Abstract
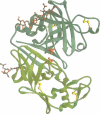
Two crystal structures are described for the lysosomal aspartic protease cathepsin D (EC 3.4.23.5). The molecular replacement method was used with X-ray diffraction data to 3 A resolution to produce structures for human spleen cathepsin D and for bovine liver cathepsin D complexed with the 6-peptide inhibitor pepstatin A. The lysosomal targeting region of cathepsin D defined by previous expression studies [Barnaski et al. (1990) Cell, 63, 281-219] is located in well defined electron density on the surface of the molecules. This region includes the putative binding site of the cis-Golgi phosphotransferase which is responsible for the initial sorting step for soluble proteins destined for lysosomes by phosphorylating the carbohydrates on these molecules. Carbohydrate density is visible at both expected positions on the cathepsin D molecules and, at the best defined position, four sugar residues extend towards the lysosomal targeting region. The active site of the protease and the active site cleft substrate binding subsites are described using the pepstatin inhibited structure. The model geometry for human cathepsin D has rms deviations from ideal of bonds and angles of 0.013 A and 3.2 degrees respectively. For bovine cathepsin D the corresponding figures are 0.014 A and 3.3 degrees. The crystallographic residuals (R factors) are 16.1% and 15.8% for the human and inhibited bovine cathepsin D models respectively. The free R factors, calculated with 10% of the data reserved for testing the models and not used for refinement, are 25.1% and 24.1% respectively.
Full text
PDF



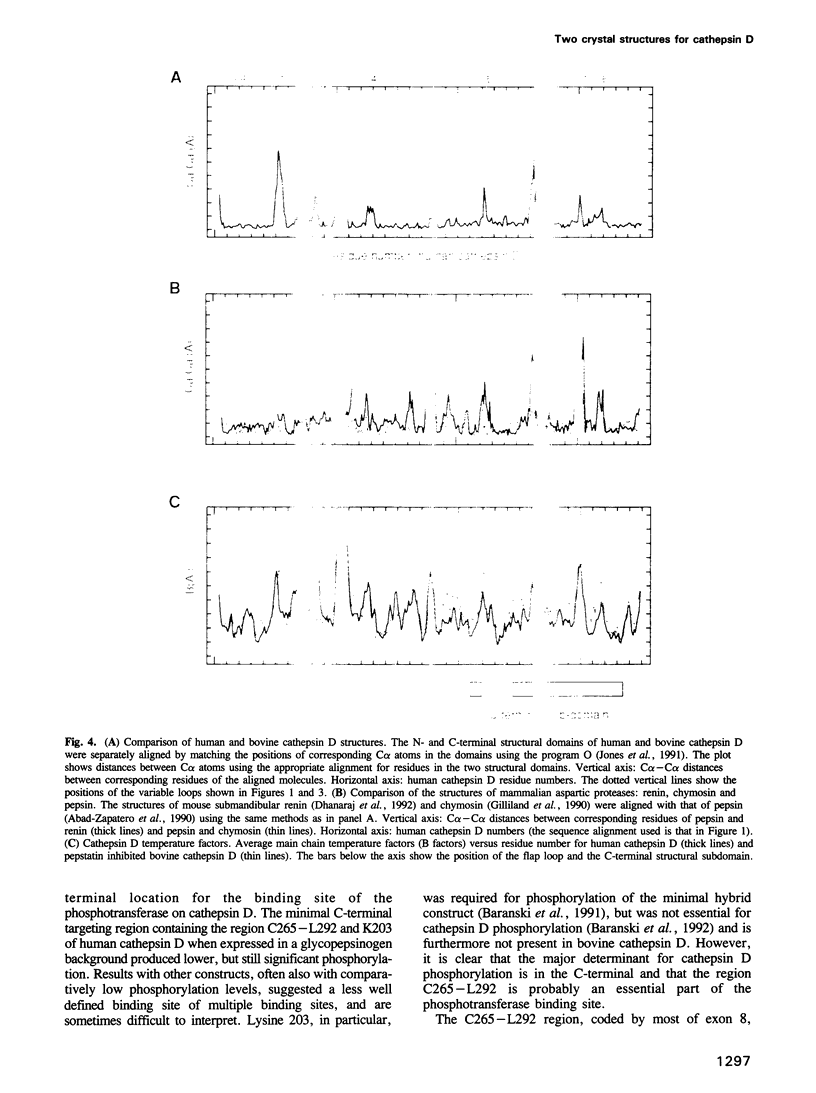


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abad-Zapatero C., Rydel T. J., Erickson J. Revised 2.3 A structure of porcine pepsin: evidence for a flexible subdomain. Proteins. 1990;8(1):62–81. doi: 10.1002/prot.340080109. [DOI] [PubMed] [Google Scholar]
- Baranski T. J., Cantor A. B., Kornfeld S. Lysosomal enzyme phosphorylation. I. Protein recognition determinants in both lobes of procathepsin D mediate its interaction with UDP-GlcNAc:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase. J Biol Chem. 1992 Nov 15;267(32):23342–23348. [PubMed] [Google Scholar]
- Baranski T. J., Faust P. L., Kornfeld S. Generation of a lysosomal enzyme targeting signal in the secretory protein pepsinogen. Cell. 1990 Oct 19;63(2):281–291. doi: 10.1016/0092-8674(90)90161-7. [DOI] [PubMed] [Google Scholar]
- Baranski T. J., Koelsch G., Hartsuck J. A., Kornfeld S. Mapping and molecular modeling of a recognition domain for lysosomal enzyme targeting. J Biol Chem. 1991 Dec 5;266(34):23365–23372. [PubMed] [Google Scholar]
- Bieber F., Brachvogel V., Arni R., Fusek M., Metcalf P. Crystallization and initial crystallographic results for pepstatin A inhibited bovine cathepsin D. J Mol Biol. 1992 Oct 20;227(4):1265–1268. doi: 10.1016/0022-2836(92)90539-v. [DOI] [PubMed] [Google Scholar]
- Bond J. S., Butler P. E. Intracellular proteases. Annu Rev Biochem. 1987;56:333–364. doi: 10.1146/annurev.bi.56.070187.002001. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Cantor A. B., Baranski T. J., Kornfeld S. Lysosomal enzyme phosphorylation. II. Protein recognition determinants in either lobe of procathepsin D are sufficient for phosphorylation of both the amino and carboxyl lobe oligosaccharides. J Biol Chem. 1992 Nov 15;267(32):23349–23356. [PubMed] [Google Scholar]
