Abstract
Light signaling by phytochrome B in long days inhibits flowering in sorghum by increasing expression of the long day floral repressors PSEUDORESPONSE REGULATOR PROTEIN (SbPRR37, Ma1) and GRAIN NUMBER, PLANT HEIGHT AND HEADING DATE 7 (SbGHD7, Ma6). SbPRR37 and SbGHD7 RNA abundance peaks in the morning and in the evening of long days through coordinate regulation by light and output from the circadian clock. 58 M, a phytochrome B deficient (phyB-1, ma3R) genotype, flowered ∼60 days earlier than 100 M (PHYB, Ma3) in long days and ∼11 days earlier in short days. Populations derived from 58 M (Ma1, ma3R, Ma5, ma6) and R.07007 (Ma1, Ma3, ma5, Ma6) varied in flowering time due to QTL aligned to PHYB/phyB-1 (Ma3), Ma5, and GHD7/ghd7-1 (Ma6). PHYC was proposed as a candidate gene for Ma5 based on alignment and allelic variation. PHYB and Ma5 (PHYC) were epistatic to Ma1 and Ma6 and progeny recessive for either gene flowered early in long days. Light signaling mediated by PhyB was required for high expression of the floral repressors SbPRR37 and SbGHD7 during the evening of long days. In 100 M (PHYB) the floral activators SbEHD1, SbCN8 and SbCN12 were repressed in long days and de-repressed in short days. In 58 M (phyB-1) these genes were highly expressed in long and short days. Furthermore, SbCN15, the ortholog of rice Hd3a (FT), is expressed at low levels in 100 M but at high levels in 58 M (phyB-1) regardless of day length, indicating that PhyB regulation of SbCN15 expression may modify flowering time in a photoperiod-insensitive manner.
Introduction
Flowering time has a significant impact on plant adaptation to agro-ecological environments, biomass accumulation and grain yield [1]. Floral initiation is regulated by plant development, photoperiod, shading, temperature, nutrient status, and many other factors [2]–[5]. Signals from many input pathways are integrated in the shoot apical meristem (SAM) through regulation of the meristem identity genes LEAFY (LFY) and APETALA1 (AP1), which are activated during transition of the SAM from a vegetative meristem to a floral meristem. Long day (LD) plants, such as Arabidopsis, flower earlier in LD compared to short days (SD). In contrast, SD plants, such as rice and sorghum, show delayed floral initiation under LD conditions. Photoperiod regulated flowering is mediated by light signaling from photoreceptors and output from the endogenous circadian clock consistent with external coincidence models of flowering time regulation [6]. Photoperiod sensitive Sorghum bicolor genotypes delay floral initiation when grown under LD conditions. Sorghum genotypes with reduced photoperiod sensitivity have been identified and used by breeders because they flower early and at similar times in both long and short days, enhancing grain production [7]. In contrast, bioenergy sorghum is highly photoperiod sensitive, flowering in long day environments only after an extended phase of vegetative growth, thereby increasing biomass accumulation and nitrogen use efficiency [1], [8].
Photoperiod regulated flowering requires perception of light and signaling by plant photoreceptors such as the red/far-red light sensing phytochromes (Phy), blue light/ultraviolet wavelength sensing cryptochromes (Cry), phototropins, and Zeitlupes [9], [10]. Phytochromes play an important role in flowering time regulation in most plants including rice [11], barley [12], and sorghum [13]. The sorghum genome encodes three phytochrome genes, PHYA, PHYB and PHYC. Quail et al. (1994) established a standard nomenclature for phytochrome where PHY corresponds to phytochrome apoproteins, while phytochrome or phy indicates presence of the holoprotein, the fully assembled chromoprotein with chromophore covalently attached to the apoprotein [14]. Since all phytochrome proteins referred in this study are presumed to be holoproteins, Phy is used to represent wild type holoprotein, while phy is used to represent mutant versions of the holoprotein. Inactivation of PhyB results in early flowering in long days [13]. Phytochromes are soluble chromoproteins that contain an N-terminal photosensory domain and a C-terminal dimerization moiety. There are three sub-domains in the N-terminal moiety: PAS (PER, ARNT and SIM), GAF (cGMP phosphodiesterase, adenylate cyclase, Fh1A) and PHY (phytochrome-specific GAF-related), which form a unique structure, the “light-sensing knot” [15]. The PAS/GAF domains transduce light signals and the C-terminal domain, consisting of two PAS and HKRD (histidine-kinase-related domain), is responsible for dimerization and nuclear localization.
The central oscillators of the plant circadian clock are encoded by TIMING OF CAB EXPRESSION 1 (TOC1), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) [16]. Rhythmic expression of these central oscillators modulates the expression of GIGANTEA (GI), an output gene of the circadian clock. GI, in concert with other factors, activates expression of CONSTANS (CO), a zinc-finger transcription factor that plays an essential role in photoperiod regulation of flowering time in Arabidopsis [17], rice [18] and sorghum [19]. In Arabidopsis, CO is stabilized and accumulates during the evening of long days through the action of Cry1, Cry2 and PhyA, where it activates expression of FT and flowering. In SD, CO is not stabilized during the evening because CO expression occurs in darkness [20]. FT is produced in leaves and translocated to the SAM where it binds to FD. In Arabidopsis, FT together with SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1), promotes expression of meristem identity gene LFY and AP1, leading to floral transition [20].
The core of photoperiod regulatory pathway GI-CO-FT is present in Arabidopsis, a LD plant, and the SD plants rice and sorghum. In rice, OsGI, HEADING DATE 1 (Hd1), and HEADING DATE 3a (Hd3a) are orthologs of GI, CO, and FT, respectively [21]. Hd1 (OsCO) delays flowering time in LD in rice and activates flowering in SD. In addition, Itoh et al. [22] identified a pair of genes in rice, EARLY HEADING DATE 1 (EHD1) and GRAIN NUMBER, PLANT HEIGHT AND HEADING DATE 7 (GHD7) that regulate flowering in response to day length by modifying expression of Hd3a (florigen). EHD1 activates Hd3a expression and induces floral transition. In contrast, GHD7, a homolog of wheat VRN2 [23], represses flowering in LD by down-regulating EHD1 and Hd3a. In maize, 25 FT-like homologs were identified and designated as Zea mays CENTRORADIALIS (ZCN) genes. ZCN8 was identified as a source of florigen [24]. SbCN8 (ortholog of ZCN8) and SbCN12 (ortholog of ZCN12) have been proposed to encode florigens in sorghum [19], [25], [26]. In sorghum, CO activates flowering in SD by inducing expression of SbEHD1, SbCN8 and SbCN12, whereas in LD, CO activity is inhibited by SbPRR37 [19].
More than 40 flowering time QTL have been identified in sorghum [27] and maturity loci Ma1–Ma6, modify photoperiod sensitivity [7], [28], [29]. Dominance at Ma1–Ma6 delays floral initiation in long days. Ma3 encodes phytochrome B, indicating that light signaling through this photoreceptor is required for photoperiod sensitive variation in flowering time [13]. Ma6 was identified as SbGHD7, a repressor of flowering in long days [26]. In LD, SbGhd7 increases photoperiod sensitivity by inhibiting expression of the floral activators SbEHD1, SbCN12 and SbCN8. Ma1 was identified as SbPRR37, a floral repressor that acts in LD [25]. The orthologs of SbPRR37 in wheat and barley, PHOTOPERIOD 1 (Ppd1, Ppd-H1, Ppd-D1a) [30], [31] and rice OsPRR37 [32], also modulate flowering time in response to photoperiod. In LD, SbPRR37 inhibits expression of SbEHD1, SbCN12, and SbCN8, resulting in repression of flowering [25]. Moreover, SbPRR37 modulates photoperiod sensitivity and floral repression in an additive fashion together with SbGHD7 [26]. Expression of SbPRR37 and SbGHD7 is regulated by the circadian clock and light, suggesting common upstream regulation [26].
The current study focused on elucidating how phytochrome B regulates flowering time in response to day-length in sorghum. We report that PHYB is required for light activation of SbPRR37 and SbGHD7 expression in the evening of long days, resulting in repression of SbEHD1, SbCN12, SbCN8 and floral initiation.
