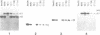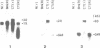Abstract
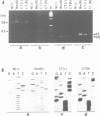
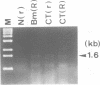
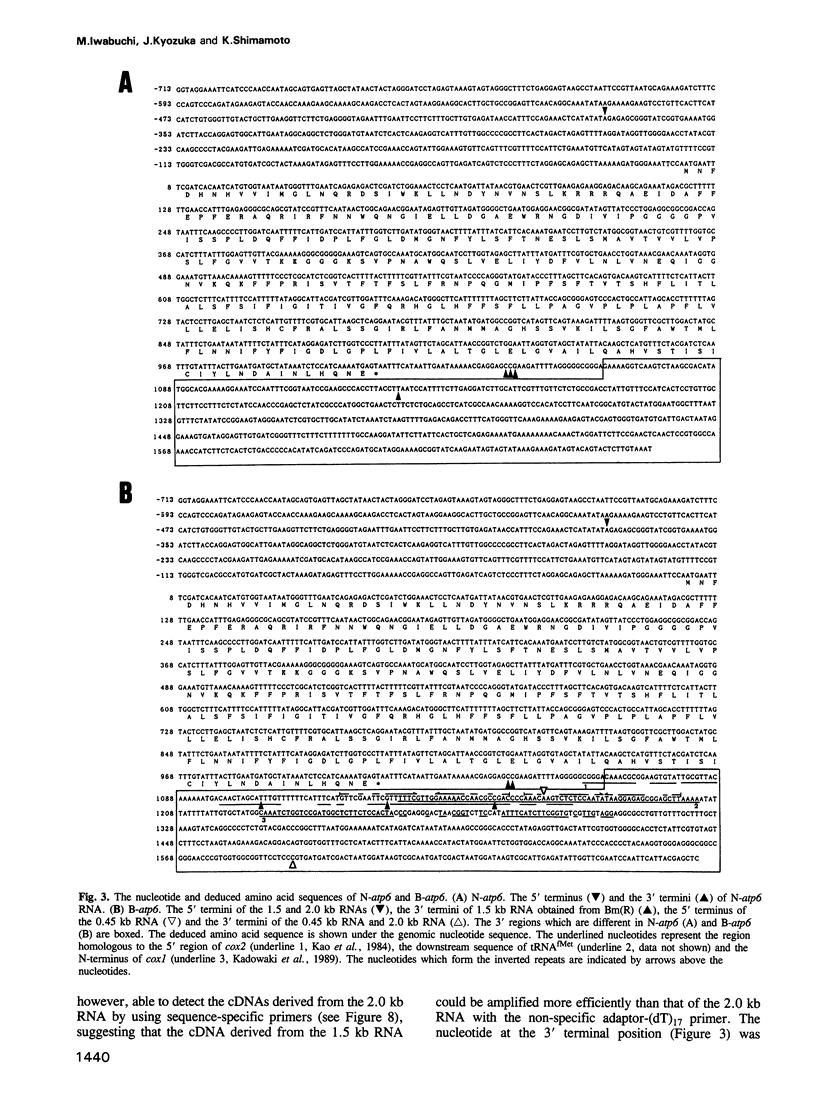
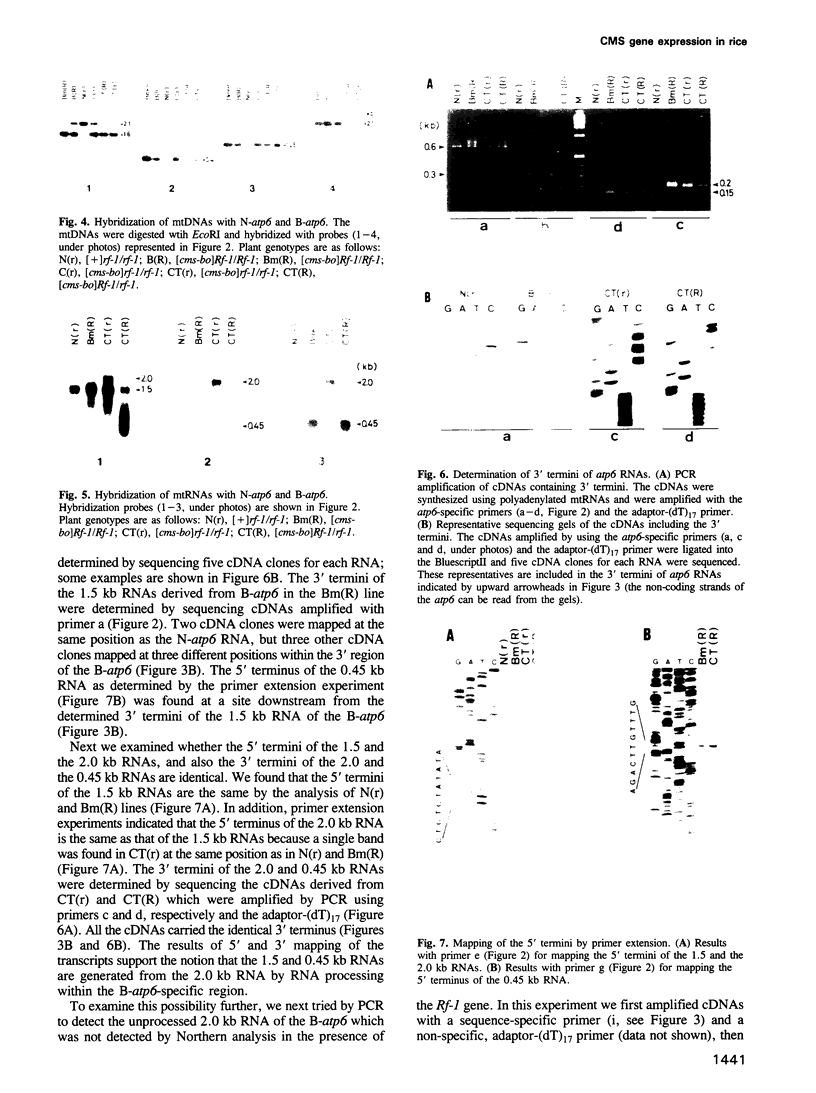
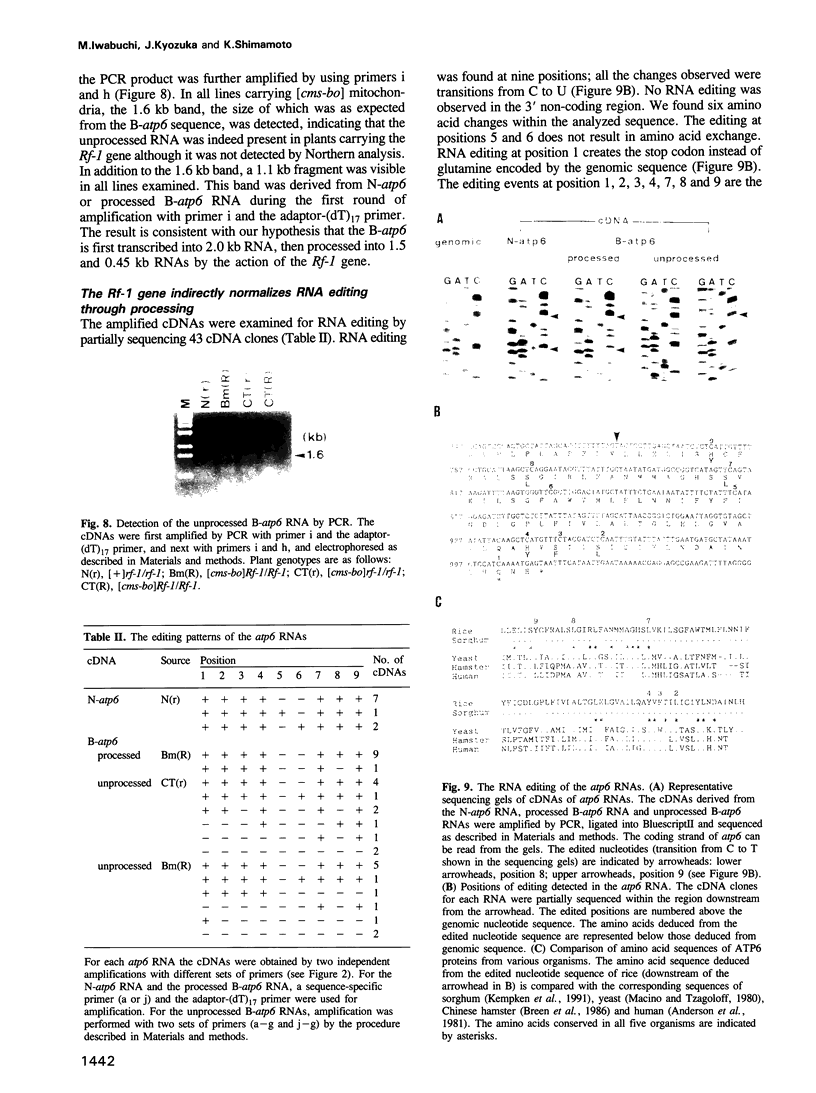
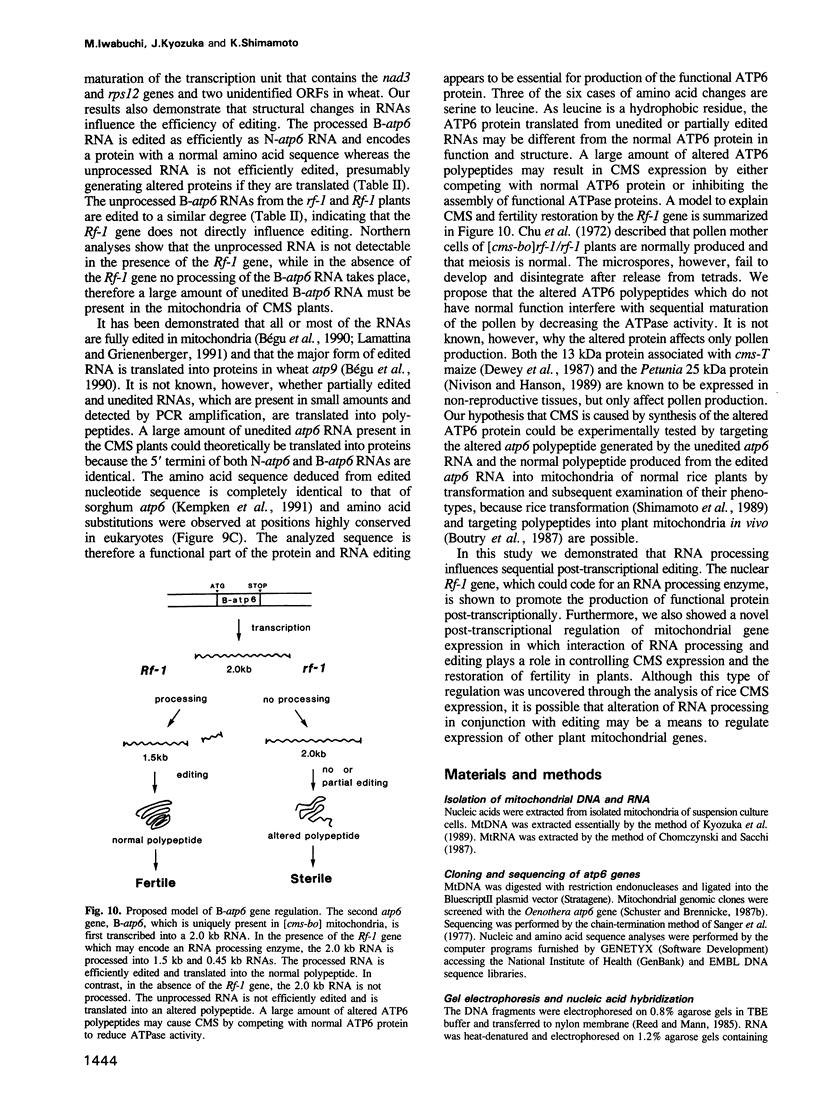
Two atp6 genes were found in the mitochondrial genome of cytoplasmic male sterile (CMS) rice carrying the [cms-bo] cytoplasm. One (N-atp6) was identical to the normal cytoplasmic gene, while the second (B-atp6) was identified as a candidate CMS gene by Southern analysis of the mitochondrial genome of CMS cybrid rice. The coding sequence of B-atp6 was identical to the normal N-atp6 gene but its 3'-flanking sequence was different starting at 49 bases downstream from the stop codon. Northern analysis showed that B-atp6 is transcribed into a 2.0 kb RNA in the absence of the Rf-1 gene, whereas two discontinuous RNAs, of approximately 1.5 and 0.45 kb, were detected in the presence of the Rf-1 gene. Determination of the 3' and 5' ends of these RNAs suggested that the two discontinuous RNAs were generated from the 2.0 kb RNA by RNA processing at sites within the B-atp6-specific sequences by the action of the Rf-1 gene. Sequence analysis of cDNA clones derived from the N-atp6 RNA and the processed and unprocessed RNAs of B-atp6 indicated that the processed B-atp6 RNAs were edited as efficiently as the N-atp6, whereas unedited and partially edited RNAs were detected among unprocessed RNAs. RNA processing by Rf-1 thus influences the sequential post-transcriptional editing of the B-atp6 RNAs. Because the unprocessed RNAs of B-atp6 are possibly translated into altered polypeptides, our results suggest that interaction of RNA processing and editing plays a role in controlling CMS expression and the restoration of fertility in rice.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Apirion D. RNA processing in a unicellular microorganism: implications for eukaryotic cells. Prog Nucleic Acid Res Mol Biol. 1983;30:1–40. doi: 10.1016/s0079-6603(08)60682-0. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Hanson D. K., Fox T. D., Leaver C. J. Mitochondrial genome rearrangement leads to extension and relocation of the cytochrome c oxidase subunit I gene in sorghum. Cell. 1986 Nov 21;47(4):567–576. doi: 10.1016/0092-8674(86)90621-5. [DOI] [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J. P., Sloof P., Van Boom J. H., Tromp M. C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986 Sep 12;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Boutry M., Nagy F., Poulsen C., Aoyagi K., Chua N. H. Targeting of bacterial chloramphenicol acetyltransferase to mitochondria in transgenic plants. Nature. 1987 Jul 23;328(6128):340–342. doi: 10.1038/328340a0. [DOI] [PubMed] [Google Scholar]
- Breen G. A., Miller D. L., Holmans P. L., Welch G. Mitochondrial DNA of two independent oligomycin-resistant Chinese hamster ovary cell lines contains a single nucleotide change in the ATPase 6 gene. J Biol Chem. 1986 Sep 5;261(25):11680–11685. [PubMed] [Google Scholar]
- Bégu D., Graves P. V., Domec C., Arselin G., Litvak S., Araya A. RNA editing of wheat mitochondrial ATP synthase subunit 9: direct protein and cDNA sequencing. Plant Cell. 1990 Dec;2(12):1283–1290. doi: 10.1105/tpc.2.12.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Y., Martin N. C. Biosynthesis of tRNA in yeast mitochondria. An endonuclease is responsible for the 3'-processing of tRNA precursors. J Biol Chem. 1988 Sep 25;263(27):13677–13682. [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cooper P., Butler E., Newton K. J. Identification of a maize nuclear gene which influences the size and number of cox2 transcripts in mitochondria of perennial ++teosintes. Genetics. 1990 Oct;126(2):461–467. doi: 10.1093/genetics/126.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. RNA editing in plant mitochondria. Nature. 1989 Oct 19;341(6243):662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- Dawson A. J., Jones V. P., Leaver C. J. The apocytochrome b gene in maize mitochondria does not contain introns and is preceded by a potential ribosome binding site. EMBO J. 1984 Sep;3(9):2107–2113. doi: 10.1002/j.1460-2075.1984.tb02098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey R. E., Levings C. S., 3rd, Timothy D. H. Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm. Cell. 1986 Feb 14;44(3):439–449. doi: 10.1016/0092-8674(86)90465-4. [DOI] [PubMed] [Google Scholar]
