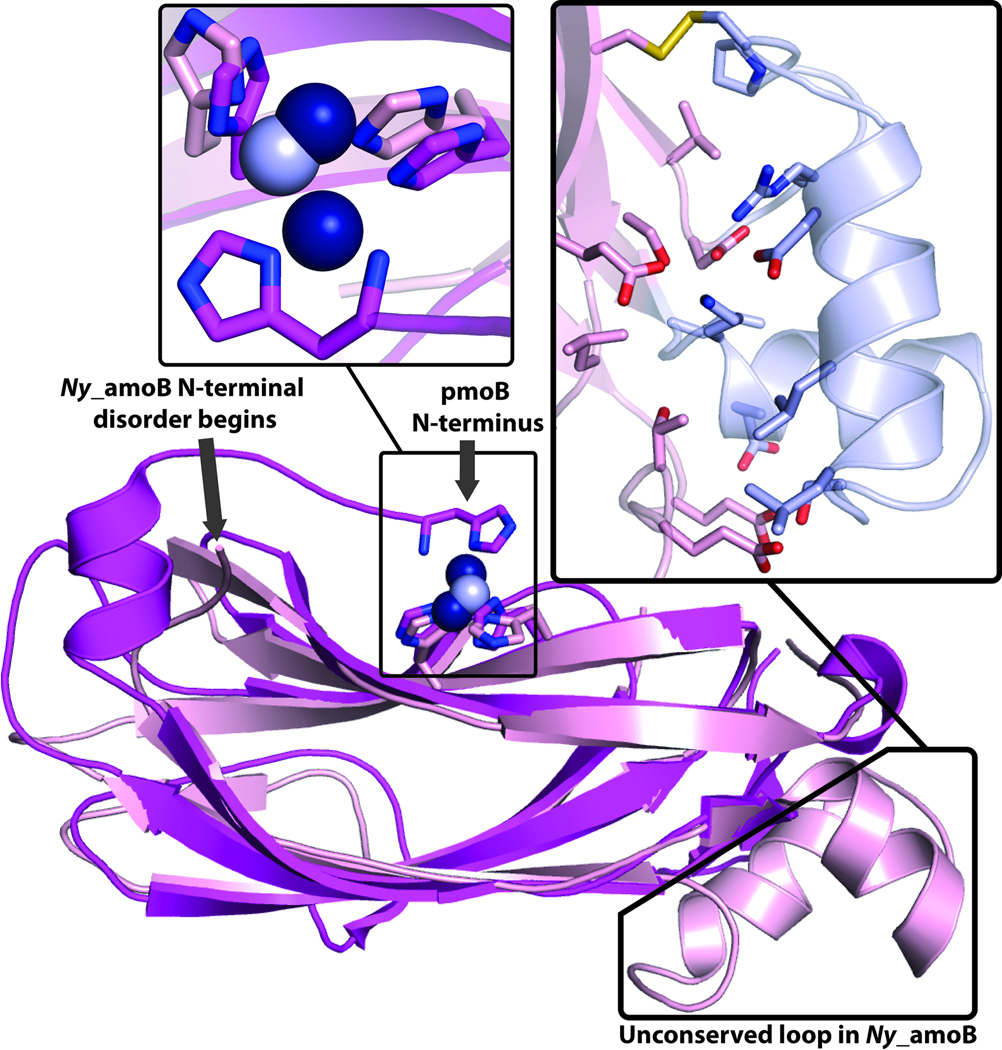
The final model includes residues 36–112 and 119 to 186, one copper ion, two sulfate ions, and 260 water molecules. 5 amino acids at the N-terminus (excluding the putative signal peptide) and 6 amino acids in the loop between β-strands 3 and 4 were not visible in the electron density map and could not be modeled. Ny_amoB is a monomer in the crystal structure as well as in solution. Comparison of the Ny_amoB sequence with that of the N-terminal cupredoxin domains of the pmoB subunits from Methylococcus capsulatus (Bath) (PDB accession code 3RGB), Methylosinus trichosporium OB3b (PDB accession code 3CHX), and Methylocystis sp. strain M (PDB accession code 3RFR) gives sequence identities of 24.1, 19.5, and 20.1%, respectively. Despite this low sequence identity, superposition of Ny_amoB onto the N-terminal cupredoxin domain of pmoB from M. capsulatus (Bath) yields an rmsd value of 1.5 Å (averaged over the three copies of pmoB in the pMMO trimer). Similar values of 1.6 Å and 1.5 Å are obtained for the M. trichosporium OB3b, and Methylocystis sp. strain M pmoB structures, respectively. The core cupredoxin domains align nearly perfectly (Fig. 2).