Materials and Methods
Phenotypic analysis of sorghum flowering time
The maturity loci and flowering dates of all sorghum lines used in this study are listed in Table S1. To characterize the difference in flowering time between different genotypes and day-length, 100 M and 58 M were planted in Metro-Mix 200 (Sunshine MVP; Sun Gro Horticulture) and grown in a greenhouse in LD (14 h light/10 h dark) and SD (10 h light/14 h dark) conditions. Days to mid-anthesis were recorded and plants were photographed. 100 M plants (n = 5) and 58 M plants (n = 9) were grown in LD and phenotyped for days to anthesis (Figure 1A). The mean days to flowering for 100 M was 126 days (±4 days) and 62 days (±3 days) for 58 M, a significant difference in flowering times for these genotypes (p-value<<0.001, Welch two sample t-test). Under SD, 100 M plants (n = 7) and 58 M plants (n = 5) were used for analysis of flowering time (Figure 1B). The mean days to flowering for 100 M was 59 days (±4 days) and for 58 M, 48 days (±1 days), a significant differences in days to flowering (p-value<<0.001). To establish the interaction between PhyB and photoperiod, factorial ANOVA was run with photoperiod and PhyB alleles as factors. The significance of the effects of PhyB alleles, day-length and PhyB:day-length interaction were detected (p-value<<0.001). All statistics were run in R 3.1.0. The two-way interaction graphs were plotted using the “HH” package in R.
Figure 1. Photographs of the sorghum lines 100 M and 58 M for flowering time phenotype.
(A) Photograph of 100 M (left) and 58 M (right) grown for 109 days in LD (14 h light/10 h dark). 100 M and 58 M flowered after 126 days and 62 days respectively. (B) Photograph of 100 M (left) and 58 M (right) grown in a greenhouse in SD for 53 days (10 h light/14 h dark). 100 M flowered after 59 days and 58 M flowered after 48 days. LD: long days. SD: short days. DTF = number of days to flowering time. Scale bar is 8.6 cm.
Sequencing of PHYB alleles
To identify coding alleles in the PHYB gene, the full-length genomic sorghum PHYB genes from historical sorghum cultivars were amplified as three overlapping segments by PCR (Phusion High-Fidelity DNA polymerase, New England BioLabs, Inc). The amplified PCR products were cleaned and concentrated (QIAquick PCR Purification kit, QIAGEN). PCR products were separated by electrophoresis on 1% agarose gels. Specific PCR products were excised and purified (QIAquick Gel Extraction Kit, QIAGEN). The purified PCR products were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and the Applied Biosystems 3130xl Genetic Analyzer. All primers used for sequencing were designed using PrimerQuestSM software (Integrated DNA Technologies, Inc) and are shown in Table S2. Sequencher v4.8 (Gene Codes) was used for sequence assembly and alignment with the BTx623 whole genome sequence of Sorghum bicolor (version 1.4) downloaded from Phytozome v8.0 (http://www.phytozome.net/). The SIFT (sorting intolerant from tolerant) program (http://sift.jcvi.org/) was utilized to predict whether an amino acid substitution affects protein function, based on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences.
QTL analysis of PHYB action
The sorghum cultivar 58 M (Ma1Ma2ma3RMa4Ma5ma6) was crossed to R.07007 (Ma1ma2Ma3Ma4ma5Ma6) to generate a population for QTL analysis. F1 generation plants were self-pollinated to produce F2 populations from which F3 populations were derived by self pollination. F2 and F3 populations were planted in the greenhouse and grown under long day conditions (14 h light/10 h dark). Days to mid-anthesis of panicles of plants from the F2 and F3 populations were recorded. The median, standard error, and range of Days to Flowering and the number of plants of each genotype analyzed from the F2 and F3 populations are shown in Table3. For analysis of epistatic interaction, three-way ANOVA was run to detect the effect of allelic variation in three maturity genes (Ma3, Ma5 and Ma6) and three two-way interactions (Ma3:Ma5, Ma3:Ma6, Ma5:Ma6). The significance of the effects of single genes and genetic interactions were detected (p-value<<0.001). All statistics were run in R 3.1.0. The two-way interaction graph was plotted using the “HH” package in R.
For genotyping, genomic DNA of 86 F2 individuals and 132 F3 individuals was extracted from leaf tissue using the FastDNA Spin Kit (MP Biomedicals). Template for sequencing on an Illumina GAIIx sequencer was generated following the standard Digital Genotyping (DG) protocol [33]. Genotypes of all individuals from both populations were identified. The genetic map was constructed using the Kosambi mapping function in MAPMAKER v3.0 with 285 markers from the F2 population and 653 markers from the F3 population. QTL were mapped using the genetic map and the Composite Interval Mapping (CIM) function in WinQTL Cartographer v2.5 [34]. Significant LOD thresholds for QTL detection were calculated based on experiment specific permutations with 1000 permutations and α = 0.05 [35].
Gene Expression Assays
Sorghum genotypes 100 M and 58 M were planted and grown in a greenhouse under long day conditions (14 h light/10 h dark) for 32 days and then transferred to growth chambers under either LD (14 h light/10 h dark) or SD (10 h light/14 h dark) conditions for seven days for entrainment prior to collection of leaf tissue. In the growth chamber, daytime (lights on) temperature was set at 30°C with a light intensity of ∼300 µmol·s−1·m−2 and night (lights off) temperature was set at 23°C. Relative humidity was ∼50% throughout the experiment. At day 39, leaf segments from the top three expanded leaves from three individual plants of each genotype and treatment were collected every 3 hours through one 24 h light-dark cycle and 48 h of continuous light. The leaf tissues at each time point were subjected to total RNA extraction using TRI Reagent (MRC) with the protocol for samples with high levels of polysaccharides. RNA was further purified using the RNeasy Mini kit (QIAGEN), including removal of DNA contamination by on-column DNase I digestion before reverse transcription. RNA integrity was examined on 1% MOPS gels. First-strand cDNA synthesis was performed using the SuperScript III First-Strand Synthesis System (Invitrogen) with oligo dT and random hexamer primer mix. After first-strand cDNA synthesis, the reactions were diluted to 10 ng/µl of the initial total RNA. Gene-specific qPCR reactions were carried out using Power SYBR Green PCR Master Mix (Applied Biosystems). 18S rRNA was selected as the internal control reference and the reactions were performed using the TaqMan Universal PCR Master Mix Protocol with rRNA Probe (VIC Probe) and rRNA Forward/Reverse Primer. All reactions were run on the 7900HT Fast Real-Time PCR System with SDS v2.3 software (Applied Biosystems). The specificity of each gene specific primer set was validated by melting temperature curve analysis. Amplification efficiency of each primer sets was determined by the serial dilution method [36] (Table S3). Relative expression was determined by the comparative cycle threshold (ΔΔCt) method [36] with calibration from most highly expressed samples. The calculated primer efficiencies were used to adjust data for relative quantification by the efficiency correction method [37]. Each relative expression value was derived from an average of three technical replicates and three biological replicates. The individual expression data points presented as 2−ΔCt [38]. The significance (p-values) of the difference in expression between genotypes were detected using Welch two sample t-test in R 3.1.0 based on three technical replicates and three biological replicates. P-values were calculated either for certain time points of the day or all time points of the day.
Results
PHYB alleles in diverse sorghum lines
Sorghum genotype 58 M, a photoperiod insensitive early flowering line, has the genotype ma3Rma3R, corresponding to the phyB-1 allele [13]. This allele contains a frame shift mutation that results in a prematurely terminated PhyB lacking regions of the protein necessary for dimerization and biological activity. To confirm and extend prior analysis of PHYB diversity in sorghum, alleles from several sorghum lines that vary in photoperiod sensitivity were sequenced and compared. The coding sequence of PHYB from BTx623 and 100 M (both Ma3) was 7285 bp in length consisting of four exons encoding a protein with 1178 amino acid residues. PHYB sequences from R.07007, Hegari, Tx7000, BTx642, SC56, Shallu and BTx3197 were identical to BTx623 and 100 M (Ma3). The PHYB sequence from 58 M (ma3R), referred to as phyB-1 (Table 1), contains a mutation that renders the gene inactive [13]. No coding mutations were identified in 90 M, a line that encodes the weak allele ma3 [28]. IS3620C encodes a different allele, designated phyB-2, which differs from PHYB by one INDEL and two SNPs, resulting in one amino acid deletion and two amino acid substitutions (Table 1). The first substitution in phyB-2 could alter function because it produces an Asp308Gly change in the GAF domain of PhyB. The SIFT prediction score of this Asp308Gly substitution is 0.1, indicating moderate intolerance.
