Abstract
Our investigations show that nonlethal concentrations of nitric oxide (NO) abrogate the antibiotic activity of β-lactam antibiotics against Burkholderia pseudomallei, Escherichia coli and nontyphoidal Salmonella enterica serovar Typhimurium. NO protects B. pseudomallei already exposed to β-lactams, suggesting that this diatomic radical tolerizes bacteria against the antimicrobial activity of this important class of antibiotics. The concentrations of NO that elicit antibiotic tolerance repress consumption of oxygen (O2), while stimulating hydrogen peroxide (H2O2) synthesis. Transposon insertions in genes encoding cytochrome c oxidase-related functions and molybdenum assimilation confer B. pseudomallei a selective advantage against the antimicrobial activity of the β-lactam antibiotic imipenem. Cumulatively, these data support a model by which NO induces antibiotic tolerance through the inhibition of the electron transport chain, rather than by potentiating antioxidant defenses as previously proposed. Accordingly, pharmacological inhibition of terminal oxidases and nitrate reductases tolerizes aerobic and anaerobic bacteria to β-lactams. The degree of NO-induced β-lactam antibiotic tolerance seems to be inversely proportional to the proton motive force (PMF), and thus the dissipation of ΔH+ and ΔΨ electrochemical gradients of the PMF prevents β-lactam-mediated killing. According to this model, NO generated by IFNγ-primed macrophages protects intracellular Salmonella against imipenem. On the other hand, sublethal concentrations of imipenem potentiate the killing of B. pseudomallei by NO generated enzymatically from IFNγ-primed macrophages. Our investigations indicate that NO modulates the antimicrobial activity of β-lactam antibiotics.
Author Summary
β-lactam drugs that inhibit peptidoglycan biosynthesis are often used in the treatment of bacterial infections, including melioidosis. Independent of their antibiotic activity, we have noted that submicromolar concentrations of β-lactams potentiate the killing of intracellular B. pseudomallei supported by NO generated by IFNγ-primed macrophages. The production of NO can nonetheless be a double-edged sword, as indicated by our observations that sublethal concentrations of nitric oxide (NO), a diatomic radical produced by phylogenetically diverse organisms to regulate neurotransmission, vascular tone and host defense, tolerize B. pseudomallei, nontyphoidal Salmonella and E. coli against the antimicrobial activity of β-lactams. Accordingly, NO produced in the inflammatory response of macrophages protects nontyphoidal Salmonella against β-lactam antibiotics. NO mediates bacterial tolerance to β-lactam antibiotics by inhibiting the electrochemical gradient supported by terminal cytochrome oxidases of the respiratory chain, rather than by decreasing oxidative stress as previously thought.
Introduction
B. pseudomallei are endemic in tropical areas of Southeast Asia, Northern Australia and equatorial countries [1]. This Gram-negative, opportunistic pathogen is a saprophyte that inhabits water and soil, becoming infectious to humans and animals if inoculated through cutaneous abrasions, ingested in contaminated food and water, or inhaled through the respiratory mucosa. Melioidosis can present as an acute, chronic or latent infection [2]. Pneumonia accounts for about 50% of all the cases of B. pseudomallei infection [3], [4], whereas septic shock, often a fulminant complication of septicemia, kills 40% of melioidosis patients receiving therapy and 95% of those untreated.
Despite recent advances in antibacterial therapy, management of melioidosis remains a challenge [4]. Antibacterial treatment of melioidosis often spans 20 weeks and requires combined antibiotic therapy. Ceftazidime is often used in the intensive phase, whereas trimethoprim-sulfamethoxazole (TMP-SMX) is used during the eradication phase of treatment [5]. Regardless of intense and vigorous treatment regimes, about 10% of melioidosis patients suffer from relapses [6]. B. pseudomallei are intrinsically resistant to most classes of antibacterials [7]. For example, B. pseudomallei growing in biofilms are phenotypically tolerant to doxicycline, ceftazidime, imipenem and TMP-SMX [8], [9]. The efflux pumps BpeAB-OprB, BpeEF-OprC and AmrAB-OprA further increase the resistance of this opportunistic pathogen to β-lactams, aminoglycosides, macrolides, fluoroquinolones, chloramphenicol and polymyxins [10]–[12]. Class A and D β-lactamases add to the arsenal of enzymatic systems that protect B. pseudomallei against ampicillin, carbenicillin, ceftazidime and imipenem [13]–[15]. In addition to these well-characterized mechanisms of antibiotic resistance, changes in bacterial physiology in response to host environmental conditions may promote resistance to antibiotics. For example, anaerobiosis, which is normally attained in the hepatic, splenic and prostate abscesses of melioidosis patients, induces a population of B. pseudomallei remarkably refractory to several classes of clinically important antibacterials [16].
In addition to being an intrinsic component of the antimicrobial arsenal of vertebrate hosts [17], the signaling properties of NO have been co-opted by prokaryotic and eukaryotic organisms. NO produced endogenously by bacterial NO synthase protects Bacillus subtilis against a wide spectrum of antibiotics [18]. This adaptive response of Bacillus might lessen the bactericidal activity of antibiotics produced by saprophytic microorganisms populating the soil. Modification of drugs and potentiation of antioxidant defenses have been evoked as mechanisms underlying the NO-induced antibiotic resistance of Bacillus [18]. NO produced in the inflammatory response has also been shown to shield Gram-positive and –negative bacteria against the antimicrobial activity of bactericidal antibiotics. Salmonella enterica survives exposure to members of the aminoglycoside family in response to the NO generated intracellularly by IFNγ-activated macrophages [19], a situation that had previously been noted for Listeria with ampicillin [20]. Given the recently described role of NO in inducing resistance of phylogenetically diverse bacteria to different classes of antibiotics and the recent controversy attributing oxidative stress as the mechanism of action of bactericidal antibiotics [18], [21]–, we tested whether NO generated chemically or enzymatically modifies the antimicrobial activity of β-lactams against B. pseudomallei and two representative members of the enterobacteriaceae family.
Methods
Bacterial strains and growth conditions
Strain K96243, a clinical isolate of B. pseudomallei [24], was grown in the BSL3 laboratory of the Department of Microbiology at the University of Colorado School of Medicine. This facility has been certified by the CDC for work with select agents. E. coli strain 3110 and S. enterica serovar Typhimurium strain 14028 s were also used in the course of these investigations. Where indicated, Salmonella strains AV0468, AV07140 and AV07141 deficient in the flavohemoprotein hmp, acetate kinase ackA or phosphotransacetylase pta, respectively, were used. The bacteria were grown overnight to stationary phase in LB broth supplemented with 4% (v/v) glycerol (LBG) at 37°C and 315 RPM in a shaker incubator (New Brunswick Innova, Edison, NJ). Where indicated, the bacteria were grown to log phase to OD600 of 0.6.