Figure 2.
Structure of N-terminal cupredoxin domain of N. yellowstonii amoB subunit (Ny_amoB). Center: Overall structure (pink with copper ions as gray spheres) with analogous domain of M. capsulatus (Bath) pmoB (magenta with copper ions as dark blue spheres) superimposed. Upper-left: Zoomed-in view of insertion region with residues 55–88 shown in gray and residues at interface with core domain shown as sticks. Upper-right: Zoomed-in view of copper binding sites in M. capsulatus (Bath) pmoB and Ny_amoB.
There are two regions of Ny_amoB that differ from the pmoB structures. First, the region between β strand 1 and β strand 2 formed by residues 55–88 is 30 amino acids longer than the analogous loop in pmoB (residues 61–63 in M. capsulatus (Bath) pmoB). This extended region is composed of two helices and two loop regions and interacts extensively with the core cupredoxin domain, with more than 20 hydrogen bonds and a disulfide bond between Cys 81 and Cys 146 (Fig. 2). This structural feature thus tethers all the β strands together and may impart additional stability that accounts for the thermotolerance of Ny_amoB. Interestingly, the insertion is only present in Ny_amoB sequence (Fig. 1C).
The other structural difference between Ny_amoB and pmoB occurs at the N-terminus. In all three pmoB structures, the N-terminal residue, which is a conserved histidine, is adjacent to a conserved HXH motif located in β strand 7 of the cupredoxin fold. These three histidines provide the ligands to the copper active site.13–16 In Ny_amoB, the 5 N-terminal residues could not be modeled, and a single copper ion, modeled at an occupancy of 0.7 and likely derived from the inclusion of 0.5 mM CuSO4 in the growth medium, is coordinated by His 156 and His 158 from the HXH motif (Figs. 2). The disorder at this location is due to a crystal contact, which precludes packing of the N-terminus adjacent to the HXH motif and may explain the binding of only one copper ion at less than full occupancy.
Also relevant to the apparent flexibility of the Ny_amoB N-terminus is the absence of a C-terminal cupredoxin domain. In all the pMMO structures, the pmoB N- and C-terminal cupredoxin domains interact extensively, likely stabilizing the N-terminus and positioning the histidine ligands for copper binding. It is not clear how a similar effect could be achieved in Ny_amoB. One possibility is that a conserved hypothetical transmembrane protein found in the AOA AMO gene cluster (amoHYP, Fig. 1B), but not in other CuMO gene clusters, may interact directly with amoB. AMOs, at least those from ammonia oxidizing bacteria, can oxidize methane to methanol,2 but Ny_amoB does not exhibit methane oxidation activity in contrast to previously characterized soluble domain constructs of M. capsulatus (Bath) pmoB.16 The lack of activity may be due to incomplete assembly or lability of the catalytic site as suggested by the structural comparison. Ammonia oxidation was not assayed because the presence of primary amines appeared to increase copper lability. Nevertheless, the presence of a copper binding site coordinated by the HXH motif is consistent with the location of the dicopper active site in pmoB.
