Abstract
We present a novel persistent homological sparse network analysis framework for characterizing white matter abnormalities in tensor-based morphometry (TBM) in magnetic resonance imaging (MRI). Traditionally TBM is used in quantifying tissue volume change in each voxel in a massive univariate fashion. However, this obvious approach cannot be used in testing, for instance, if the change in one voxel is related to other voxels. To address this limitation of univariate-TBM, we propose a new persistent homological approach to testing more complex relational hypotheses across brain regions. The proposed methods are applied to characterize abnormal white matter in maltreated children. The results are further validated using fractional anisotropy (FA) values in diffusion tensor imaging (DTI).
1 Introduction
Traditionally tensor-based morphometry (TBM) in magnetic resonance imaging (MRI) has been massively univariate in that response variables are fitted using a linear model at each voxel producing massive number of test statistics (Figure 1). However, univariate approaches are ill-suited for testing more complex hypotheses about multiple anatomical regions. For example, the univariate-TBM cannot answer how the volume increase in one voxel is related to other voxels. To address this type of more complex relational hypothesis across different brain regions, we propose a new persistent homological approach.
Fig. 1.
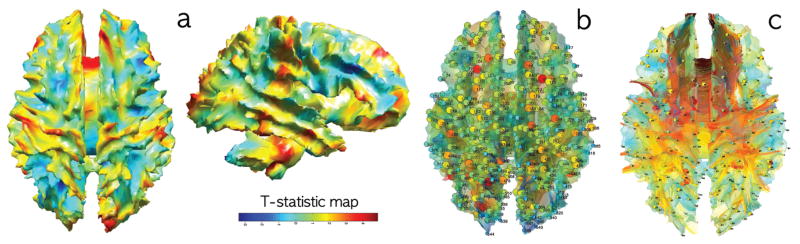
(a) T-statistic map of group differences (PI-controls) on Jacobian determinants. (b) 548 uniformly sampled nodes in MRI where the persistent homology is applied. The nodes are sparsely sampled in the template to guarantee there is no spurious high correlation due to proximity between nodes. (c) The same nodes are taken in DTI to check the consistency against the MRI results.
The Jacobian determinant is the most often used volumetric mesurement in TBM. We propose to correlate the Jacobian determinant across different voxels and quantify how the volume change in one voxel is correlated to the volume changes in other voxels. However, existing multivariate statistical methods exhibit serious defects in applying to the whole brain regions due to the small-n large-p problem [2]. Specifically, the number of voxels p are substantially larger than the number of subjects n so the often used maximum likelihood estimation (MLE) of the covariance matrix shows the rank deficiency and it is no longer positive definite. In turn, the estimated correlation matrix is not considered as a good approximation to the true correlation matrix. The small-n large-p problem can be addressed by regularizing the ill-conditioned correlation or covariance matrices by sparse regularization terms.
Sparse model
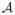 is usually parameterized by a tuning parameter λ that controls the sparsity of the representation. Increasing the sparse parameter makes the representation more sparse. Instead of performing statistical inference at one fixed λ that may not be optimal, we propose to quantify how the topology of sparse solution changes over the increasing λ using the persistent homology. Then it is possible to obtain additional characterization of a population that cannot be obtained in the univariate-TBM.
is usually parameterized by a tuning parameter λ that controls the sparsity of the representation. Increasing the sparse parameter makes the representation more sparse. Instead of performing statistical inference at one fixed λ that may not be optimal, we propose to quantify how the topology of sparse solution changes over the increasing λ using the persistent homology. Then it is possible to obtain additional characterization of a population that cannot be obtained in the univariate-TBM.
The proposed framework is applied in characterizing abnormal white matter alterations in children who experienced maltreatment while living in post-institutional (PI) settings before being adopted by families in the US. The main contributions of the paper are (i) the introduction of a novel persistent homological approach to characterizing white matter abnormality and (ii) its application to MRI and DTI showing consistent results between the modalities.
2 Motivation
Let Jn×p = (Jij) be the matrix of Jacobian determinant for subject i at voxel position j. The subscripts denote the dimension of matrix. There are p voxels of interest and n subjects. The Jacobian determinants of all subjects at the j-th voxel is denoted as xj = (J1j, ···, Jnj)′. The Jacobian determinants of all voxels for the i-th subject is denoted as yi = (Ji1, ···, Jip)′. xj is the j-th column and yi is the i-th row of the data matrix J. The covariance matrix of yi is given by Cov (yi) = Σp×p = (σkl) and estimated using the sample covariance matrix S via MLE. To remedy this small–n and large-p problem, the likelihood is regularized with L1-penalty:
| (1) |
where || · ||1 is the sum of the absolute values of the elements. L is maximized over all possible symmetric positive definite matrices. (1) is a convex problem and it is usually solved using the graphical-lasso (GLASSO) algorithm [4,6].