Susceptibility of B. pseudomallei to antibiotics
Log phase B. pseudomallei was grown from overnight cultures in a shaker incubator at 37°C in LBG broth to an OD600 of 0.6. Log and stationary phase B. pseudomallei cultures were diluted to OD600 of 0.012 in 1 ml of LBG broth in 14 ml polypropylene tubes containing sterile stirrer magnets. The killing activity of imipenem was assessed in bacterial cultures at the indicated concentrations. The tubes were loosely capped and placed on a magnetic stirrer in a 37°C cell culture incubator. The anti-B. pseudomallei activity of ceftazidime was tested in 250 ml flasks as previously described [16]. Both antibiotics were purchased from Sigma-Aldrich, St. Louis, MO. Selected cultures were co-treated with spermine NONOate or DETA NONOate, which generate NO with half-lives of 39 min and 20 h, respectively, at 37°C, pH 7.4. In selected experiments the susceptibility of log phase E. coli and Salmonella was also tested. Where indicated, E. coli and Salmonella were grown in EG medium [i.e., E salts (0.2 g/L MgSO4, 2 g/L C6H8O7-H2O, 10 g/L K2HPO4, 3.5 g/L Na(NH4)HPO4-4H2O) supplemented with 0.4% glucose]. The number of surviving bacteria after antibiotic treatment was determined after culture on LB agar plates, and the fraction of bacteria that survived antibiotic treatment was calculated as (cfu tn/cfu t0)×100.
Construction of the transposon mutant pool and deep-sequencing analysis
A mini-Mariner transposon [25], [26] expressed from the suicide plasmid pTBurk1 was electroporated into B. pseudomallei strain K96243. This plasmid contains the Himar1 transposable element with a kanamycin cassette flanked by inverted repeats (IR). The Himar1 transposase was chosen because of its TA dinucleotide specificity. Bacteria with an integrated kanamycin cassette were selected on LB agar plates containing 50 µg/ml kanamycin and 100 µg/ml zeocin. The sequencing libraries were quantified using the Agilent Bioanalyzer DNA7500 chip, multiplexed, cluster amplified, and sequenced on the Illumina MiSeq platform. Sequencing reads containing the Himar1 IR sequence and the adjacent TA were isolated from the raw fastq file. The IR sequence was removed from the analysis. The processed reads were mapped onto the B. pseudomallei K96243 reference genome using the program Bowtie2 with local alignment settings and a k value of 1 [27]. Annotation of TA sites was accomplished using seqanno, a custom set of Python scripts (https://github.com/brwnj/seqanno) used to characterize specific genomic sequences. The publicly available code was used to quantify reads over a given sequence, annotate those counts at the gene level, compare results between samples, and annotate using a UniProt flat file for B. pseudomallei.
Determination of endogenous H2O2 synthesis
B. pseudomallei grown overnight in LBG broth were subcultured 1∶100 in LBG broth at 37°C with shaking. The generation of H2O2 by B. pseudomallei was measured in stationary phase bacteria diluted to an OD600 of 0.5. Selected bacterial cultures were treated for 1 h at 37°C with 12.5 µg/ml imipenem in the presence or absence of 100 µM spermine NONOate or 500 µM KCN. The cultures were continuously agitated with a magnetic stir-bar. The specimens were placed into a sealed, temperature-controlled chamber (World Precision Instruments, Inc., Sarasota, FL) containing a small magnetic stir bar. To prevent artifacts associated with a possible loss of viability, the experiments were carried out for 1 h after exposure to imipenem before the onset of killing took place. The H2O2 accumulated in the bacterial cultures after 1 h incubation was measured pollarographically for about 2 min using an ISO-H2O2 sensor attached to an APOLLO 4000 free radical analyzer (World Precision Instruments). The concentration of H2O2 produced by the bacterial cultures was calculated by regression analysis of a standard curve generated with known concentrations of H2O2.
O2 consumption
B. pseudomallei was grown overnight in LBG broth at 37°C with shaking. Overnight cultures were diluted to OD600 of 0.5 in a volume of 1 ml. Where indicated, 100 µM spermine NONOate or 500 µM KCN were added to the bacterial cultures. The samples were transferred to a multiport temperature-controlled chamber, and the consumption of O2 by the bacteria was measured over a 5 min period using an ISO-OXY-2 O2 sensor attached to an APOLLO 4000 free radical analyzer. To ensure the even distribution of gases in the chamber, the samples were placed on a chamber containing a magnetic stir-bar. The data are expressed as µM of O2.
Nitrate reductase activity
Nitrate reductase enzymatic activity was monitored by measuring the accumulation of NO2 − in the cultures. Bacterial pellets of B. pseudomallei grown overnight in LBG broth diluted to an OD600 of 0.6 were moved into the anaerobic chamber, where they are aliquoted into 1 ml volumes in LBG broth or LBG broth supplemented with 50 mM NaNO3 −. The O2 in the LBG broth had been eliminated by culturing the media in the anaerobic chamber for at least 24 h. The bacterial cells were allowed to reduce NO3 − to NO2 − for 2.5 h. NO2 − concentrations were measured spectrophotometrically at 550 nm after mixing with an equal volume of Griess reagent (0.5% sulfanilamide and 0.05% N-1-naphthylethylenediamide hydrochloride in 2.5% phosphoric acid). NO2 − concentrations were calculated by regression analysis using standard curves prepared with NaNO2.
Proton motive force
The membrane potential of S. Typhimurium grown in LB broth and EG medium to OD600 of 0.5 was measured with the fluorescent probe DiSC3(5) (Molecular Probes, Eugene, OR). The pellet of 1 mL of cells grown to log phase in LB broth or EG medium was resuspended in 5 mM HEPES, pH 7.2, supplemented with 5 mM casamino acids or 5 mM glucose, respectively. Samples were treated in 1 ml aliquots with 750 µM spermine NONOate for 15 minutes at 37°C. DiSC3(5) was added to a final concentration of 1 µM from a stock solution made in DMSO. DiSC3(5) was allowed to equilibrate in the cells before fluorescence measurements were collected in a Synergy 2 microtiter plate reader (BioTek, Winooski, VT) using excitation and emission wavelengths of 590 and 680 nm, respectively.
Macrophage assays
J774 murine macrophage-like cells (clone ATCC TIB-67) were grown in RPMI medium supplemented with 10% fetal bovine serum (BioWhittaker, Walkersville, MD), 15 mM Hepes, 2 mM L-glutamine, 1 mM sodium pyruvate (Sigma-Aldrich, St. Louis, MO), and 100 U·ml−1/100 mg·ml−1 of penicillin/streptomycin (Cellgro). The macrophages were treated with 200 U/ml of recombinant murine IFNγ (Peprotech, Rocky Hill, NJ) 16 h before infection. The macrophages were infected for 2.5 h with B. pseudomallei at an MOI of 4, after which the media was exchanged with RPMI+ containing 350 µg/ml kanamycin. One hour later, B. pseudomallei-infected cells were incubated for 4 h in fresh culture media containing 250 µg/ml kanamycin. The media was then replaced with fresh media containing increasing concentrations of imipenem in the presence or absence of 500 µM of the iNOS inhibitor aminoguanidine. In parallel experiments, macrophages were infected for 25 min with S. Typhimurium at an MOI of 2. Extracellular Salmonella were killed after treatment for 1 h with 50 µg/ml gentamicin. The Salmonella-infected macrophages were then incubated in fresh RPMI media containing 10 µg/ml gentamicin in the presence or absence of aminoguanidine, which was maintained in the culture media for the rest of the experiment. After 8 h, the Salmonella-infected cells were washed and fresh media containing imipenem was added to the cultures. The B. pseudomallei and Salmonella burden in the cultures was determined 12–14 h after exposure to imipenem. The amount of nitrite, a terminal oxidative product of NO, synthesized by the macrophages was estimated by the Griess reaction.
Statistical analysis
The data were analyzed using a Student's paired t test. Determination of statistical significance between multiple comparisons was achieved using one-way analysis of variance (ANOVA) followed by a Bonferroni post-test. Data were considered statistically significant when p<0.05.