- Casciola-Rosen L. A., Renfrew C. A., Hubbard A. L. Lumenal labeling of rat hepatocyte endocytic compartments. Distribution of several acid hydrolases and membrane receptors. J Biol Chem. 1992 Jun 15;267(17):11856–11864. [PubMed] [Google Scholar]
- Cho W. L., Raikhel A. S. Cloning of cDNA for mosquito lysosomal aspartic protease. Sequence analysis of an insect lysosomal enzyme similar to cathepsins D and E. J Biol Chem. 1992 Oct 25;267(30):21823–21829. [PubMed] [Google Scholar]
- Church W. B., Swenson L., James M. N., Mahuran D. Crystallization of human beta-hexosaminidase B. J Mol Biol. 1992 Sep 20;227(2):577–580. doi: 10.1016/0022-2836(92)90911-3. [DOI] [PubMed] [Google Scholar]
- Davies D. R. The structure and function of the aspartic proteinases. Annu Rev Biophys Biophys Chem. 1990;19:189–215. doi: 10.1146/annurev.bb.19.060190.001201. [DOI] [PubMed] [Google Scholar]
- Dhanaraj V., Dealwis C. G., Frazao C., Badasso M., Sibanda B. L., Tickle I. J., Cooper J. B., Driessen H. P., Newman M., Aguilar C. X-ray analyses of peptide-inhibitor complexes define the structural basis of specificity for human and mouse renins. Nature. 1992 Jun 11;357(6378):466–472. doi: 10.1038/357466a0. [DOI] [PubMed] [Google Scholar]
- Diment S., Leech M. S., Stahl P. D. Cathepsin D is membrane-associated in macrophage endosomes. J Biol Chem. 1988 May 15;263(14):6901–6907. [PubMed] [Google Scholar]
- Dunn B. M., Jimenez M., Parten B. F., Valler M. J., Rolph C. E., Kay J. A systematic series of synthetic chromophoric substrates for aspartic proteinases. Biochem J. 1986 Aug 1;237(3):899–906. doi: 10.1042/bj2370899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust P. L., Kornfeld S., Chirgwin J. M. Cloning and sequence analysis of cDNA for human cathepsin D. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4910–4914. doi: 10.1073/pnas.82.15.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusek M., Baudys M., Metcalf P. Purification and crystallization of human cathepsin D. J Mol Biol. 1992 Jul 20;226(2):555–557. doi: 10.1016/0022-2836(92)90968-p. [DOI] [PubMed] [Google Scholar]
- Gilliland G. L., Winborne E. L., Nachman J., Wlodawer A. The three-dimensional structure of recombinant bovine chymosin at 2.3 A resolution. Proteins. 1990;8(1):82–101. doi: 10.1002/prot.340080110. [DOI] [PubMed] [Google Scholar]
- Gopalan P., Dufresne M. J., Warner A. H. Thiol protease and cathepsin D activities in selected tissues and cultured cells from normal and dystrophic mice. Can J Physiol Pharmacol. 1987 Feb;65(2):124–129. doi: 10.1139/y87-025. [DOI] [PubMed] [Google Scholar]
- Gulnik S., Baldwin E. T., Tarasova N., Erickson J. Human liver cathepsin D. Purification, crystallization and preliminary X-ray diffraction analysis of a lysosomal enzyme. J Mol Biol. 1992 Sep 5;227(1):265–270. doi: 10.1016/0022-2836(92)90696-h. [DOI] [PubMed] [Google Scholar]
- Hasilik A., Neufeld E. F. Biosynthesis of lysosomal enzymes in fibroblasts. Phosphorylation of mannose residues. J Biol Chem. 1980 May 25;255(10):4946–4950. [PubMed] [Google Scholar]