- Dewey R. E., Timothy D. H., Levings C. S. A mitochondrial protein associated with cytoplasmic male sterility in the T cytoplasm of maize. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5374–5378. doi: 10.1073/pnas.84.15.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escote-Carlson L. J., Gabay-Laughnan S., Laughnan J. R. Nuclear genotype affects mitochondrial genome organization of CMS-S maize. Mol Gen Genet. 1990 Sep;223(3):457–464. doi: 10.1007/BF00264454. [DOI] [PubMed] [Google Scholar]
- Fox T. D., Leaver C. J. The Zea mays mitochondrial gene coding cytochrome oxidase subunit II has an intervening sequence and does not contain TGA codons. Cell. 1981 Nov;26(3 Pt 1):315–323. doi: 10.1016/0092-8674(81)90200-2. [DOI] [PubMed] [Google Scholar]
- Grivell L. A. Nucleo-mitochondrial interactions in yeast mitochondrial biogenesis. Eur J Biochem. 1989 Jul 1;182(3):477–493. doi: 10.1111/j.1432-1033.1989.tb14854.x. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Bonnard G., Lamattina L., Grienenberger J. M. Expression of the wheat mitochondrial nad3-rps12 transcription unit: correlation between editing and mRNA maturation. Plant Cell. 1991 Oct;3(10):1109–1120. doi: 10.1105/tpc.3.10.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto J. M., Lamattina L., Bonnard G., Weil J. H., Grienenberger J. M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature. 1989 Oct 19;341(6243):660–662. doi: 10.1038/341660a0. [DOI] [PubMed] [Google Scholar]
- Hanic-Joyce P. J., Gray M. W. Processing of transfer RNA precursors in a wheat mitochondrial extract. J Biol Chem. 1990 Aug 15;265(23):13782–13791. [PubMed] [Google Scholar]
- Hiesel R., Schobel W., Schuster W., Brennicke A. The cytochrome oxidase subunit I and subunit III genes in Oenothera mitochondria are transcribed from identical promoter sequences. EMBO J. 1987 Jan;6(1):29–34. doi: 10.1002/j.1460-2075.1987.tb04714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesel R., Wissinger B., Schuster W., Brennicke A. RNA editing in plant mitochondria. Science. 1989 Dec 22;246(4937):1632–1634. doi: 10.1126/science.2480644. [DOI] [PubMed] [Google Scholar]