Results
NO protects B. pseudomallei from antibiotics that target peptidoglycan biosynthesis
The β-lactam antibiotic imipenem has been used in the clinic to treat people with melioidosis [7], [28]. Under the experimental conditions tested, stationary B. pseudomallei did not grow 2.5 h after subculture in LB broth supplemented with 4% glycerol (figure 1A). Despite this lack of growth, 1 µg/ml imipenem reduced the viability of stationary phase B. pseudomallei by ∼1,000-fold (figure 1B). These findings contrast with those reported earlier by Eng et al who found poor antimicrobial activity of imipenem against nongrowing bacteria [29]. Differences in bacterial species might account for these discrepancies. As expected, imipenem effectively killed log phase B. pseudomallei (figure 1B), a population that double in numbers 2.5 h after culture in fresh LBG broth (figure 1A). Together, our investigations indicate that imipenem can be equally efficient at killing both replicating and non-replicating B. pseudomallei. Our investigations also indicate that the imipenem-dependent inhibition of both peptidoglycan remodeling in stationary phase bacteria and de novo peptidoglycan biosynthesis in growing B. pseudomallei can exert profound antimicrobial activity. The NO donor spermine NONOate, which has a half-life of 39 min, was used to test whether this diatomic radical abrogates the antimicrobial activity of imipenem. Given that B. pseudomallei is extraordinarily susceptible to the antimicrobial activity of NO [30], spermine NONOate was titrated in order to find conditions in which the viability of B. pseudomallei was not affected upon NO treatment. The addition of 100–200 µM spermine NONOate, which generates 2 moles of NO per mole of parent compound, failed to kill B. pseudomallei under the experimental conditions used in the course of these investigations. The concentrations of NO used in these experiments effectively inhibited growth of log phase B. pseudomallei (not shown). The addition of 100 µM spermine NONOate completely abrogated killing by imipenem against both stationary and log phase B. pseudomallei (figure 1C). We noticed that the colonies of B. pseudomallei treated simultaneously with imipenem and NO took even longer to grow that those of NO-treated controls, indicating that NO does not prevent imipenem from poisoning penicillin-binding proteins in the cell wall. The protective effects appear to be explained by the NO released by spermine NONOate and not the polyamine base, since spermine did not affect the imipenem-dependent killing of B. pseudomallei (not shown).
Figure 1. Effect of NO on the anti-B. pseudomallei activity of β-lactam antibiotics.
Growth of log and stationary phase B. pseudomallei in LBG broth 2.5 h after culture (A). B. pseudomallei grown to log (log) or stationary (sta) phase were treated for 2.5 h with increasing concentrations of imipenem (B). Panel C shows the effect of 100 µM spermine NONOate (sNO) on the anti-B. pseudomallei activity of 25 µg/ml imipenem (IM). The protection afforded by 2.5 mM DETA NONOate (dNO) against 64 µg/ml ceftazidime (CT) is shown in D. The data are the mean ± SD from 3 observations collected in 2 separate days. p<0.001 compared to the antibiotic-treated group.
We also tested the effects of NO on the anti-B. pseudomallei activity of ceftazidime, which is the β-lactam antibiotic of choice in the acute phase of treatment of melioidosis [31]. Ceftazidime failed to kill B. pseudomallei under the same experimental conditions under which imipenem exerted profound bactericidal activity (not shown). Therefore we adopted a system of long-term exposure to a high concentration of ceftazidime that has been shown to sustain cytotoxicity against the seemingly resistant strain of B. pseudomallei used in our studies [16]. B. pseudomallei was effectively killed 6 h after the addition of 64 µg/ml ceftazidime (figure 1D). We used this in vitro culture system to test the effects of NO on ceftazidime-mediated killing of B. pseudomallei. To ensure long-term release of NO, these investigations made use of the slow NO donor DETA NONOate, which has an estimated half-life of 20 h at 37°C, pH 7.4. The addition of 2.5 mM DETA NONOate abrogated most of the anti-B. pseudomallei activity associated with ceftazidime treatment (figure 1D). Together, these findings indicate that chemically-generated NO protects B. pseudomallei against β-lactam antibiotics.
NO induces antibiotic tolerance
The following experiments were performed in order to determine whether NO tolerizes Burkholderia against the cytotoxic actions of antibiotics or stimulates long-lasting genetic resistance. Imipenem was used to test these two models because 1) this β-lactam antibiotic is endowed with potent anti-Burkholderia activity, 2) imipenem-mediated killing occurs within a few hours of exposure, and 3) NO induces excellent protection against this drug. Two independent experimental approaches tested whether NO tolerizes Burkholderia or induces long-lasting genetic resistance. First, 100 µM spermine NONOate was added to the cultures 1 h after exposure to 25 µg/ml imipenem. As seen with Burkholderia co-exposed to NO and imipenem, the addition of NO 1 h after imipenem treatment abrogated killing of stationary phase B. pseudomallei (figure 2A). Second, bacterial cultures were pretreated with spermine NONOate and imipenem for 1 h and then washed by centrifugation. The bacterial cells were then resuspended with fresh LBG broth containing 25 µg/ml imipenem. Again, these cultures were as protected as cultures receiving NO and imipenem during the full 2.5 h of challenge. These findings indicate that the protective actions afforded by NO are immediate and can occur after the bacteria have been exposed to imipenem. To test the duration of the protective effects associated with NO treatment, B. pseudomallei were treated with NO for 1 h, and then placed in fresh media containing 12.5 µg/ml imipenem for up to 5 additional hours. The protective effects associated with NO treatment were lost over time (figure 2B). For instance, imipenem killed more than 99.99% of the bacteria in the population 5 h after NO was removed from the cultures. Cumulatively, our investigations indicate that the protective effects afforded by NO against imipenem are transitory and are best observed in cells actively undergoing nitrosative stress.
Figure 2. NO tolerizes B. pseudomallei to the antimicrobial activity of imipenem.
(A) All bacteria in groups I-III were treated with 25 µg/ml imipenem for 2.5 h. B. pseudomallei in group I were treated with 100 µM spermine NONOate (sNO) 1 h after exposure to imipenem; sNO was removed from group II after 1 h of treatment; sNO was maintained in group III during the 2.5 h of challenge. The survival of control bacteria (C) treated with 25 µg/ml of imipenem is shown for comparison. Panel B shows the fraction of B. pseudomallei that survived 25 µg/ml imipenem at the indicated times. Selected specimens (dark symbols) were treated with 100 µM sNO during the first hour of exposure to imipenem. The data are the mean ± SEM from 5 observations collected on 2 separate days. p<0.001 compared to groups I–III.
Identification of loci associated with NO-induced tolerance to imipenem
To identify loci that may be associated with the NO-induced tolerance to imipenem, a B. pseudomallei mutant library was constructed using a mini-mariner transposon with an insertion specificity for a TA dinucleotide [25], [26]. Sequencing of genomic DNA at the transposon-chromosome junctions allowed us to determine the coverage of the library. Overall, the library consists of 35,075 independent clones encompassing 28,543 and 6,532 intragenic and intergenic insertions, respectively. On average, each gene harbors 3–4 independent transposons. Disrupted genes were defined as those that sustained 6 or more sequence-reads per site within the internal 5–80% of gene length. Essential genes in Burkholderia were defined as those sustaining fewer than 6 sequence-reads within 3–97% of the open reading frame length. We estimate that 590 genes are essential for growth of B. pseudomallei under the experimental conditions tested (table S1). The estimated essential genes encode functions such as DNA replication, chromosome maintenance, lipid metabolism, translation, cell division, and energy and nucleic acid metabolism (figure S1).