Table 1. Sequence analysis of PHYB coding alleles in different sorghum lines.
| Exon 1 | Exon 1 | Exon 3 | Exon 4 | Sorghum Genotypes | |
| Nucleotide Variation | CAC>… | A>G | A>. | C>G | |
| Protein Modification | His>… | Asp>Gly | Premature stop codon | Leu>Val | |
| Mutation Position (AA #) | 31 | 308 | 1023 | 1113 | |
| Alignment with PHYB inArabidopsis (AA #) | 32 | 293 | 1007 | 1096 | |
| Phytochrome Domain | GAF(N) | ||||
| PHYB ( Ma3 or ma3 ) | − | − | − | − | BTx623, 100 M, 90 M, R.07007, Hegari, Tx7000, BTx642, SC56, Shallu, BTx3197 |
| phyB-1 ( ma3R ) | − | − | + | − | 58 M |
| phyB-2 | + | + | − | + | IS3620C |
PhyB affects flowering time in LD and SD
The sorghum maturity standards, 100 M and 58 M, were constructed from Milo genotypes that contain alleles of Ma1 and Ma3 that modify flowering time [28]. The sorghum maturity standard 100 M is photoperiod sensitive with a maturity genotype Ma1Ma2Ma3Ma4Ma5ma6 [26]. The genotype 58 M is photoperiod insensitive, flowers early in LD and SD, and has the genotype Ma1Ma2ma3RMa4Ma5ma6 [26]. Genotype 58 M contains null alleles of Ma3 (ma3R, phyB-1) and Ma6 (ghd7-1). When grown in a greenhouse under 14 h LD during the summer, 58 M plants were spindly and flowered in ∼62 days (±3 days), whereas 100 M flowered in ∼126 days (±4 days) due to the repressing action of SbPRR37 (Ma1) (Figure 1A). This result confirmed that loss of PhyB activity in 58 M reduces the ability of Ma1 to inhibit flowering in LD (p-value<<0.001) [13]. When grown in a greenhouse in 10 h SD during December–February at lower light intensity, 100 M flowered in ∼59 days (±4 days) while 58 M flowered in ∼48 days (±1 days) (Figure 1B). Therefore in sorghum, PhyB has a smaller but still significant effect on flowering time in SD (p-value<<0.001). The factorial ANOVA with photoperiod and PHYB alleles as factors indicated the effects of PhyB, day-length and PhyB:day-length interaction are all significant (p-value<<0.001) (Figure S1-A).
PHYB is epistatic to Ma1 (SbPRR37) and Ma6 (SbGHD7)
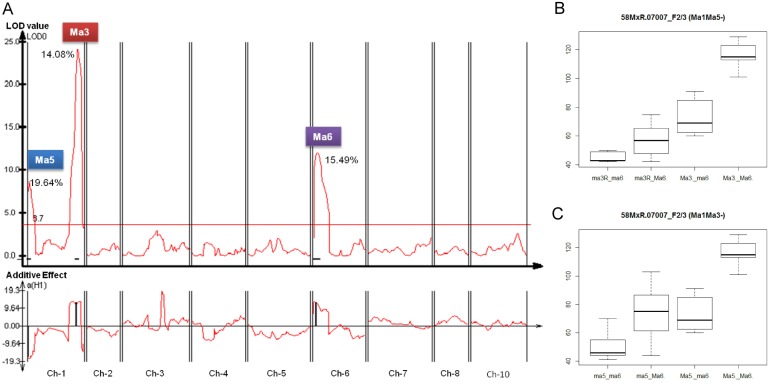
In sorghum, SbPRR37 (Ma1) and SbGHD7 (Ma6) are primary determinants of photoperiod sensitivity in Ma3 backgrounds acting in an additive fashion to inhibit flowering in LD [26]. Expression of both genes is induced by light, although the photoreceptor or photoreceptors that mediate light signaling were not known prior to the current study [25], [26]. To examine how PHYB (Ma3), SbPRR37 (Ma1), and SbGHD7 (Ma6) co-regulate the timing of floral initiation, F2 and F3 populations were derived from a cross of R.07007 (Ma1Ma3ma5Ma6) and 58 M (Ma1ma3RMa5ma6). These populations segregated for a wide range of flowering times (∼85 days) when planted in July and grown in a greenhouse in 14 h LD. Digital genotyping [33] was employed to generate DNA markers for genetic map construction. The genetic map spanned all of the ten sorghum chromosomes, although the long arms of SBI02 and SBI09 in its entirety were deficient in DNA markers. QTL analysis identified three significant QTL (LOD score>3.7) for days to anthesis in LD using the F2 population (n = 86), which together explained ∼50% of the phenotypic variance for flowering time (Figure 2A). The QTL with the highest LOD score (LOD = 24.2), spanned DNA on chromosome 1 from 60,402,909–61,604,749 bp which encompasses PHYB (chromosome_1∶60,915,677–60,917,553) (Table 2). Recessive ma3R alleles from 58 M associated with this QTL caused early flowering time phenotypes. The flowering time QTL on chromosome 6 spanning a physical interval from 203,707–1,716,581 bp (1 LOD interval) aligned with SbGHD7 [26]. The recessive ghd7-1 null allele from 58 M was associated with early flowering in LD. The third flowering time QTL near the proximal end of chromosome 1 (chromosome 1:6,139,583–9,077,991) had a LOD score of 8.7 and explained 19.6% percent of phenotype variance. This QTL was tentatively identified as Ma5 because R.07007 was reported to be recessive for Ma5, a rare allele in sorghum [29]. No QTL aligned with Ma1 as expected because both 58 M and R.07007 contain dominant alleles of Ma1 (SbPRR37). The three flowering time QTL were also identified in the corresponding F3 population (data not shown).
Figure 2. Flowering time QTL and analysis of epistasis in populations derived from 58MxR.07007.
(A) Flowering time QTL labeled Ma3, Ma5 and Ma6, were identified through analysis of flowering time variation in LD in the F2 population derived from 58MxR.07007. LOD values are shown on the Y-axis and sorghum chromosome numbers on the X-axis. The percent of the variance explained by each QTL is noted. The additive plot is shown in the lower portion of 2A where a positive value corresponds to alleles from R.07007 that delay flowering time. (B) Boxplot of flowering time distribution in the subset of the population with Ma1Ma5- genotypes but varying for alleles of Ma3/ma3R and Ma6/ma6. (C) Boxplot of flowering time distribution in the subset of the population having Ma1Ma3- genotypes but varying for Ma5/ma5 and Ma6/ma6. Median values for flowering time are represented by horizontal lines within boxes.
Table 2. Information on flowering time QTL identified in the 58MxR.07007 F2 population.
| QTL | Maturity Locus | Chromosome Number | Position (cM)a | LOD score | Physical Intervalb | Additive Effectc | Dominant Effectd | R2e |
| 1 | Ma5 | Ch_1 | 1.8 | 8.66 | 6139583–9077991 | −17.09 | 18.19 | 0.1964 |
| 2 | Ma3 | Ch_1 | 99.4 | 24.21 | 60402909–61604749 | 12.55 | 16.09 | 0.1408 |
| 3 | Ma6 | Ch_6 | 7.2 | 12.09 | 203707–1716581 | 12.83 | 5.81 | 0.1549 |
| Total | 49.21% |
Position of likelihood peak (highest LOD score).
Physical Interval: physical coordinate interval spanning 1 LOD interval across the likelihood peak.