- Hoch B., Maier R. M., Appel K., Igloi G. L., Kössel H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature. 1991 Sep 12;353(6340):178–180. doi: 10.1038/353178a0. [DOI] [PubMed] [Google Scholar]
- Hollingsworth M. J., Martin N. C. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Mol Cell Biol. 1986 Apr;6(4):1058–1064. doi: 10.1128/mcb.6.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki K., Suzuki T., Kazama S. A chimeric gene containing the 5' portion of atp6 is associated with cytoplasmic male-sterility of rice. Mol Gen Genet. 1990 Oct;224(1):10–16. doi: 10.1007/BF00259445. [DOI] [PubMed] [Google Scholar]
- Kadowaki K., Suzuki T., Kazama S., Oh-fuchi T., Sakamoto W. Nucleotide sequence of the cytochrome oxidase subunit I gene from rice mitochondria. Nucleic Acids Res. 1989 Sep 25;17(18):7519–7519. doi: 10.1093/nar/17.18.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao T., Moon E., Wu R. Cytochrome oxidase subunit II gene of rice has an insertion sequence within the intron. Nucleic Acids Res. 1984 Oct 11;12(19):7305–7315. doi: 10.1093/nar/12.19.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempken F., Mullen J. A., Pring D. R., Tang H. V. RNA editing of sorghum mitochondrial atp6 transcripts changes 15 amino acids and generates a carboxy-terminus identical to yeast. Curr Genet. 1991 Nov;20(5):417–422. doi: 10.1007/BF00317071. [DOI] [PubMed] [Google Scholar]
- Köhler R. H., Horn R., Lössl A., Zetsche K. Cytoplasmic male sterility in sunflower is correlated with the co-transcription of a new open reading frame with the atpA gene. Mol Gen Genet. 1991 Jul;227(3):369–376. doi: 10.1007/BF00273925. [DOI] [PubMed] [Google Scholar]