Since the different choice of parameter λ will produce different solutions, we propose to use the collection of Σ−1(λ) for every possible value of λ for the subsequent statistical inference. This avoids the problem of identifying the optimal sparse parameter that may not be optimal in practice. The question is then how to use the collection of Σ−1(λ) in a coherent mathematical fashion.
Consider a sparse model
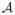 (λ), which gets more sparse as λ increases. Then under some condition, it is possible to have
(λ), which gets more sparse as λ increases. Then under some condition, it is possible to have
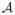 (λ1) ⊃
(λ1) ⊃
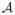 (λ2) ⊃
(λ2) ⊃
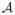 (λ3) ⊃ ··· for λ1 ≤ λ2 ≤ ···. Within the persistent homological framework [5],
(λ3) ⊃ ··· for λ1 ≤ λ2 ≤ ···. Within the persistent homological framework [5],
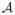 (λ) is said to be persistent if it has this type of nested subset structure. The collection of the nested subsets is called filtration.
(λ) is said to be persistent if it has this type of nested subset structure. The collection of the nested subsets is called filtration.
3 Persistent Structures for Sparse Network Models
Sparse Correlations
We assume the measurement vector xj at the j-th node is centered with zero mean and unit variance. These condition is achieved by centering and normalizing data. Let Γ = (γjk) be the correlation matrix, where γjk is the correlation between the nodes j and k. Sparse correlation Γ is then estimated as
| (2) |
where β = (βjk). When λ = 0, the sparse correlation is simply given by the sample correlation, i.e. . As λ increases, the correlation becomes more sparse. Using the sparse solution (2), we will explicitly construct a persistent structure on Γ̂(λ) over changing λ.
Let A = (ajk) be the adjacency matrix defined using the sparse correlation:
Let
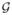 (λ) be the graph induced from the adjacency matrix A. It can be algebraically shown that the induced graph is persistent and from a filtration:
(λ) be the graph induced from the adjacency matrix A. It can be algebraically shown that the induced graph is persistent and from a filtration:
| (3) |
for λ1 ≤ λ2 ≤ λ3. The proof follows by simplifying the adjacency matrix A into a simpler but equivalent adjacency matrix B = (bjk):
| (4) |
Then it is not difficult to see the graph induced from the adjacency matrix B should be persistent. Hence, the filtration on
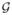 (λ) can be constructed by simply thresholding the sample correlation
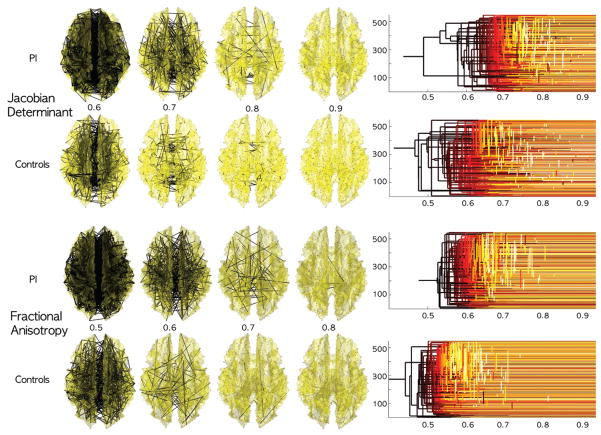
for each λ without solving the optimization problem (2). Figure 2 shows filtrations obtained from sparse correlations between Jacobian determinants on preselected 548 nodes in the two groups showing group difference. It is not necessary to perform filtrations for infinitely many possible filtration values. For an n-node network, it can be algebraically shown that at most n − 1 increments are sufficient to obtain a unique filtration.
(λ) can be constructed by simply thresholding the sample correlation
for each λ without solving the optimization problem (2). Figure 2 shows filtrations obtained from sparse correlations between Jacobian determinants on preselected 548 nodes in the two groups showing group difference. It is not necessary to perform filtrations for infinitely many possible filtration values. For an n-node network, it can be algebraically shown that at most n − 1 increments are sufficient to obtain a unique filtration.
Fig. 2.