Additive Effect: A positive value means the delay of flowering time due to R.07007 allele. A negative value means the delay of flowering time due to 58 M allele.
Dominant Effect: A positive value means dominance for the delay of flowering time.
R2 (coefficient of determination): percentage of phenotypic variance explained by the QTL.
Plants from the F2/3 population are homozygous for Ma1, a repressor of flowering in LD, but varied in alleles of Ma3, Ma5 and Ma6. Three-way ANOVA was used to analyze the effect of allelic variation in three maturity genes (Ma3, Ma5 and Ma6) on flowering time, and three two-way interactions (Ma3:Ma5, Ma3:Ma6, Ma5:Ma6) showed that allelic variation of the three Ma genes and three two-way interactions were significant (p-values<<0.001). The three two-way interaction graphs between Ma3:Ma5, Ma3:Ma6 and Ma5:Ma6 are shown in Figure S1-B–D. Progeny with the genotypes Ma3_Ma5_Ma6_ and Ma3_Ma5_ma6ma6 flowered later than genotypes that were homozygous recessive for ma3R, showing that PHYB is epistatic to the floral repressors encoded by Ma1 and/or Ma6 (Figure 2B; Figure S1-B,C). Progeny with the genotype Ma3_Ma5_Ma6_ (101–129 days) flowered later than plants with the genotype Ma3_Ma5_ma6ma6 (60–91 days), consistent with increased floral repression due to Ma6 in Ma1 dominant backgrounds. The effect of Ma6 was delay flowering with varying extents in different genetic backgrounds ranging from 14 days in ma3Rma3RMa5_, ∼29 days in Ma3_ma5ma5, and ∼9 days in ma3Rma3Rma5ma5. Furthermore, it was noted that progeny lacking PhyB with a dominant Ma6 allele showed a significant range of flowering times (42–75 days), suggesting that additional genes and/or environmental factors affect Ma6 action in this genetic background (Figure 2B; Table 3). A similar wide range of flowering time (59 days) was observed among plants with the genotype Ma3_ma5ma5Ma6_ (Figure 2C; Table 3). In addition, plants with the genotype Ma3_Ma5_Ma6_ flowered later in LD than plants with the genotypes Ma3ma5ma5Ma6_ or Ma3ma5ma5ma6ma6 (Figure 2C; Figure S1-D). This shows that Ma5 is also required for late flowering in LD in Ma1Ma3 backgrounds and that Ma5 is epistatic to Ma1 and Ma6. Plants with the genotype ma3Rma3RMa5_ma6ma6 and Ma3_ma5ma5ma6ma6 flowered early and in a similar number of days as genotypes that are homozygous recessive for both ma3R and ma5 (ma3Rma3Rma5ma5ma6ma6) indicating that the products of both Ma3 and Ma5 are required in LD for delayed flowering mediated by Ma1 (SbPRR37).
Table 3. Flowering time of F2/F3 progeny from 58MxR.07007 in LD.
| Genotype (All plants = Ma1Ma1) | Days to Flowering: median (±SE) | Days to Flowering: range | Number of plants | ||
| Ma3_ | Ma5_ | Ma6_ | 115 (±5) | 101–129 | 42 |
| Ma3_ | Ma5_ | ma6ma6 | 69 (±8) | 60–91 | 19 |
| ma3Rma3R | Ma5_ | Ma6_ | 57 (±8) | 42–75 | 15 |
| ma3Rma3R | Ma5_ | ma6ma6 | 43 (±2) | 42–50 | 6 |
| Ma3_ | ma5ma5 | Ma6_ | 75 (±12) | 44–103 | 52 |
| Ma3_ | ma5ma5 | ma6ma6 | 46 (±6) | 41–70 | 30 |
| ma3Rma3R | ma5ma5 | Ma6_ | 53 (±6) | 42–76 | 24 |
| ma3Rma3R | ma5ma5 | ma6ma6 | 44 (±6) | 39–68 | 17 |
The requirement for both PhyB and the product of Ma5 to observe delayed flowering in LD led us to examine the Ma5 locus for candidate genes that might explain this interaction. The Ma5 locus is located on SBI-01 and spans a large number of genes including several genes known to affect flowering time in other plants, including AP1, CK2, and PHYC. PHYC appeared to be the best candidate gene for Ma5 because PhyC modifies flowering time in rice specifically in LD, similar to Ma5 in sorghum [39], PhyB stabilizes PhyC, and PhyB:PhyC act as heterodimers in both Arabidopsis [40], [41] and rice [39], consistent with the co-dependence observed between PHYB and Ma5 in this study. Comparison of PHYC sequences from BTx623 (Ma5), 100 M (Ma5), and R.07007 (ma5) revealed four differences in PhyC amino acid sequence between BTx623 and R.07007, and two differences between 58 M/100 M and R.07007 (Table 4). The latter amino acid variants occur in the PAS domain (Gly:Val) and HKRD domain (Glu:Asp) and SIFT analysis [42] indicated these changes could affect the function of PhyC. These results are consistent with PHYC as the candidate gene for Ma5. Further analysis is underway to test this assignment.
Table 4. Sequence analysis of PHYC coding alleles in different sorghum lines.
| Exon 1 | Exon 1 | Exon 1 | Exon 2 | Sorghum Genotypes | |
| Nucleotide Variation | G>T | G>A | T>C | G>T | |
| Protein Modification | Gly>Val | Gly>Arg | Val>Ala | Glu>Asp | |
| Mutation Position (AA #) | 124 | 162 | 190 | 922 | |
| Alignment with PHYBin Arabidopsis (AA #) | 160 | 198 | 226 | 954 | |
| Phytochrome Domain | PAS(N) | PAS(N) | PAS-GAF Loop | HKRD(C) | |
| PHYC-1 ( Ma5 ) | − | − | − | − | BTx623 |
| PHYC-2 | − | + | + | − | 100 M, 90 M |
| phyC-1 ( ma5 ) | + | + | + | + | R.07007 |
PhyB modulates expression of SbPRR37 and SbGHD7 in long days
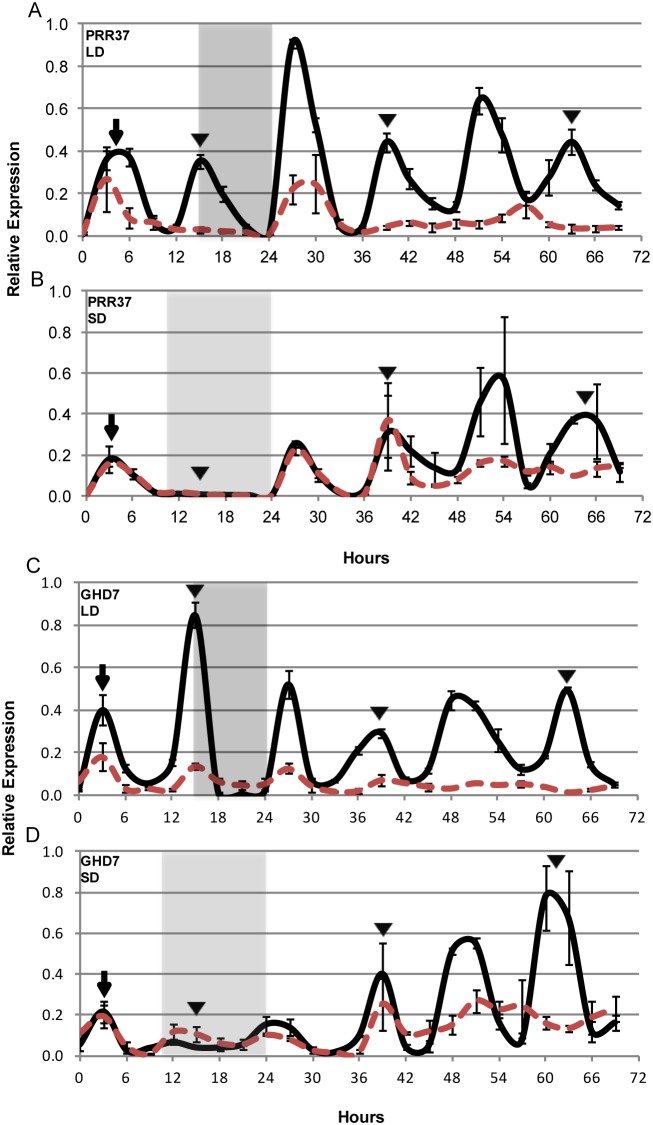
Expression of SbPRR37 and SbGHD7 in leaves is regulated by light and gating by the circadian clock [25], [26]. The influence of PhyB on SbPRR37 and SbGHD7 expression was analyzed using 100 M (PHYB) and 58 M (phyB-1) plants grown for 32 days in LD then entrained for 7 days in LD or SD (Figure 3). Following entrainment, leaf samples were collected from plants for one 24 h LD or SD light-dark cycle, then from plants exposed to continuous light and temperature for an additional 48 h. In leaves of 100 M, SbPRR37 and SbGHD7 expression peaked in the morning (arrow) and evening (arrowhead) in LD as previously reported [25], [26] (Figure 3A/C, solid lines). SbPRR37 and SbGHD7 RNA abundance continued to oscillate with peaks in the morning and evening when 100 M plants were transferred to continuous light and temperature consistent with regulation by the circadian clock (Figure 3, 24–72 h). In leaves of 58 M in LD (Figure 3A/C, dashed red lines), SbPRR37 and SbGHD7 showed an increase in RNA abundance in the morning (arrow) but only a small increase in expression in the evening (arrowhead) compared to 100 M (Figure 3A, p-value<0.1; Figure 3C, p-value<0.05). These results indicate that PhyB is required for elevated evening expression of SbPRR37 and SbGHD7 in LD in 100 M.