- Lamattina L., Grienenberger J. M. RNA editing of the transcript coding for subunit 4 of NADH dehydrogenase in wheat mitochondria: uneven distribution of the editing sites among the four exons. Nucleic Acids Res. 1991 Jun 25;19(12):3275–3282. doi: 10.1093/nar/19.12.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver H. K., Reynolds S. J., Moneger F., Leaver C. J. Mitochondrial genome organization and expression associated with cytoplasmic male sterility in sunflower (Helianthus annuus). Plant J. 1991 Sep;1(2):185–193. doi: 10.1111/j.1365-313x.1991.00185.x. [DOI] [PubMed] [Google Scholar]
- Levings C. S., 3rd, Brown G. G. Molecular biology of plant mitochondria. Cell. 1989 Jan 27;56(2):171–179. doi: 10.1016/0092-8674(89)90890-8. [DOI] [PubMed] [Google Scholar]
- Levings C. S., 3rd The Texas cytoplasm of maize: cytoplasmic male sterility and disease susceptibility. Science. 1990 Nov 16;250(4983):942–947. doi: 10.1126/science.250.4983.942. [DOI] [PubMed] [Google Scholar]
- Macino G., Tzagoloff A. Assembly of the mitochondrial membrane system: sequence analysis of a yeast mitochondrial ATPase gene containing the oli-2 and oli-4 loci. Cell. 1980 Jun;20(2):507–517. doi: 10.1016/0092-8674(80)90637-6. [DOI] [PubMed] [Google Scholar]
- Makaroff C. A., Apel I. J., Palmer J. D. The atp6 coding region has been disrupted and a novel reading frame generated in the mitochondrial genome of cytoplasmic male-sterile radish. J Biol Chem. 1989 Jul 15;264(20):11706–11713. [PubMed] [Google Scholar]
- Morikami A., Nakamura K. The pea mitochondrial ATPase subunit 9 gene is located upstream of the ATPase alpha-subunit gene. Nucleic Acids Res. 1987 Jun 11;15(11):4692–4692. doi: 10.1093/nar/15.11.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan R. M., Maloney A. P., Walbot V. RNA processing and multiple transcription initiation sites result in transcript size heterogeneity in maize mitochondria. Mol Gen Genet. 1988 Mar;211(3):373–380. doi: 10.1007/BF00425688. [DOI] [PubMed] [Google Scholar]
- Nivison H. T., Hanson M. R. Identification of a mitochondrial protein associated with cytoplasmic male sterility in petunia. Plant Cell. 1989 Nov;1(11):1121–1130. doi: 10.1105/tpc.1.11.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L. M., Wallis S. C., Pease R. J., Edwards Y. H., Knott T. J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987 Sep 11;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Pruitt K. D., Hanson M. R. Transcription of the Petunia mitochondrial CMS-associated Pcf locus in male sterile and fertility-restored lines. Mol Gen Genet. 1991 Jul;227(3):348–355. doi: 10.1007/BF00273922. [DOI] [PubMed] [Google Scholar]