Networks
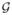 (λ) obtained by thresholding sparse correlations for the Jacobian determinant from MRI and fractional anisotropy from DTI at different λ values. The collection of the thresholded graphs forms a filtration. PI shows more dense network at a given λ value. Since PI is more homogenous, in the white matter region, there are more dense high correlations between nodes. The filtration is visualized using the equivalent dendrogram [5], which also shows more dense linkages for PI at high correlations.
(λ) obtained by thresholding sparse correlations for the Jacobian determinant from MRI and fractional anisotropy from DTI at different λ values. The collection of the thresholded graphs forms a filtration. PI shows more dense network at a given λ value. Since PI is more homogenous, in the white matter region, there are more dense high correlations between nodes. The filtration is visualized using the equivalent dendrogram [5], which also shows more dense linkages for PI at high correlations.
Sparse Likelihood
The identification of a persistent homological structure out of the inverse covariance Σ̂−1(λ) in (1) is similar. However, it is more involved than the sparse correlation case. Let A = (aij) be the adjacency matrix
| (5) |
The adjacency matrix A induces a graph
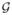 (λ) consisting of κ(λ) number of partitioned subgraphs
(λ) consisting of κ(λ) number of partitioned subgraphs
where Vl and El are vertex and edge sets of the subgraph Gl respectively. Unlike the sparse correlation case, we do not have full persistency on the induced graph
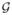 . The partitioned graphs can be proven to be partially nested in a sense that only the partitioned node sets are persistent [4,6], i.e.
. The partitioned graphs can be proven to be partially nested in a sense that only the partitioned node sets are persistent [4,6], i.e.
| (6) |
for λ1 ≤ λ2 ≤ λ3 and all l. Subsequently the collection of partitioned vertex set is also persistent. On the other hand, the edge sets El may not be persistent. The identification of the vertex filtration can be fairly time consuming since it requires solving the convex optimization problem (1) for multiple λ values. However, it can be easily obtained by identifying a simpler adjacency matrix B that gives the identical vertex sets Vl (Figure 3).
Fig. 3.
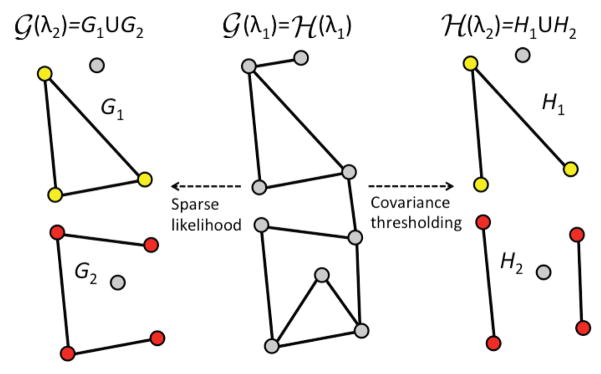
Schematic of graph filtrations obtained by sparse-likelihood (5) and sample covariance thresholding (7). The vertex set of
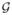 (λ1) =
(λ1) =
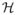 (λ1) consists of gray nodes. For the next filtration value λ2,
(λ1) consists of gray nodes. For the next filtration value λ2,
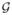 (λ1) ≠
(λ1) ≠
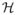 (λ1). However, the partitioned vertex sets (yellow and red) of
(λ1). However, the partitioned vertex sets (yellow and red) of
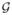 (λ1) and
(λ1) and
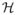 (λ1) match.
(λ1) match.
Let B(λ) = (bij) be another adjacency matrix given by
| (7) |
where ŝij is the sample covariance matrix. The adjacency matrix B similarly induces the graph
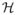 with τ(λ) disjoint subgraphs:
with τ(λ) disjoint subgraphs:
with Hl = {Wl(λ), Fl(λ)}. Wl and Fl are vertex and edge sets of the subgraph Hl respectively. Then trivially the node set Wl forms a filtration over the sparse parameter:
| (8) |
It can be further shown that κ(λ) = τ(λ) and Vl(λ) = Wl(λ) for all λ > 0 [6]. Hence, the filtration on the vertex set Vl(λ) is constructed by simply thresholding the sample covariance ŝij(λ) for each λ without solving the time consuming optimization problem (2). Figure 3 shows the schematic of constructing the filtration on sparse likelihood by the covariance thresholding.