Figure 3. Relative expression of SbPRR37 and SbGHD7 in 100 M (Ma3/PHYB) and 58 M (ma3R/phyB-1) in LD and SD.
100 M (solid black line) and 58 M (dashed red line) plants were entrained LD (14 h light/10 h dark) or SD (10 h light/14 h dark) and sampled for one 24 h cycle, followed by 48 h in LL (continuous light and temperature). The grey background corresponds to time when plants are in darkness. Relative gene expression was determined every 3 hours by qRT-PCR. Arrows represent morning peaks of expression and arrowheads represent evening peaks of expression. (A) In LD, the second peak (arrowhead) of SbPRR37 expression in the evening (∼15 h) is missing in the phyB deficient line, 58 M. (B) In SD, the second peak (arrowhead) of SbPRR37 is absent in both 100 M and 58 M. (C) In LD, the second peak (arrowhead) of SbGHD7 expression in the evening (∼15 h) is attenuated in 58 M. (D) In SD, the second peak of SbGHD7 is attenuated in both 100 M and 58 M. Each data point of relative expression was based on data from three technical replicates and three biological replicates. Error bars indicate SEM.
When 100 M and 58 M plants were entrained and assayed in SD, the morning peak of SbPRR37 expression was of similar amplitude in both genotypes and expression of SbPRR37 was low during the evening (Figure 3B). Similarly, SbGHD7 expression in SD was highest in the morning, reaching similar levels in 100 M and 58 M, and lower in the evening when compared to expression levels measured in LD (Figure 3D). These results indicate that in SD, PhyB has a limited effect on SbPRR37 and SbGHD7 expression. When 100 M plants entrained in SD were exposed to continuous light, the evening peak of SbPRR37 and SbGHD7 expression observed in LD reappeared on the first subjective day and expression levels were also elevated in the second subjective day (Figure 3B/D). In 58 M, the evening peak of SbPRR37 and SbGHD7 reappeared during the first subjective day, however overall expression was attenuated relative to 100 M during the second subjective day.
PhyB modulates expression of CO, Ehd1, SbCN8, SbCN12 and SbCN15
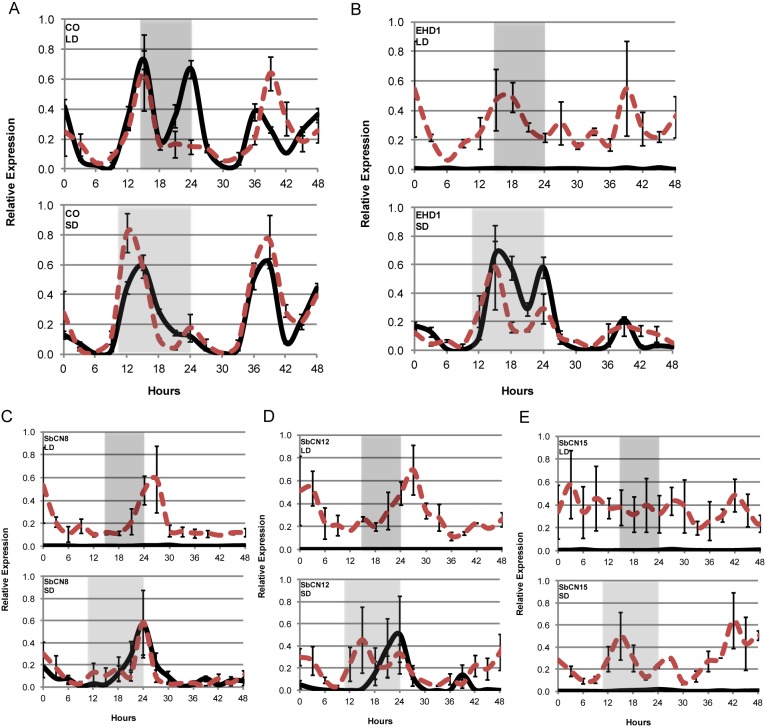
In 100 M entrained to LD, the sorghum ortholog of CONSTANS (SbCO) shows peaks of expression at dawn (24 h) and in the evening (15 h) that are regulated by SbPRR37, the circadian clock, and day length [25]. In 58 M entrained and sampled in LD, the amplitude of the peak of SbCO expression at dawn (24 h) was reduced compared to 100 M (Figure 4A, p-value<0.05). The peak of SbCO expression at dawn was also reduced and of similar amplitude in plants entrained and sampled in SD (Figure 4A, lower). These results show that the peak of SbCO expression at dawn is dependent on PhyB, most likely because expression of SbPRR37 in the evening of LD is dependent on PhyB (Figure 3A). In contrast, the evening peak (15 h) of SbCO expression was similar in both LD and SD in 100 M and 58 M indicating that PhyB does not significantly modulate SbCO expression at this time (15 h) of day.
Figure 4. Expression of SbCO, SbEhd1, SbCN8/12/15 in 100 M (Ma3/PHYB) and 58 M (ma3R/phyB-1) in LD and SD.
Relative RNA levels in leaves of 100 M (solid black lines) and 58 M (dashed red lines) entrained and sampled in LD (14 h light/10 h dark) or SD (10 h light/14 h dark) for 24 h followed by 24 h in LL (continuous light and temperature). Relative expression levels were determined every 3 hours by qRT-PCR analysis. The gray shaded areas represent the dark periods. (A) SbCO, (B) SbEHD1, (C) SbCN8, (D) SbCN12, (E) SbCN15. Each data point of relative expression is based on three technical replicates and three biological replicates. Error bars indicate SEM.
EHD1 is an activator of Hd3a, one of the florigens in rice [43]. The sorghum ortholog of Hd3a is SbCN15. Expression of SbEHD1 increases when 100 M is transferred from LD to SD in parallel with increased expression of SbCN8 (ortholog of ZCN8 [24]) and SbCN12 (ortholog of ZCN12) that have been proposed to encode florigens in sorghum [19], [25], [26]. SbPRR37 and SbGHD7 repress expression of SbEHD1 in 100 M entrained in LD [25], [26]. Therefore SbEHD1 expression in 58 M and 100 M was quantified and compared to determine if PhyB modulates SbEHD1 expression. In LD, SbEHD1 RNA abundance peaked in the evening and was up to ∼100-fold higher in 58 M relative to 100 M throughout the time course (Figure 4B, upper; Figure S2-A, p-value<<0.001). In SD, expression of SbEHD1 was high in both genotypes and peaked during the night (Figure 4B, lower; Figure S2-A).