- Rasmussen J., Hanson M. R. A NADH dehydrogenase subunit gene is co-transcribed with the abnormal Petunia mitochondrial gene associated with cytoplasmic male sterility. Mol Gen Genet. 1989 Jan;215(2):332–336. doi: 10.1007/BF00339738. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster W., Brennicke A. Nucleotide sequence of the Oenothera ATPase subunit 6 gene. Nucleic Acids Res. 1987 Nov 11;15(21):9092–9092. doi: 10.1093/nar/15.21.9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siculella L., Palmer J. D. Physical and gene organization of mitochondrial DNA in fertile and male sterile sunflower. CMS-associated alterations in structure and transcription of the atpA gene. Nucleic Acids Res. 1988 May 11;16(9):3787–3799. doi: 10.1093/nar/16.9.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L., Shaw J. RNA editing and the mitochondrial cryptogenes of kinetoplastid protozoa. Cell. 1989 May 5;57(3):355–366. doi: 10.1016/0092-8674(89)90911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton C. A., Conklin P. L., Pruitt K. D., Hanson M. R. Editing of pre-mRNAs can occur before cis- and trans-splicing in Petunia mitochondria. Mol Cell Biol. 1991 Aug;11(8):4274–4277. doi: 10.1128/mcb.11.8.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. M., Lamb R. A., Paterson R. G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988 Sep 9;54(6):891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A. J., Mulligan R. M. RNA editing intermediates of cox2 transcripts in maize mitochondria. Mol Cell Biol. 1991 Aug;11(8):4278–4281. doi: 10.1128/mcb.11.8.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E. G., Hanson M. R. A fused mitochondrial gene associated with cytoplasmic male sterility is developmentally regulated. Cell. 1987 Jul 3;50(1):41–49. doi: 10.1016/0092-8674(87)90660-x. [DOI] [PubMed] [Google Scholar]