We used this transposon library to identify transposon mutants with increased resistance to imipenem. The transposon library was treated for 2.5 h with 12.5 µg/ml imipenem and/or 750 µM spermine NONOate at an OD600 of 0.5 in LBG broth. The specimens were then subcultured in 25 ml of fresh LBG broth until the bacteria reached an OD600 of 0.6. The frequency of transposons in genomic DNA isolated from B. pseudomallei treated with either spermine NONOate, or spermine NONOate and imipenem was quantified by Illumina deep-sequencing as described previously for Heamophilus [32]. Sequencing data from 3 independent experiments were averaged, the fold change between samples calculated, and false discovery rate analysis determined. Sixteen genes with several transposons were found to be enriched in the group treated with imipenem and spermine NONOate (table 1). Among the positively selected genes were mutants with transposons in loci encoding cytochrome c oxidase function. In addition, the moaC and mogA genes involved in molybdenum utilization were also positively selected. Molybdenum is a common cofactor of enzymes such as nitrate reductases that allow bacteria to grow using NO3 − for respiration. LBG broth and the autoxidation of the NO generated from spermine NONOate are likely sources of the terminal electron acceptor NO3 − in our system. Together, the positive selection of clones bearing mutations in cytochromes and molybdenum utilization genes suggest that disruption of the electron transport chain provides B. pseudomallei a selective advantage against the killing of imipenem. In addition, the disruption of several genes associated with nucleotide metabolism, tRNA synthesis, β-lactamase processing and transcriptional regulation appear to provide a selective advantage to B. pseudomallei against the antimicrobial activity of imipenem.
Table 1. Positively selected genes bearing transposon mutations that increase the tolerance of B. pseudomallei to imipemen.
| Genes | Description | Fold change1 |
| BPSL0453 | cytochrome c oxidase | 3.25 |
| BPSL3181 | cytochrome c | 4.37 |
| moaC | molybdenum cofactor biosynthesis protein | 6.06 |
| mogA | molybdenum cofactor biosynthesis protein | 3.03 |
| oxa | β-lactamase precursor | 8.09 |
| obgE | GTPase | 6.67 |
| amn | AMP nucleosidase | 3.10 |
| gmk | guanylate kinase | 7.68 |
| glnB1 | nitrogen regulatory protein P-II 1 | 4.59 |
| trmB | tRNA (guanine-N-(7)-)methyltransferase | 4.51 |
| clpA | ATP-dependent Clp protease | 3.53 |
| BPSS1056 | CopG Family transcriptional regulator | 8.12 |
| BPSL3313 | HNS-like transcriptional regulator | 3.70 |
| BPSL0428 | Hypothetical protein | 34.00 |
| BPSL1511 | Hypothetical protein | 4.08 |
| BPSL2413 | Hypothetical protein | 13.12 |
The fold change represents the ratio of the number of sequence reads in the imipenem + spermine NONOate sample over spermine NONOate control.
Inhibition of bacterial respiration protects B. pseudomallei against imipenem
Given the selectivity of NO for metal prosthetic groups in the terminal oxidases of the electron transport chain and the fact that mutations in components of the respiratory chain provided a competitive advantage to Burkholderia in response to imipenem (table 1), it is possible that the antibiotic tolerance elicited in response to NO is associated with a loss in respiratory function. To test this hypothesis, we measured whether the addition of classical antagonists of terminal oxidases of the electron transport chain affects the susceptibility of B. pseudomallei to imipenem. As seen with NO, the respiratory inhibitor potassium cyanide (KCN) prevented the imipenem-dependent killing of B. pseudomallei (figure 3A). To determine whether the concentrations of NO and KCN that protect B. pseudomallei against imipenem affect the respiratory activity of B. pseudomallei, we measured the consumption of O2. Compared to untreated controls grown in LBG broth saturated with O2 (figure 3B), bacterial cultures treated with 100 µM spermine NONOate or 500 µM KCN had reduced respiratory activity. Under the conditions tested, neither 100 µM spermine NONOate nor 500 µM KCN killed B. pseudomallei. These findings suggest that terminal cytochromes of the electron transport chain are critical molecular targets of NO-induced antibiotic resistance. Our investigations also suggest that β-lactam antibiotics require an active electron transport chain to exert their antimicrobial activity. This model is supported further by the fact that the antimicrobial activity of ceftizidime and imipenem was dramatically reduced in anaerobic cultures (figure 3C).
Figure 3. Respiratory activity of B. pseudomallei undergoing nitrosative stress.
Survival of B. pseudomallei after 2.5 h of treatment with 12.5 µg/ml of imipenem (A). Some of the specimens were co-treated with either 500 µM potassium cyanide (KCN) or 100 µM spermine NONOate (sNO). The percent survival was determined as described in figure 1. The data are the mean ± SD from 3 observations collected on 3 separate days. The ability of B. pseudomallei to consume O2 was monitored polarographically (B). The data shown in panel B are representative from 3 independent experiments. B. pseudomallei were adapted for 5 h in an anaerobic chamber and the % survival scored 2.5 h after the addition of 64 µg/ml ceftazidime (CT) or 25 µg/ml imipenem (IM) (C). The results in C are expressed as % survival over controls that were not exposed to antibiotics. B. pseudomallei controls in the anaerobic chamber neither grew nor lost viability in the time frame tested. p<0.01 compared to sNO- and KCN-treated groups.
Inhibitors of PMF prevent imipenem-mediated killing
The enzymatic activity of terminal cytochrome oxidases and nitrate reductases of the electron transport chain help maintain an electrochemical gradient across the cytoplasmic membrane [33]. It is therefore possible that decreases of PMF in response to NO could mediate antibiotic tolerance. To test this idea, we evaluated the effect that collapsing the PMF has on the anti-B. pseudomallei activity of imipenem. The protonophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP) and the ionophore valinomycin were chosen for these investigations, because these drugs dissipate the ΔH+ and ΔΨ components of the PMF by facilitating the transport of H+ and K+ down an electrochemical potential gradient. Remarkably, CCCP and valinomycin protected B. pseudomallei against the antimicrobial activity of imipenem (figure 4A), supporting the model that the antimicrobial activity of this β-lactam antibiotic depends on a functional PMF.
Figure 4. Effect of the PMF on the antimicrobial activity of imipenem.
Effect of 50 µM CCCP or 25 µM valinomycin on the anti-Burkholderia activity of 12.5 µg/ml imipenem (IM) (A). Stationary phase B. pseudomallei controls grown overnight in LBG broth were treated with 0.25% DMSO for 2.5 h. Killing of S. Typhimurium (STM) and E. coli (Eco) grown to log phase in LBG broth (B, C, D) or EG medium (E, F) by IM. Selected samples were treated with 750 µM spermine NONOate (sNO) or 50 µM CCCP. The samples in F were treated with 10 µM CCCP ± 250 µM sNO. p<0.01 compared to the CCCP, valinomycin- or sNO-treated groups. The PMF was estimated by measuring the accumulation of DiSC3(5) in STM (G). The data are expressed as arbitrary fluorescent units (A.U.). p<0.001 compared to the sNO-treated EG group. The data are the mean ± SEM from 3–5 independent observations collected on 2 separate days.