In 58 M entrained and analyzed in LD, expression of SbCN8 (Figure 4C, upper) and SbCN12 (Figure 4D, upper) peaked early in the morning and the relative abundance of RNA derived from these genes was elevated more than ∼100-fold relative to their levels in 100 M (Figure S2-B/C, p-values<<0.001). In SD, SbCN8 (Figure 4C, lower) and SbCN12 (Figure 4D, lower) expression was similar in both genotypes. Similarly, SbCN15 (Hd3a) expression was increased up to ∼60-fold in 58 M compared to 100 M in LD and SD (Figure 4E; Figure S2-D, p-values<<0.001) at all time points assayed, indicating that PhyB mediated repression of SbCN15 expression occurs regardless of photoperiod.
PhyB could be inducing SbPRR37 and SbGHD7 expression directly, and/or indirectly by altering output from the circadian clock. To determine if allelic variation in PHYB affected clock gene expression, TOC1 and LHY/CCA1, the central oscillators, and GI, a mediator of clock output were examined (Figure S3). In LD and SD, TOC1, LHY and GI expression in 58 M and 100 M peaked at similar times and most of these genes showed similar amplitude of expression, although expression of GI was approximately 2-fold lower in 58 M. Although three biological replications at the indicated time points may not be sufficient to detect all biologically significant variation present, the small fold differences of circadian clock genes do not appear sufficient to explain the large variation in SbPRR37 and SbGHD7 expression observed in Ma3 vs. ma3R backgrounds. PHYB and PHYC RNA levels were similar in 100 M and 58 M plants in LD and SD (data not shown).
Discussion
Sorghum genotypes used for grain production are typically photoperiod insensitive and flower in 55–75 days when planted in April in locations such as College Station, Texas where day lengths increase during the early portion of the growing season. Early flowering in grain sorghum helps avoid adverse weather and insect pressure during the reproductive phase, thereby enhancing yield. In contrast, highly photoperiod sensitive energy sorghum genotypes planted in this same location will not initiate flowering for 175 days until mid-September when day lengths decrease to less than 12.2 h [1], [29]. Delayed flowering results in long duration of vegetative growth of energy sorghum, increasing biomass yield [8] and nitrogen use efficiency [8]. The importance of optimal flowering time for sorghum productivity led us to investigate the genetic and molecular basis of variation in this trait in sorghum.
Variation of flowering time of sorghum germplasm grown in LD environments is caused principally by differences in photoperiod sensitivity, although shading, GA, temperature, length of the juvenile phase among other factors also affect this trait [7]. A model summarizing information about photoperiod regulation of flowering time in sorghum is shown in Figure 5. In LD, flowering is delayed in photoperiod sensitive sorghum by the additive action of the floral repressors, SbPRR37 (Ma1) and SbGhd7 (Ma6) [25], [26], [28], [29]. SbPRR37 and SbGhd7 repress expression of the grass specific floral activator, SbEHD1. In addition, SbPRR37 inhibits the activity of CO, another activator of flowering in sorghum [19]. The floral activators, SbEhd1 and SbCO, induce expression of SbCN8 and SbCN12, the proposed sources of FT in sorghum. SbCN15, the ortholog of Hd3a and a source of florigen in rice [21], may also be a source of florigen in sorghum. The circadian clock is shown regulating expression of SbGI, SbCO, SbPRR37 and SbGHD7, and light regulating expression of SbGHD7 and SbPRR37 as shown in previous studies [25], [26].
Figure 5. Model of the photoperiod flowering time pathway in sorghum.
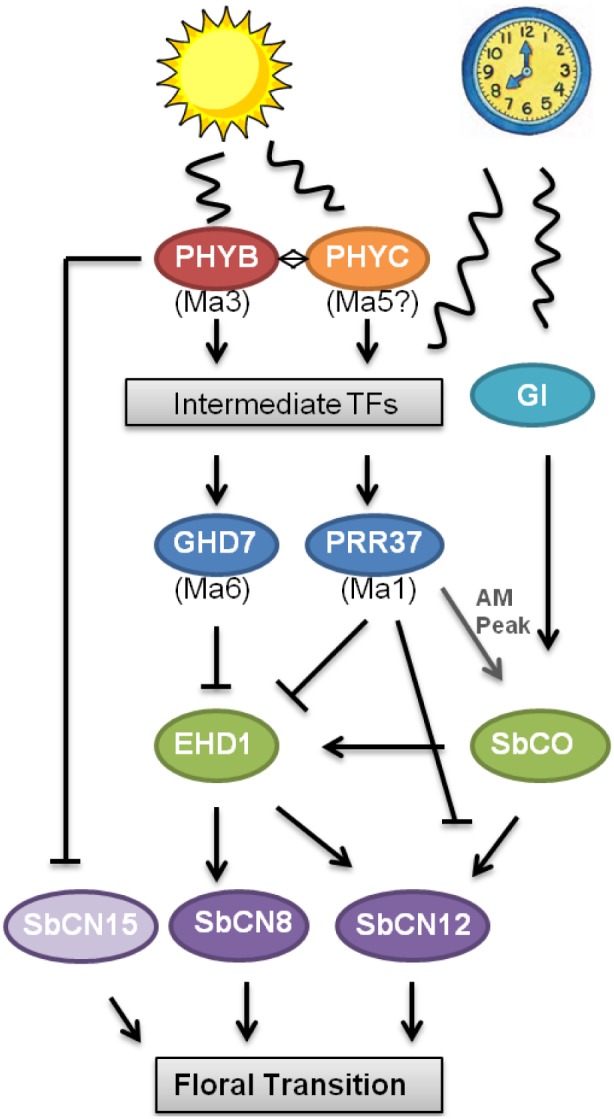
Phytochrome B (PhyB) is mediates light signaling that modulates flowering time in response to photoperiod in sorghum. In LD, PhyB up-regulates the expression of PRR37 and GHD7, two central floral repressors, during the evening phase of LD but with minimal influence in SD. Induction at this time of day is also dependent on output from the circadian clock. PhyB may stabilize and interact with PhyC, a candidate gene for Ma5 a locus that also contributes to photoperiod regulation of flowering time. SbPRR37 activates SbCO expression peaking at dawn. SbPRR37 and SbGhd7 repress expression of the floral inductors SbEHD1, SbCN8, SbCN12 and SbCN15, leading to delayed flowering in long days. In SD or 58 M (phyB-1), expression of the floral repressors SbPRR37 and SbGHD7 is reduced which results in floral initiation once plants have satisfied other requirements for flowering. PhyB was found to mediate repression of SbCN15 regardless of day length.
Photoperiod has minimal impact on flowering time in sorghum genotypes such as SM100 that encode null versions of SbPRR37 and SbGHD7 [25], [26]. Presence of functional alleles of either gene increases photoperiod sensitivity and a further delay in flowering is observed when both genes are present in dominant Ma3Ma5 backgrounds. Expression of SbPRR37 and SbGHD7 is regulated by light and the circadian clock. Both genes show peaks of RNA abundance in the morning and again in the evening in LD and both peaks of RNA are attenuated in darkness. Importantly, the evening peak of expression is attenuated in SD when this phase occurs in darkness, indicating a requirement for light signaling during the evening to maintain sufficiently high levels expression of SbPRR37 and SbGHD7 to inhibit flowering. The morning and evening peaks of SbPRR37 and SbGHD7 expression observed in sorghum in LD is a pattern of expression first observed in photoperiod versions of this C4 grass. In Arabidopsis, PRR7, the ortholog of SbPRR37, shows a single peak of clock-regulated expression during the morning [44]. In rice, SbGHD7 shows a single peak of clock-gated expression in the morning of LD [22]. It will be interesting to determine if the dual peak pattern of PRR37 and GHD7 expression observed in sorghum is found in other related C4 grasses such as pearl millet, Miscanthus and sugarcane.