- Hasilik A. The early and late processing of lysosomal enzymes: proteolysis and compartmentation. Experientia. 1992 Feb 15;48(2):130–151. doi: 10.1007/BF01923507. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Horst M., Hasilik A. Expression and maturation of human cathepsin D in baby-hamster kidney cells. Biochem J. 1991 Jan 15;273(Pt 2):355–361. doi: 10.1042/bj2730355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto T., Okazaki K., Koga H., Yamada H. Specificity of rat liver cathepsin D. J Biochem. 1987 Mar;101(3):575–580. doi: 10.1093/jb/101.3.575. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Jupp R. A., Dunn B. M., Jacobs J. W., Vlasuk G., Arcuri K. E., Veber D. F., Perlow D. S., Payne L. S., Boger J., de Laszlo S. The selectivity of statine-based inhibitors against various human aspartic proteinases. Biochem J. 1990 Feb 1;265(3):871–878. doi: 10.1042/bj2650871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A., Achord D. T., Sly W. S. Phosphohexosyl components of a lysosomal enzyme are recognized by pinocytosis receptors on human fibroblasts. Proc Natl Acad Sci U S A. 1977 May;74(5):2026–2030. doi: 10.1073/pnas.74.5.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketcham C. M., Kornfeld S. Characterization of UDP-N-acetylglucosamine:glycoprotein N-acetylglucosamine-1-phosphotransferase from Acanthamoeba castellanii. J Biol Chem. 1992 Jun 5;267(16):11654–11659. [PubMed] [Google Scholar]
- Ketcham C. M., Kornfeld S. Purification of UDP-N-acetylglucosamine:glycoprotein N-acetylglucosamine-1-phosphotransferase from Acanthamoeba castellanii and identification of a subunit of the enzyme. J Biol Chem. 1992 Jun 5;267(16):11645–11653. [PubMed] [Google Scholar]
- Kornfeld S., Mellman I. The biogenesis of lysosomes. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- Kornfeld S. Structure and function of the mannose 6-phosphate/insulinlike growth factor II receptors. Annu Rev Biochem. 1992;61:307–330. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- Kornfeld S. Trafficking of lysosomal enzymes in normal and disease states. J Clin Invest. 1986 Jan;77(1):1–6. doi: 10.1172/JCI112262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang L., Reitman M., Tang J., Roberts R. M., Kornfeld S. Lysosomal enzyme phosphorylation. Recognition of a protein-dependent determinant allows specific phosphorylation of oligosaccharides present on lysosomal enzymes. J Biol Chem. 1984 Dec 10;259(23):14663–14671. [PubMed] [Google Scholar]
- Ludwig T., Griffiths G., Hoflack B. Distribution of newly synthesized lysosomal enzymes in the endocytic pathway of normal rat kidney cells. J Cell Biol. 1991 Dec;115(6):1561–1572. doi: 10.1083/jcb.115.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre G. F., Erickson A. H. Procathepsins L and D are membrane-bound in acidic microsomal vesicles. J Biol Chem. 1991 Aug 15;266(23):15438–15445. [PubMed] [Google Scholar]
- Musil D., Zucic D., Turk D., Engh R. A., Mayr I., Huber R., Popovic T., Turk V., Towatari T., Katunuma N. The refined 2.15 A X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity. EMBO J. 1991 Sep;10(9):2321–2330. doi: 10.1002/j.1460-2075.1991.tb07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermann M. K., Chlebowski J. F., Bond J. S. Action of cathepsin D on fructose-1,6-bisphosphate aldolase. Biochem J. 1983 Jun 1;211(3):529–534. doi: 10.1042/bj2110529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri J., Factorovich Y. Selective inhibition of antigen presentation to cloned T cells by protease inhibitors. J Immunol. 1988 Nov 15;141(10):3313–3317. [PubMed] [Google Scholar]