Next, we tested whether NO abrogates killing of other Gram-negative bacteria by imipenem. Surprisingly, exposure of S. enterica serovar Typhimurium or E. coli grown in LBG broth to 750 µM spermine NONOate had a small effect on the antimicrobial activity of imipenem (figure 4B). To investigate whether the failure of NO to protect Salmonella against imipenem is associated with antinitrosative defenses, we compared the susceptibility of wild-type and an hmp mutant that lacks the main mechanism of NO detoxification known in Salmonella and E. coli [34], [35]. NO induced remarkable levels of protection against imipenem in hmp-deficient Salmonella (figure 4C). It should be noted that about 90% of the hmp mutants were still killed by imipenem, raising the possibility that, in addition to antinitrosative defenses, the metabolic pliability of enteric bacteria could prevent the protective effects associated with NO. We reasoned that the incomplete protection afforded by NO to Salmonella and E. coli might be related to the fact that these facultative anaerobes can energize the PMF using alternative electron acceptors. If this were the case, then we would predict that 1) mutations that favor metabolism through the TCA cycle could allow for a more complete NO-induced tolerance to imipenem, 2) classical PMF inhibitors may induce imipenem tolerance under conditions that NO fails to do so, 3) growth of Salmonella and E. coli with glucose as the sole carbon source might estimulate NO-induced antibiotic tolerance, and 4) NO may have different effects on the PMF according to the carbon source used for growth. The following experiments were performed to test these predictions. 1) We tested ackA and pta mutants unable to ferment pyruvate to acetate, thus forcing Salmonella to more fully utilize the TCA cycle and oxidative phosphorylation. Remarkably, NO completely protected ackA and pta mutants against the antimicrobial activity of imipenem (figure 4D). 2) In contrast to NO, the addition of 50 µM CCCP protected Salmonella and E. coli grown in LBG broth against 12.5 µg/ml imipenem (figure 4B), demonstrating that inhibition of PMF induces tolerance to imipenem under conditions that NO is unable to do so. 3) E. coli and Salmonella grown in E salts medium supplemented with 0.4% glucose (i.e., EG medium) were efficiently killed by imipenem (figure 4E). Moreover, the addition of 750 µM spermine NONOate or 500 µM KCN similarly protected most Salmonella and E. coli grown in EG medium against the bactericidal activity of 12.5 µg/ml imipenem. Salmonella exposed to suboptimal concentrations of spermine NONOate and CCCP (i.e., 250 and 10 µM, respectively) became fully tolerant to imipenem (figure 4F). Lastly, 4) we measured the PMF in the BSL2 pathogen S. Typhimurium with 3,3′-dipropylthiadicarbocyanine iodide [DiSC3(5)], the fluorescence of which is inversely proportional to the PMF [36]. As expected, valinomycin increased DiSC3(5)-mediated fluorescence (figure S2). DiSC3(5)-mediated fluorescence increased (p<0.001) in both Salmonella grown to OD600 of 0.5 in LBG broth or EG medium after exposure to 750 µM spermine NONOate (figure 4G), suggesting that NO inhibits the PMF under both conditions. It should be noted, however, that DiSC3(5) fluorescence was significantly (p<0.001) lower in NO-treated Salmonella grown in LBG broth than NO-treated controls grown in EG medium, suggesting that NO is less efficient at inhibiting the PMF in cells grown in LBG broth. Our investigations indicate that the protection afforded by NO against imipenem is dependent on the degree of PMF inhibition. Together, these findings suggest that both metabolic activity and antinitrosative defenses modulate NO-mediated tolerance to imipenem.
The protective effects of NO cannot be explained by reduced oxidative stress
Our investigations demonstrate that NO induces tolerance to β-lactams by collapsing the PMF. It remains possible that the protection afforded by NO against β-lactam antibiotics could emanate from its ability to promote antioxidant defenses [18]. Following this line of reasoning, the limited antimicrobial activity of imipenem against anaerobic B. pseudomallei could be interpreted as a sign that oxidative stress is required for killing. Consequently, we tested whether imipenem induces oxidative stress in B. pseudomallei, and whether NO affects this response. Membrane soluble H2O2, which arises by the spontaneous or enzymatic dismutation of O2 −, was used as readout of overall production of reactive oxygen species. B. pseudomallei treated with sublethal concentrations of imipenem produced consistently lower concentrations of H2O2 than untreated controls (figure 5A). The addition of 100 µM spermine NONOate to the bacterial cultures increased the amount of H2O2 generated by 3-fold. Similar to NO-treated cells, KCN-treated B. pseudomallei generated about 15 µM H2O2. The addition of a sublethal concentration of imipenem significantly (p<0.05) reduced the amount of H2O2 generated by B. pseudomallei in response to NO or KCN. These findings are consistent with those reported by Liu and Imlay, who conjectured that the effects of β-lactams on respiration could reflect damage of the cell envelope and dissipation of the back pressure of the proton motive force [22]. Our investigations indicate that NO-mediated antibiotic tolerance cannot be explained by diminished oxidative stress; nor does the imipenem-mediated killing of B. pseudomallei appear to be dependent on the elicitation of oxidative stress. Collectively, these data support recent investigations that have questioned oxidative stress as the mode of action of bactericidal antibiotics [22], [23]. Our data also suggest that NO-induced antibiotic resistance takes place independently of its effects on antioxidant defenses.
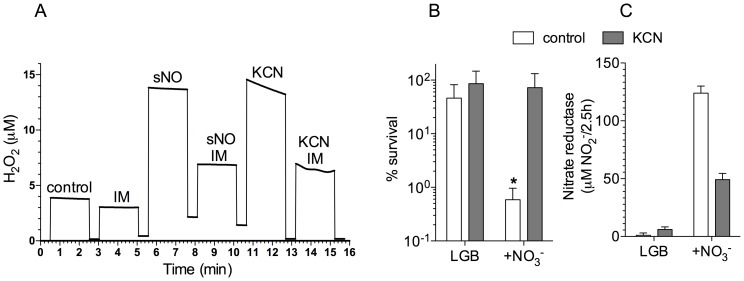
Figure 5. Effect of imipenem and nitrosative stress on H2O2 synthesis.
The production of H2O2 by B. pseudomallei diluted to OD600 of 0.5 was measured polarographically (A). Some of the specimens were treated with 12.5 µg/ml imipenem (IM), and where indicated the bacterial cells were treated with 100 µM spermine NONOate (sNO) or 500 µM KCN. Untreated cells are shown as controls. The H2O2 probe was washed with fresh LB broth between individual specimens. The data are representative of 3 observations collected on 3 separate days. The killing of anaerobic B. pseudomallei by 50 µg/ml IM was tested in LBG broth in the presence or absence of 50 mM NO3 − (B). p<0.01 compared to the KCN-treated group. Nitrate reductase activity was monitored by measuring the accumulation of NO2 − by the Griess reaction (C).
According to our proposed model that the NO-dependent collapse of the PMF induces tolerance to β-lactams, the poor antibiotic activity of imipenem against anaerobic B. pseudomallei could reflect a lack of respiratory activity. To shed light into this possibility, the antimicrobial activity of imipenem was tested in anaerobic B. pseudomallei grown in LBG broth supplemented with 50 mM of the terminal electron acceptor NO3 −. Anaerobic cells respiring NO3 − became susceptible to 50 µg/ml imipenem (figure 5B). We tried to determine the effects of NO treatment on the killing of anaerobic Burkholderia by imipenem but, as previously noted [16], anaerobic bacteria were found to be extraordinarily susceptible to 100 µM spermine NONOate. Therefore, KCN was used instead. The addition of KCN abrogated the antibiotic activity of imipenem against anaerobic B. pseudomallei cultured in LBG broth supplemented with 50 mM NO3 − (figure 5B). The concentrations of KCN that elicited protection also inhibited nitrate reductase activity (figure 5C). These findings indicate that imipenem has antibiotic activity in the absence of O2 and derived reactive oxygen species if the electron transport chain is energized by the reduction of alternative electron acceptors such as NO3 −.