The current study focused on characterizing the light-signaling pathway that regulates SbPRR37 and SbGHD7 expression in response to day length. Previous studies showed that sorghum genotypes lacking PHYB (58 M, phyB-1) flower earlier in LD compared to near isogenic genotypes (100 M) expressing PHYB, demonstrating that light signaling through this photoreceptor is required for photoperiod sensitive variation in flowering time [13]. The current study showed that PhyB (Ma3) is epistatic to genes encoding the floral repressors SbPRR37 and SbGhd7 and that PhyB is required for photoperiod-regulated expression of these genes. Moreover, 58 M, a genotype lacking functional PhyB, showed attenuated expression of SbPRR37 and SbGHD7 during the evening of LD compared to 100 M (PhyB). In SD, expression of the floral repressors was similar in 58 M and 100 M. Taken together, these results indicate that in sorghum PhyB is required for light signaling in LD that results in elevated expression of SbPRR37 and SbGHD7 during the evening.
The molecular basis of PhyB induced expression of SbPRR37 and SbGHD7 during the evening of long days is unknown but could involve other photoreceptors and intermediary transcription factors such as PIFs [45]. Detailed studies in rice showed that PhyA, PhyB and PhyC modulate flowering time [39]. PhyC in particular plays a role in natural variation of flowering time in pearl millet [46], Arabidopsis [47], and wheat [48]. In Arabidopsis, a long day plant, PhyB destabilizes CO, an action countered by Cry, PhyA and SPA in LD, leading to floral induction [20]. In rice, phyB mutants flower early in LD and SD similar to sorghum. Interestingly, rice phyC mutants flower early only in LD [39]. In addition, in rice, both PhyB and PhyC are required to induce GHD7 expression, where PhyB alone causes some repression of GHD7 mRNA levels [49]. This indicates that in rice PhyB regulates floral induction in both LD and SD, while PhyC modifies flowering time selectively in LD. The stability of PhyC is reduced in the absence of PhyB in rice and Arabidopsis [40]. PhyB increases PhyC stability, and chromophore-containing PhyB:PhyC heterodimers are required for PhyC activity [41]. Therefore, in sorghum the requirement for PhyB in photoperiod sensitive flowering time may be because PhyB increases PhyC stability and through formation of PhyB:PhyC heterodimers.
Genetic analysis of the role of PHYB in sorghum was examined using a population dominant for Ma1 (SbPRR37) and segregating for alleles of PHYB (Ma3), Ma5, and SbGHD7 (Ma6). The presence of Ma1 in all progeny of the population caused delayed flowering in LD unless the expression or activity of Ma1 (and in some genotypes Ma1 and Ma6) was altered by recessive alleles of Ma3 or Ma5. The analysis showed that plants homozygous for null alleles of PHYB (phyB-1) in Ma5_ backgrounds had reduced photoperiod sensitivity and flowered earlier in LD compared to plants encoding PhyB. Similarly, progeny homozygous for recessive alleles of Ma5, in Ma3_ backgrounds, showed reduced photoperiod sensitivity and flowered earlier in LD. The results indicated that both PHYB and Ma5 are epistatic to Ma1 and Ma6. Progeny recessive for either gene flowered earlier in LD, but showed a range of flowering times, indicating that other genes and/or environmental factors affected flowering time in these backgrounds, although with reduced response to photoperiod. Interestingly, PHYB and Ma5 appear to be co-dependent or acting at a similar point in the regulatory pathway because allelic differences at Ma5 did not affect flowering time significantly in phyB-1 backgrounds and vice versa. R.07007 (Ma3ma5) and 58 M (ma3RMa5) show attenuated expression of SbPRR37 and SbGHD7 in the evening of LD ([25] and this study) indicating that both Ma3 (PhyB) and Ma5 are required for elevated expression of the sorghum floral repressors during the evening of LD. In searching for an explanation for this co-dependence, we found the Ma5 locus spans several genes known to affect flowering time including PHYC and that the sequence of PhyC in R.07007 (ma5) contained amino acid changes that could potentially modify the function of this protein. The hypothesis that Ma5 corresponds to PHYC is consistent with studies showing that PhyC modifies flowering in an LD specific manner in rice, similar to Ma5 [39]. In addition, PhyC stability is dependent in part on PhyB and PhyC activity requires the formation of functional heterodimers with PhyB (and other phytochromes) [41]. If sorghum PhyC is regulated by PhyB in a manner similar to their counterparts in rice, this would explain why Ma5 (presumptive PHYC) activity is not observed in phyB-1 backgrounds. Experiments designed to test this hypothesis are currently underway.
In Arabidopsis, CO expression peaks once per day in the evening and the amplitude of CO expression is regulated by blue light/GI-FKF1-ZTL mediated turnover of CDF1, a repressor of CO expression [50]. PRR7 also modifies CO expression through repression of CDF1 expression [51]. In sorghum, SbCO expression peaks twice each day, at dawn and again in the evening in LD. The peak of SbCO expression at dawn is attenuated in SD ([25] and this study) and in genetic backgrounds lacking SbPRR37 [19]. It is possible that SbPRR37 modulates SbCO expression by repressing sorghum orthologs of CDF1 as occurs in Arabidopsis [51]. The peak of SbCO expression at dawn in LD was not observed in the sorghum genotype lacking PhyB (58 M). Since PhyB is required for elevated SbPRR37 expression in the evening of LD, and SbPRR37 has been shown to induce elevated expression of SbCO at dawn, it is likely that lack of PhyB induced expression of SbPRR37 during the evenings of LD explains the observed expression of SbCO in 58 M.
In rice, Hd3a, a member of the PEBP gene family, encodes an FT protein that acts as a florigen [52]. In maize, ZCN8 and possibly ZCN12 are sources of florigen [24], [53]. Sorghum encodes orthologs of Hd3a (SbCN15), ZCN8 (SbCN8) and ZCN12 (SbCN12). SbCN8 and SbCN12 expression is regulated by day length and by alleles of SbPRR37, SbGHD7, and PHYB in a manner consistent with these genes being sources of florigen in sorghum. In prior studies, SbCN15 expression was modulated to only a small extent by variation in photoperiod and in mutants of SbPRR37 and SbGHD7 that affect flowering time, suggesting that this gene was not an important target of photoperiod regulation [25], [26]. In the current study, expression of SbCN15 was found to be ∼60-fold higher in leaves of 58 M (phyB-1) compared to 100 M (PHYB) in both LD and SD. If SbCN15 functions as a source of florigen as in rice, photoperiod independent repression of SbCN15 expression by PhyB suggests that this gene may be responsible for early flowering induced by shading [7]. 58 M plants exhibit shade avoidance responses including longer leaf blades and sheaths, fewer tillers, narrower leaf blades, less leaf area, and more rapid stem elongation [7]. In Arabidopsis, light signaling through PhyB represses shade avoidance responses, and PhyB deficient mutants have elongated stems and an early flowering phenotype associated with “constitutive shade avoidance” [54]. Information on photoperiod regulated flowering time in sorghum described in this paper will hopefully facilitate analysis of flowering time variation caused by shading and other environmental factors.
Supporting Information
ANOVA interaction graphs showing (A) Day-length:PhyB (Day:Genotype) interaction. (B–D) Three two-way interactions (Ma3:Ma5, Ma3:Ma6, Ma5:Ma6) in the 58MxR.07007 F2/F3 population.
(TIF)
Fold differences of SbEHD1, SbCN8, SbCN12 and SbCN15 RNA abundance at peaks of expression in 100 M and 58 M grown in LD (14 h light/10 h dark) or SD (10 h light/14 h dark). Positive fold difference values indicate higher mRNA levels detected in 58 M. (A) SbEHD1, (B) SbCN8, (C) SbCN12, (D) SbCN15. The time point corresponding to peak expression is shown below each graph.
(TIF)
Relative expression levels of circadian clock genes and GI in 100 M (black solid line) and 58 M (red dashed line) under either LD (14 h light/10 h dark) or SD (10 h light/14 h dark) conditions. The gray shaded area represents the dark period. The first 24 h covers one light-dark cycle, followed by 24 h of continuous light. (A) GI. (B) TOC1. (C) LHY. Each data point of relative expression corresponds to three technical replicates and three biological replicates. Error bars indicates SEM.
(TIF)
Genotypes and flowering dates of sorghum lines.