In sum, the Ny_amoB structure reveals that the fold of the N-terminal domain of the B subunit in the AMO/pMMO superfamily is very well conserved as is the presence of an N-terminal copper binding site. Additional structural elements in Ny_amoB likely account for its thermotolerance, whereas the absence of a C-terminal domain may impact the stability of the copper binding site. Further studies of the intact N. yellowstonii AMO are needed to assess the physiological implications of these findings.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Grant GM070473. The authors would like to thank José R. de la Torre (San Francisco State University) for providing genomic DNA and the staff at the Life Sciences Collaborative Access Team (LS-CAT) beamlines, especially Zdzislaw Wawrzak, for assistance with data collection. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U. S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U. S. DOE under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
REFERENCES
- 1.Tavormina PL, Orphan VJ, Kalyuzhnaya MG, Jetten MSM, Klotz MG. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ Microbiol Rep. 2011;3(1):91–100. doi: 10.1111/j.1758-2229.2010.00192.x. [DOI] [PubMed] [Google Scholar]
- 2.Arp DJ, Sayavedra-Soto LA, Hommes NG. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch Microbiol. 2002;178:250–255. doi: 10.1007/s00203-002-0452-0. [DOI] [PubMed] [Google Scholar]
- 3.Hakemian AS, Rosenzweig AC. The biochemistry of methane oxidation. Ann Rev Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 4.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koops H-P, Purkhold U, Pommerening-Röser A, Timmermann G, Wagner M. The lithoautotrophic ammonia-oxidizing bacteria. Prokaryotes. 2006;5:778–811. [Google Scholar]
- 6.Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, Brochier-Armanet C, Chain PSG, Chan PP, Gollabgir A, Hemp J, Hugler M, Karr EA, Konneke M, Shin M, Lawton TJ, Lowe T, Martens-Habbena W, Sayavedra-Soto LA, Lang D, Sievert SM, Rosenzweig AC, Manning G, Stahl DA. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107(19):8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Torre JR, Walker CB, Ingalls AE, Konneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10(3):810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 8.Sayavedra-Soto LA, Hamamura N, Liu CW, Kimbrel JA, Chang JH, Arp DJ. The membrane-associated monooxygenase in the butane-oxidizing Gram-positive bacterium Nocardioides sp. strain CF8 is a novel member of the AMO/PMO family. Environ Microbiol Rep. 2011;3(3):390–396. doi: 10.1111/j.1758-2229.2010.00239.x. [DOI] [PubMed] [Google Scholar]
- 9.Coleman NV, Le NB, Ly MA, Ogawa HE, McCarl V, Lwilson N, Holmes AJ. Hydrocarbon monooxygenase in Mycobacterium: recombinant expression of a member of the ammonia monooxygenase superfamily. ISME J. 2012;6(1):171–182. doi: 10.1038/ismej.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman NV, Yau S, Wilson NL, Nolan LM, Migocki MD, Ly MA, Crossett B, Holmes AJ. Untangling the multiple monooxygenases of Mycobacterium chubuense strain NBB4, a versatile hydrocarbon degrader. Environ Microbiol Rep. 2011;3(3):297–307. doi: 10.1111/j.1758-2229.2010.00225.x. [DOI] [PubMed] [Google Scholar]
- 11.Redmond MC, Valentine DL, Sessions AL. Identification of novel methane-, ethane-, and propane-oxidizing bacteria at marine hydrocarbon seeps by stable isotope probing. Appl Environ Microbiol. 2010;76(19):6412–6422. doi: 10.1128/AEM.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki T, Nakamura T, Fuse H. Isolation of two novel marine ethylene-assimilating bacteria, Haliea species ETY-M and ETY-NAG, containing particulate methane monooxygenase-like genes. Microbes and Environments. 2012;27(1):54–60. doi: 10.1264/jsme2.ME11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman RL, Rosenzweig AC. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 14.Hakemian AS, Kondapalli KC, Telser J, Hoffman BM, Stemmler TL, Rosenzweig AC. The metal centers of particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry. 2008;47:6793–6801. doi: 10.1021/bi800598h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith SM, Rawat S, Telser J, Hoffman BM, Stemmler TL, Rosenzweig AC. Crystal structure and characterization of particulate methane monooxygenase from Methylocystis species strain M. Biochemistry. 2011;50(47):10231–10240. doi: 10.1021/bi200801z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balasubramanian R, Smith SM, Rawat S, Stemmler TL, Rosenzweig AC. Oxidation of methane by a biological dicopper centre. Nature. 2010;465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Culpepper MA, Cutsail GE, Hoffman BM, Rosenzweig AC. Evidence for oxygen binding at the active site of particulate methane monooxygenase. J Am Chem Soc. 2012;134(18):7640–7643. doi: 10.1021/ja302195p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Culpepper MA, Rosenzweig AC. Architecture and active site of particulate methane monooxygenase. Crit Rev Biochem Mol Biol. 2012;47(6):483–492. doi: 10.3109/10409238.2012.697865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Op den Camp HJ, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland N-K, Pol A, Dunfield PF. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep. 2009;1:293–306. doi: 10.1111/j.1758-2229.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Cryst. 2004;D60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 25.Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]