Effects of nitric oxide produced by IFNγ-treated macrophages on the antimicrobial activity of imipenem against intracellular B. pseudomallei and S. enterica
Our investigations indicate that chemically-generated NO protects B. pseudomallei, S. enterica and E. coli against the antimicrobial activity of β-lactams. Next, we studied whether NO generated through the enzymatic activity of NO synthases modulates the antimicrobial activity of imipenem. J744 macrophage-like cells were stimulated overnight with 200 U/ml recombinant murine IFNγ. The macrophages were infected with B. pseudomallei at an MOI of 4. The data shown in figure 6A indicate that NO produced by IFNγ-treated macrophages enhances the antimicrobial activity of 2.5 µg/ml imipenem. Our investigations identify NO as the mechanism by which recombinant IFNγ enhances the antimicrobial activity of imipenem in vivo [37]. These findings, however, contrast with the protective effects observed for NO in vitro. Because B. pseudomallei are hypersusceptible to NO [30], we tested the survival of B. pseudomallei after exposure to a bactericidal concentration of NO and a sublethal amount of imipenem. About 90% of B. pseudomallei in the cultures were killed after exposure to 500 µM spermine NONOate, whereas controls treated with 0.25 µM imipenem doubled in cell number during the 2.5 h of the experiment (figure 6C). However, the simultaneous addition of 500 µM spermine NONOate and 0.25 µM imipenem killed ∼99.9% of B. pseudomallei in the cultures. Together, these investigations suggest that β-lactams can potentiate the antimicrobial activity of host defenses such as NO. We next tested whether NO produced by IFNγ-treated macrophages can modify the killing of intracellular Salmonella by imipenem. In contrast to B. pseudomallei (figure 6A), NO produced by IFNγ-treated macrophages appears to protect Salmonella against this β-lactam (figure 6D and E), since imipenem killed Salmonella in a concentration-dependent manner in macrophages treated with the iNOS inhibitor aminoguanidine.
Figure 6. Modulation of antimicrobial activity of imipenem by NO produced by IFNγ-activated J774 cells.
Killing of B. pseudomallei (BPS) and Salmonella (STM) in IFNγ-activated J774 cells treated with increasing concentrations of imipenem (A and D, respectively). Where indicated, 500 µM of the iNOS inhibitor aminoguanidine (AG) was added to the cells. NO2 − produced by the macrophages was estimated by the Griess reaction (B and E). Panel C shows the killing of log phase B. pseudomallei treated for 2.5 h in LBG broth with 500 µM spermine NONOate (sNO) and/or 0.25 µg/ml imipenem (IM).
Discussion
Our investigations indicate that the antimicrobial activity of β-lactams can be modified by NO produced chemically or enzymatically in the inflammatory response of IFNγ-activated macrophages. At sublethal concentrations, NO protects B. pseudomallei, nontyphoidal Salmonella and E. coli from the antimicrobial activity of β-lactams, and NO generated by IFNγ-activated macrophages shields intracellular Salmonella from the cytotoxicity of imipenem. At higher concentrations, however, the bactericidal activity of NO itself against B. pseudomallei was potentiated by sublethal concentrations of imipenem.
NO has been proposed to protect bacteria against different classes of antibiotics by promoting antioxidant defenses [18]. However, our investigations suggest that the mechanism by which NO induces resistance of B. pseudomallei and other Gram-negative bacteria to β-lactam antibiotics is mediated through the collapse of the PMF. The following independent lines of evidence support the proposed model. First, the concentrations of NO that protect B. pseudomallei against β-lactams also inhibit respiratory activity. Second, high throughput sequencing of a transposon library revealed that B. pseudomallei mutants in cytochrome oxidase are hyperresistant to β-lactam killing. Third, cyanide, a classical inhibitor of cytochromes, inhibits respiration and protects B. pseudomallei, E. coli and S. enterica against β-lactams. Fourth, the degree of the PMF appears to be inversely associated with the extent of β-lactam-mediated killing. Fifth, dissipation of ΔH+ and ΔΨ components of the PMF independently protect against β-lactams. And sixth, killing by imipenem is marginal in anaerobic B. pseudomallei cultures, unless the electron transport chain is energized with terminal electron acceptors such as NO3 −. Collapse of the PMF can be added to β-lactamases, mutated penicillin-binding proteins and efflux pumps as strategies that protect bacteria against β-lactam antibiotics. The immediate antibiotic tolerance elicited in response to NO may provide a window of time required for the acquisition of mutations that mediate inheritable resistance to antibiotics.
NO, KCN, CCCP and valinomycin blocked the imipenem-mediated killing of B. pseudomallei, E. coli and S. enterica. These findings indicate that the antimicrobial activity of β-lactams requires an energized membrane. Considering the high affinity of NO for terminal cytochromes of the electron transport chain and the advantage afforded by transposons in cytochrome c or cytochrome c oxidase for the survival of B. pseudomallei in the presence of imipenem, we propose that terminal cytochromes are the likely molecular switch by which NO induces tolerance to β-lactams. This model is independently supported by the fact that the concentrations of NO that elicited antibiotic tolerance in B. pseudomallei also inhibited O2 consumption. Repression of respiratory activity has already been shown to mediate NO-induced resistance of Salmonella to aminoglycosides [19]. Our investigations with Salmonella and E. coli indicate that the ability of NO to inhibit respiration is necessary but not sufficient for β-lactam drug tolerance. Ultimately, the ability of NO to induce antibiotic tolerance seems to be associated with the degree of inhibition of the PMF. For example, NO-dependent tolerance to imipenem in E. coli and Salmonella grown in EG medium or LBG broth is inversely proportional to the PMF of the bacteria. Interestingly, NO prevented most of the antibiotic activity of imipenem against the strict aerobe B. pseudomallei under growth conditions that failed to protect Salmonella or E. coli. This is likely explained by the fact that the PMF in B. pseudomallei is preferentially maintained by the enzymatic activity of cytochrome oxidases. The heavy dependence of B. pseudomallei on cytochrome oxidases to maintain the PMF may explain why β-lactam antibiotics are most efficient during the acute phase of therapy at a time when, in the absence of abscesses and NO-mediated immunity, cytochrome oxidases are expected to be fully functional.
The NO blockage of energy-dependent drug uptake has been shown to protect Salmonella against aminoglycosides [19]. In an analogous fashion, NO could protect bacteria by interfering with the expression or function of the Omp38 porin that is required to transport β-lactams into the periplasmic space [38]. Blockage of drug uptake, however, may not explain why NO protects bacteria against β-lactam antibiotics, because B. pseudomallei treated simultaneously with NO and imipenem yielded even smaller colonies than NO-treated controls. Moreover, NO protected B. pseudomallei already exposed to imipenem. The NO-induced tolerance to β-lactams could be mediated by the negative impact that the collapse of the PMF has on metabolism and membrane function. At least three mechanisms could explain how the collapse of the PMF by NO lessens the antibiotic activity of β-lactam drugs. First, electrochemical gradients energize the transport of muropeptides across the cytoplasmic membrane [39]; thus, the negative impact of NO on the PMF may inhibit peptidoglycan biosynthesis. Second, rapid β-lactam-induced lysis requires successful assembly of the divisome [40], [41], an event that is initiated by the PMF-dependent localization of FtsA to the FtsZ septal ring [42]. FtsA serves as a scaffold for the assembly of several morphogenetic proteins, including penicillin-binding proteins [43] that are the targets of β-lactam antibiotics. Consequently, the NO-dependent inhibition of the PMF could delocalize morphogenetic proteins from the division septum, thereby contributing to resistance to β-lactams. Disassembly of the division ring could be a critical step by which NO protects rapidly growing bacteria from β-lactams, but may be of lesser importance in non-dividing stationary phase bacteria. And third, NO and the other chemical inhibitors of the electron transport chain could protect bacteria by stalling growth. According to this idea, imipenem exerted negligible antimicrobial activity against stationary phase Salmonella. However, inhibition of cell growth might not explain why sublethal concentrations of NO protect stationary phase B. pseudomallei against imipenem. We found it remarkable that the 1,000-fold reduction in viability of nongrowing B. pseudomallei was abrogated in the presence of sublethal concentrations of NO.