(DOCX)
Primer sequences used for PHYB alleles amplification and sequencing.
(DOCX)
Primer sequences and amplification efficiency for qRT-PCR.
(DOCX)
Acknowledgments
The authors would like to thank Susan Hall for help with plant management.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by Pioneer Hi-Bred International, Inc., Ceres Inc., and the Perry Adkisson Chair in Agricultural Biology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rooney WL, Blumenthal J, Bean B, Mullet JE (2007) Designing sorghum as a dedicated bioenergy feedstock. Biofuels, Bioproducts and Biorefining 1: 147–157. [Google Scholar]
- 2. Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welch SM, Dong Z, Roe JL (2004) Modelling gene networks controlling transition to flowering in Arabidopsis; 2004 26 Sep–1 Oct Brisbane, Australia. CDROM. 1–20.
- 4. Greenup A, Peacock WJ, Dennis ES, Trevaskis B (2009) The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann Bot 103: 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andres F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639. [DOI] [PubMed] [Google Scholar]
- 6. Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, et al. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. [DOI] [PubMed] [Google Scholar]
- 7.Morgan PW, Finlayson SA (2000) Physiology and Genetics of Maturity and Height. In: Smith CW, Frederiksen RA, editors. Sorghum: Origin, History, Technology, and Production. New York: Wiley Series in Crop Science. 240–242.
- 8. Olson SN, Ritter K, Rooney W, Kemanian A, McCarl BA, et al. (2012) High biomass yield energy sorghum: developing a genetic model for C4 grass bioenergy crops. Biofuels, Bioproducts and Biorefining 6: 640–655. [Google Scholar]
- 9. Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230. [DOI] [PubMed] [Google Scholar]
- 10.Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-Regulated Plant Growth and Development. Current Topics in Developmental Biology: Elsevier Inc. 29–66. [DOI] [PubMed]
- 11. Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, et al. (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16: 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hanumappa M, Pratt LH, Cordonnier-Pratt MM, Deitzer GF (1999) A photoperiod-insensitive barley line contains a light-labile phytochrome B. Plant Physiol. 119: 1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Childs KL, Miller FR, Cordonnier-Pratt MM, Pratt LH, Morgan PW, et al. (1997) The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol. 113: 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quail PH, Briggs WR, Chory J, Hangarter RP, Harberd NP, et al. (1994) Spotlight on Phytochrome Nomenclature. Plant Cell 6: 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nagatani A (2010) Phytochrome: structural basis for its functions. Curr Opin Plant Biol 13: 565–570. [DOI] [PubMed] [Google Scholar]
- 16. Pruneda-Paz JL, Kay SA (2010) An expanding universe of circadian networks in higher plants. Trends Plant Sci 15: 259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robson F, Costa MM, Hepworth SR, Vizir I, Pineiro M, et al. (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J 28: 619–631. [DOI] [PubMed] [Google Scholar]
- 18. Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, et al. (2000) Hd1, a Major Photoperiod Sensitivity Quantitative Trait Locus in Rice, Is Closely Related to the Arabidopsis Flowering Time Gene CONSTANS. Plant Cell 12: 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang S, Weers B, Morishige D, Mullet J (2014) CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biology 14: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594. [DOI] [PubMed] [Google Scholar]
- 21. Tsuji H, Taoka K, Shimamoto K (2011) Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol 14: 45–52. [DOI] [PubMed] [Google Scholar]
- 22. Itoh H, Nonoue Y, Yano M, Izawa T (2010) A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet 42: 635–638. [DOI] [PubMed] [Google Scholar]
- 23. Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, et al. (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meng X, Muszynski MG, Danilevskaya ON (2011) The FT-like ZCN8 Gene Functions as a Floral Activator and Is Involved in Photoperiod Sensitivity in Maize. Plant Cell 23: 942–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, et al. (2011) Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci U S A 108: 16469–16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy RL, Morishige DT, Brady JA, Rooney WL, Yang S, et al.. (2014) Ghd7 (Ma6) Represses Flowering in Long Days: A Key Trait in Energy Sorghum Hybrids. The Plant Genome In press.
- 27. Mace ES, Hunt CH, Jordan DR (2013) Supermodels: sorghum and maize provide mutual insight into the genetics of flowering time. Theor Appl Genet 126: 1377–1395. [DOI] [PubMed] [Google Scholar]
- 28.Quinby JR (1974) Sorghum improvement and the genetics of growth. College Station: Texas A&M Univ. Press.
- 29. Rooney W, Aydin S (1999) Genetic control of a photoperiod-sensitive response in Sorghum bicolor (L.) Moench. Crop Science 39: 397–400. [Google Scholar]
- 30. Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115: 721–733. [DOI] [PubMed] [Google Scholar]
- 31. Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034. [DOI] [PubMed] [Google Scholar]
- 32. Koo BH, Yoo SC, Park JW, Kwon CT, Lee BD, et al. (2013) Natural Variation in OsPRR37 Regulates Heading Date and Contributes to Rice Cultivation at a Wide Range of Latitudes. Mol Plant 6: 1877–1888. [DOI] [PubMed] [Google Scholar]
- 33. Morishige DT, Klein PE, Hilley JL, Sahraeian SM, Sharma A, et al. (2013) Digital genotyping of sorghum - a diverse plant species with a large repeat-rich genome. BMC Genomics 14: 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Basten CJ, Zeng Z-B (2012) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC.
- 35. Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bookout AL, Mangelsdorf DJ (2003) Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal 1: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nature Protocols 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 39. Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, et al. (2005) Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17: 3311–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Monte E, Alonso JM, Ecker JR, Zhang Y, Li X, et al. (2003) Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 15: 1962–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clack T, Shokry A, Moffet M, Liu P, Faul M, et al. (2009) Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21: 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 43. Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, et al. (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18: 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, et al. (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vigouroux Y, Mariac C, De Mita S, Pham JL, Gerard B, et al. (2011) Selection for earlier flowering crop associated with climatic variations in the Sahel. PLoS One 6: e19563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, et al. (2006) The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet 38: 711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Distelfeld A, Dubcovsky J (2010) Characterization of the maintained vegetative phase deletions from diploid wheat and their effect on VRN2 and FT transcript levels. Mol Genet Genomics 283: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Osugi A, Itoh H, Ikeda-Kawakatsu K, Takano M, Izawa T (2011) Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol 157: 1128–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297. [DOI] [PubMed] [Google Scholar]
- 51. Imaizumi T (2010) Arabidopsis circadian clock and photoperiodism: time to think about location. Curr Opin Plant Biol 13: 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. [DOI] [PubMed] [Google Scholar]
- 53. Danilevskaya ON, Meng X, Hou Z, Ananiev EV, Simmons CR (2008) A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol 146: 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ANOVA interaction graphs showing (A) Day-length:PhyB (Day:Genotype) interaction. (B–D) Three two-way interactions (Ma3:Ma5, Ma3:Ma6, Ma5:Ma6) in the 58MxR.07007 F2/F3 population.
(TIF)
Fold differences of SbEHD1, SbCN8, SbCN12 and SbCN15 RNA abundance at peaks of expression in 100 M and 58 M grown in LD (14 h light/10 h dark) or SD (10 h light/14 h dark). Positive fold difference values indicate higher mRNA levels detected in 58 M. (A) SbEHD1, (B) SbCN8, (C) SbCN12, (D) SbCN15. The time point corresponding to peak expression is shown below each graph.
(TIF)
Relative expression levels of circadian clock genes and GI in 100 M (black solid line) and 58 M (red dashed line) under either LD (14 h light/10 h dark) or SD (10 h light/14 h dark) conditions. The gray shaded area represents the dark period. The first 24 h covers one light-dark cycle, followed by 24 h of continuous light. (A) GI. (B) TOC1. (C) LHY. Each data point of relative expression corresponds to three technical replicates and three biological replicates. Error bars indicates SEM.
(TIF)
Genotypes and flowering dates of sorghum lines.
(DOCX)
Primer sequences used for PHYB alleles amplification and sequencing.
(DOCX)
Primer sequences and amplification efficiency for qRT-PCR.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.