- Redecker B., Heckendorf B., Grosch H. W., Mersmann G., Hasilik A. Molecular organization of the human cathepsin D gene. DNA Cell Biol. 1991 Jul-Aug;10(6):423–431. doi: 10.1089/dna.1991.10.423. [DOI] [PubMed] [Google Scholar]
- Rijnboutt S., Aerts H. M., Geuze H. J., Tager J. M., Strous G. J. Mannose 6-phosphate-independent membrane association of cathepsin D, glucocerebrosidase, and sphingolipid-activating protein in HepG2 cells. J Biol Chem. 1991 Mar 15;266(8):4862–4868. [PubMed] [Google Scholar]
- Rochefort H. Biological and clinical significance of cathepsin D in breast cancer. Acta Oncol. 1992;31(2):125–130. doi: 10.3109/02841869209088891. [DOI] [PubMed] [Google Scholar]
- Runeberg-Roos P., Törmäkangas K., Ostman A. Primary structure of a barley-grain aspartic proteinase. A plant aspartic proteinase resembling mammalian cathepsin D. Eur J Biochem. 1991 Dec 18;202(3):1021–1027. doi: 10.1111/j.1432-1033.1991.tb16465.x. [DOI] [PubMed] [Google Scholar]
- Sali A., Veerapandian B., Cooper J. B., Moss D. S., Hofmann T., Blundell T. L. Domain flexibility in aspartic proteinases. Proteins. 1992 Feb;12(2):158–170. doi: 10.1002/prot.340120209. [DOI] [PubMed] [Google Scholar]
- Shewale J. G., Tang J. Amino acid sequence of porcine spleen cathepsin D. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3703–3707. doi: 10.1073/pnas.81.12.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sielecki A. R., Fedorov A. A., Boodhoo A., Andreeva N. S., James M. N. Molecular and crystal structures of monoclinic porcine pepsin refined at 1.8 A resolution. J Mol Biol. 1990 Jul 5;214(1):143–170. doi: 10.1016/0022-2836(90)90153-D. [DOI] [PubMed] [Google Scholar]
- Tandon A. K., Clark G. M., Chamness G. C., Chirgwin J. M., McGuire W. L. Cathepsin D and prognosis in breast cancer. N Engl J Med. 1990 Feb 1;322(5):297–302. doi: 10.1056/NEJM199002013220504. [DOI] [PubMed] [Google Scholar]
- Ullrich K., Mersmann G., Weber E., Von Figura K. Evidence for lysosomal enzyme recognition by human fibroblasts via a phosphorylated carbohydrate moiety. Biochem J. 1978 Mar 15;170(3):643–650. doi: 10.1042/bj1700643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa S., Fujii K., Maejima Y., Tamoto K., Mori Y., Muto N. Further studies on rat cathepsin E: subcellular localization and existence of the active subunit form. Arch Biochem Biophys. 1988 Nov 15;267(1):176–183. doi: 10.1016/0003-9861(88)90021-5. [DOI] [PubMed] [Google Scholar]
- Yonezawa S., Takahashi T., Wang X. J., Wong R. N., Hartsuck J. A., Tang J. Structures at the proteolytic processing region of cathepsin D. J Biol Chem. 1988 Nov 5;263(31):16504–16511. [PubMed] [Google Scholar]
- van Noort J. M., van der Drift A. C. The selectivity of cathepsin D suggests an involvement of the enzyme in the generation of T-cell epitopes. J Biol Chem. 1989 Aug 25;264(24):14159–14164. [PubMed] [Google Scholar]
- von Figura K. Molecular recognition and targeting of lysosomal proteins. Curr Opin Cell Biol. 1991 Aug;3(4):642–646. doi: 10.1016/0955-0674(91)90035-w. [DOI] [PubMed] [Google Scholar]