NO did not select for intrinsically resistant populations, because the surviving bacteria remained susceptible to imipenem. Constant NO fluxes were required for the elicitation of tolerance to imipenem, and the effects were short-lived. The inhibition of metal centers in terminal oxidases of the electron transport chain, with the consequent reduction in PMF, provides a reasonable model for the fast and transient adaptation of B. pseudomallei, E. coli and Salmonella to β-lactam antibiotics. The transient protection afforded by NO can be better understood if we consider that the k off value for the dissociation of NO from cytochrome aa 3 is 0.01 sec−1 [44]. Thus, the association of cytochrome c oxidase to NO would last about 1 min (i.e., t 1/2 of 69 sec). In other words, the fast denitrosylation of metal prosthetic groups in the terminal cytochromes of the electron transport chain could explain the transient protection noted after NO is removed from the bacterial cultures.
Oxidative stress has been proposed as a common killing mechanism of bactericidal antibiotics [21], and the antioxidant defenses elicited by NO are thought to mediate resistance of B. subtilis to several classes of antibacterials [18]. Our investigations indicate that NO prevents antibiotic killing despite increasing the rate of H2O2 synthesis. The stasis of electrons that follows the nitrosylation of terminal cytochrome oxidases of the electron transport chain can facilitate the adventitious reduction of O2 by flavin cofactors or Q sites of NADH dehydrogenases [45], [46]. The O2 − generated in this process spontaneously or enzymatically dismutates to H2O2. This model may explain why imipenem, similar to other β-lactam antibiotics that increase bacterial respiratory rates [22], diminishes H2O2 synthesis. The increased H2O2 synthesis noted in NO-treated bacteria challenges the notion that NO enhances antibiotic resistance by eliciting antioxidant defenses [18]. Our investigations are consistent with recent work that has reported that bactericidal antibiotics can kill microorganisms in the absence of oxidative stress [22], [23]. This idea is further substantiated by the fact that imipenem can kill B. pseudomallei in anaerobic cultures given that the bacteria are actively respiring the terminal electron acceptor NO3 −.
Although sublethal concentrations of NO reversed β-lactam-mediated killing of B. pseudomallei, NO produced by IFNγ-primed macrophages synergized with imipenem in killing intracellular B. pseudomallei. Our investigations identify NO as the mechanism by which recombinant IFNγ enhances the antimicrobial activity of imipenem in vivo [47]. We find it remarkable that inos, which is just one of over 150 loci regulated by IFNγ [48], made such a difference in the outcome of imipenem treatment. Cumulatively, these investigations support the widely accepted concept that immunocompetent hosts respond better to antibiotic therapy than immunodeficient controls. A closer look at our investigations indicate, however, that imipenem potentiates the killing activity of NO in vivo and not vice versa. In fact, sublethal concentrations of imipenem augmented the bactericidal activity of NO in an exponential fashion.
Weakening of the cell wall by β-lactams increases NO-mediated killing. This observation could be explained by a model in which the outstanding killing that NO exerts against B. pseudomallei is a direct consequence of membrane dysfunction. Because B. pseudomallei draw most energy from oxidative phosphorylation, nitrosylation of terminal cytochromes of the electron transport chain could have greater deleterious actions on the energetics and membrane function of the aerobe B. pseudomallei as compared to more metabolically pliable organisms such as Salmonella that can draw significant energy from fermentation. Degree or duration of the inhibition of the respiratory chain by NO could then rationalize why sublethal concentrations of NO protect against β-lactams, whereas higher NO fluxes become more lethal in the presence of imipenem. The clinical relevance of these findings can already be inferred from the observation that mice treated with recombinant IFNγ and ceftazidime clear acute B. pseudomallei infections [37]. It remains puzzling that during the natural course of meliodosis ceftazidime is largely ineffective during the eradication phase of treatment. Reduced respiratory activity imposed by growing abscesses or the sublethal amounts of NO could contribute to the more limited use of β-lactams in the eradication phase of therapy.
Supporting Information
Functional categorization of B. pseudomallei essential genes.
(TIF)
Effect of valinomycin on the PMF as estimated fluorometrically by measuring the accumulation of DiSC3(5).
(TIFF)
List of B. pseudomallei essential genes.
(PDF)
Acknowledgments
We thank the University of Colorado Next-Generation Sequencing Development Core for their help with the MiSeq.
Funding Statement
This research was supported by NIH grants AI54959 and U54 AI-065357, the Veterans Administration grant 1I01BX002073 and the Burroughs Wellcome Fund. CA and TT were supported in part by the institutional training grants T32 AI052066 and T32 GM008730, respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yabuuchi E, Arakawa M (1993) Burkholderia pseudomallei and melioidosis: be aware in temperate area. Microbiol Immunol 37: 823–836. [DOI] [PubMed] [Google Scholar]
- 2. Wuthiekanun V, Langa S, Swaddiwudhipong W, Jedsadapanpong W, Kaengnet Y, et al. (2006) Short report: Melioidosis in Myanmar: forgotten but not gone? Am J Trop Med Hyg 75: 945–946. [PubMed] [Google Scholar]
- 3. Currie BJ, Ward L, Cheng AC (2010) The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year darwin prospective study. PLoS Negl Trop Dis 4: e900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheng AC, Currie BJ (2005) Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev 18: 383–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng AC (2010) Melioidosis: advances in diagnosis and treatment. Curr Opin Infect Dis 23: 554–559. [DOI] [PubMed] [Google Scholar]
- 6. Rajchanuvong A, Chaowagul W, Suputtamongkol Y, Smith MD, Dance DA, et al. (1995) A prospective comparison of co-amoxiclav and the combination of chloramphenicol, doxycycline, and co-trimoxazole for the oral maintenance treatment of melioidosis. Trans R Soc Trop Med Hyg 89: 546–549. [DOI] [PubMed] [Google Scholar]
- 7. Chaowagul W (2000) Recent advances in the treatment of severe melioidosis. Acta Trop 74: 133–137. [DOI] [PubMed] [Google Scholar]
- 8. Vorachit M, Lam K, Jayanetra P, Costerton JW (1993) Resistance of Pseudomonas pseudomallei growing as a biofilm on silastic discs to ceftazidime and co-trimoxazole. Antimicrob Agents Chemother 37: 2000–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sawasdidoln C, Taweechaisupapong S, Sermswan RW, Tattawasart U, Tungpradabkul S, et al. (2010) Growing Burkholderia pseudomallei in biofilm stimulating conditions significantly induces antimicrobial resistance. PLoS One 5: e9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan YY, Chua KL (2005) The Burkholderia pseudomallei BpeAB-OprB efflux pump: expression and impact on quorum sensing and virulence. J Bacteriol 187: 4707–4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar A, Chua KL, Schweizer HP (2006) Method for regulated expression of single-copy efflux pump genes in a surrogate Pseudomonas aeruginosa strain: identification of the BpeEF-OprC chloramphenicol and trimethoprim efflux pump of Burkholderia pseudomallei 1026b. Antimicrob Agents Chemother 50: 3460–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mima T, Schweizer HP (2010) The BpeAB-OprB efflux pump of Burkholderia pseudomallei 1026b does not play a role in quorum sensing, virulence factor production, or extrusion of aminoglycosides but is a broad-spectrum drug efflux system. Antimicrob Agents Chemother 54: 3113–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Livermore DM, Chau PY, Wong AI, Leung YK (1987) β-Lactamase of Pseudomonas pseudomallei and its contribution to antibiotic resistance. J Antimicrob Chemother 20: 313–321. [DOI] [PubMed] [Google Scholar]
- 14. Niumsup P, Wuthiekanun V (2002) Cloning of the class D β-lactamase gene from Burkholderia pseudomallei and studies on its expression in ceftazidime-susceptible and -resistant strains. J Antimicrob Chemother 50: 445–455. [DOI] [PubMed] [Google Scholar]
- 15. Tribuddharat C, Moore RA, Baker P, Woods DE (2003) Burkholderia pseudomallei class a beta-lactamase mutations that confer selective resistance against ceftazidime or clavulanic acid inhibition. Antimicrob Agents Chemother 47: 2082–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hamad MA, Austin CR, Stewart AL, Higgins M, Vazquez-Torres A, et al. (2011) Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei . Antimicrob Agents Chemother 55: 3313–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henard CA, Vazquez-Torres A (2011) Nitric oxide and Salmonella pathogenesis. Front Microbiol 2: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gusarov I, Shatalin K, Starodubtseva M, Nudler E (2009) Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325: 1380–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McCollister BD, Hoffman M, Husain M, Vazquez-Torres A (2011) Nitric oxide protects bacteria from aminoglycosides by blocking the energy-dependent phases of drug uptake. Antimicrob Agents Chemother 55: 2189–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ouadrhiri Y, Scorneaux B, Sibille Y, Tulkens PM (1999) Mechanism of the intracellular killing and modulation of antibiotic susceptibility of Listeria monocytogenes in THP-1 macrophages activated by gamma interferon. Antimicrob Agents Chemother 43: 1242–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ (2007) A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130: 797–810. [DOI] [PubMed] [Google Scholar]
- 22. Liu Y, Imlay JA (2013) Cell death from antibiotics without the involvement of reactive oxygen species. Science 339: 1210–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K (2013) Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339: 1213–1216. [DOI] [PubMed] [Google Scholar]
- 24. Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, et al. (2004) Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei . Proc Natl Acad Sci U S A 101: 14240–14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rholl DA, Trunck LA, Schweizer HP (2008) In vivo Himar1 transposon mutagenesis of Burkholderia pseudomallei . Appl Environ Microbiol 74: 7529–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, et al. (2006) An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103: 2833–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chadwick DR, Ang B, Sitoh YY, Lee CC (2002) Cerebral melioidosis in Singapore: a review of five cases. Trans R Soc Trop Med Hyg 96: 72–76. [DOI] [PubMed] [Google Scholar]
- 29. Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE (1991) Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob Agents Chemother 35: 1824–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones-Carson J, Laughlin JR, Stewart AL, Voskuil MI, Vazquez-Torres A (2012) Nitric oxide-dependent killing of aerobic, anaerobic and persistent Burkholderia pseudomallei . Nitric Oxide 27: 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Estes DM, Dow SW, Schweizer HP, Torres AG (2010) Present and future therapeutic strategies for melioidosis and glanders. Expert Rev Anti Infect Ther 8: 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ (2009) Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A 106: 16422–16427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moncada S, Erusalimsky JD (2002) Does nitric oxide modulate mitochondrial energy generation and apoptosis? Nat Rev Mol Cell Biol 3: 214–220. [DOI] [PubMed] [Google Scholar]
- 34. Robinson JL, Brynildsen MP (2013) A kinetic platform to determine the fate of nitric oxide in Escherichia coli . PLoS Comput Biol 9: e1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bang IS, Liu L, Vazquez-Torres A, Crouch ML, Stamler JS, et al. (2006) Maintenance of nitric oxide and redox homeostasis by the salmonella flavohemoglobin hmp . J Biol Chem 281: 28039–28047. [DOI] [PubMed] [Google Scholar]
- 36. Waggoner A (1976) Optical probes of membrane potential. J Membr Biol 27: 317–334. [DOI] [PubMed] [Google Scholar]
- 37. Propst KL, Mima T, Choi KH, Dow SW, Schweizer HP (2010) A Burkholderia pseudomallei ΔpurM mutant is avirulent in immunocompetent and immunodeficient animals: candidate strain for exclusion from select-agent lists. Infect Immun 78: 3136–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suginta W, Mahendran KR, Chumjan W, Hajjar E, Schulte A, et al. (2011) Molecular analysis of antimicrobial agent translocation through the membrane porin BpsOmp38 from an ultraresistant Burkholderia pseudomallei strain. Biochim Biophys Acta 1808: 1552–1559. [DOI] [PubMed] [Google Scholar]
- 39. Cheng Q, Park JT (2002) Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J Bacteriol 184: 6434–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. de Pedro MA, Holtje JV, Schwarz H (2002) Fast lysis of Escherichia coli filament cells requires differentiation of potential division sites. Microbiology 148: 79–86. [DOI] [PubMed] [Google Scholar]
- 41. Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, et al. (2009) Rapid β-lactam-induced lysis requires successful assembly of the cell division machinery. Proc Natl Acad Sci U S A 106: 21872–21877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Strahl H, Hamoen LW (2010) Membrane potential is important for bacterial cell division. Proc Natl Acad Sci U S A 107: 12281–12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weiss DS (2004) Bacterial cell division and the septal ring. Mol Microbiol 54: 588–597. [DOI] [PubMed] [Google Scholar]
- 44. Cooper CE, Davies NA, Psychoulis M, Canevari L, Bates TE, et al. (2003) Nitric oxide and peroxynitrite cause irreversible increases in the Km for oxygen of mitochondrial cytochrome oxidase: in vitro and in vivo studies. Biochim Biophys Acta 1607: 27–34. [DOI] [PubMed] [Google Scholar]
- 45. Boveris A, Chance B (1973) The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem J 134: 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Galkin A, Brandt U (2005) Superoxide radical formation by pure complex I (NADH:ubiquinone oxidoreductase) from Yarrowia lipolytica . J Biol Chem 280: 30129–30135. [DOI] [PubMed] [Google Scholar]
- 47. Propst KL, Troyer RM, Kellihan LM, Schweizer HP, Dow SW (2010) Immunotherapy markedly increases the effectiveness of antimicrobial therapy for treatment of Burkholderia pseudomallei infection. Antimicrob Agents Chemother 54: 1785–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Der SD, Zhou A, Williams BR, Silverman RH (1998) Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 95: 15623–15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional categorization of B. pseudomallei essential genes.
(TIF)
Effect of valinomycin on the PMF as estimated fluorometrically by measuring the accumulation of DiSC3(5).
(TIFF)
List of B. pseudomallei essential genes.
(PDF)