4 Application to Maltreated Children Study
MRI Data and Univariate-TBM
T1-weighted MRI were collected using a 3T GE SIGNA scanner for 23 children who experienced maltreatment while living in post-institutional (PI) settings before being adopted by families in the US, and age-matched 31 normal control subjects. The average age for PI is 11.26 ± 1.71 years while that of controls is 11.58 ± 1.61 years. A study specific template was constructed using the diffeomorphic shape and intensity averaging technique through Advanced Normalization Tools (ANTS) [1]. Image normalization of each individual image to the template was done using symmetric normalization with cross-correlation as the similarity metric. The 1mm deformation fields are then smoothed out with Guassian kernel with bandwidth σ = 4mm.
The computed Jacobian maps were feed into univariate-GLM at each voxel for testing the group effect while accounting for age and gender difference. Figure 1 shows the significant group difference between PI and controls. Any region above the T-statistic value of 4.86 or below −4.86 is considered significant at 0.05 (corrected). However, what the univariate-TBM can not determine is the dependency of Jacobian determinants at two different positions. It is possible that structural abnormality at one region of the brain might be related to the other regions due to interregional dependency. For this type of more complex hypothesis, we need the proposed persistent homological approach.
Inference on Barcodes
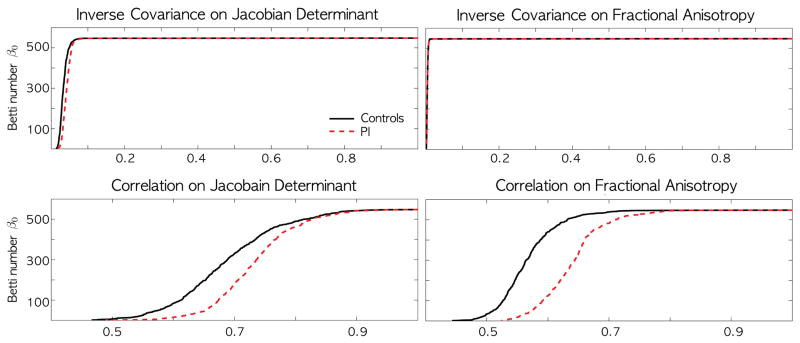
Since Jacobian determinants at neighboring voxels are highly correlated, we uniformly subsampled p = 548 number of nodes along the white matter template mesh vertices in order not to have spurious high correlation between two adjacent nodes (Figure 1). The proposed method is very robust under the change of node sizes. For the node sizes between 548 and 1856, the choice of node sizes did not affect the subsequent analysis. Following the proposed method, we constructed the filtrations on sparse correlations and inverse covariance without solving the optimizations (1) and (2). The filtrations are quantified using the barcode representation, which plots the change of Betti numbers over filtration values [5] (Figure 4). The first Betti number β0(λ) counts the number of connected components of the graph
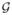 (λ) at the filtration value λ.
(λ) at the filtration value λ.
Fig. 4.
The barcodes on the sparse inverse covariance (top) and correlation (bottom) for Jacobian determinant (left) and FA (right). Unlike the inverse sparse covariance, the sparse correlation shows huge group separation between normal controls and post-institutionalized (PI) children (p-value < 0.001).
Given the barcode for group i, we tested if the barcodes were different between the groups, i.e. for some λ ∈ [0, 1]. A Kolmogorov-Smirnov (KS) like test statistic is used. Since each group produces one barcode, we used the Jackknife resampling technique for inference. For a group with n subjects, one subject is removed and the remaining n − 1 subjects are used in constructing the barcode. This process is repeated for each subject to produce n barcodes (Figure 4). The Jackknife resampling produces 23 and 31 barcodes respectively for PI and controls. In order for the permutation test to converge for our data set, it requires tens of thousands permutations and it is really time consuming. So we used a much simpler Jackknife resampling. Then the test statistic T is constructed between 23 × 31 pairs of barcodes. Under the null, T is expected to be zero. One-sample T-test on the collection of T is then subsequently performed to show huge group discrimination for sparse correlations in Figure 4 (p-value < 0.001). The barcodes for normal controls show much higher Betti numbers at the given threshold. This suggests higher non-uniformity in Jacobian determinants across the brain that causes increased disconnections in correlations. The inverse covariance was not able to discriminate the groups. The MATLAB codes for constructing barcodes and statical inference is given in http://brainimaging.waisman.wisc.edu/~chung/barcodes.
Validation Against DTI
For children who suffered early neglect and abuse, white matter microstructures are more diffusely organized [3]. So we expect less white matter variability not only in the Jacobian determinants but also in the fractional anisotropy (FA) values in DTI as well. The MRI data in this study has the corresponding DTI. The DTI acquisition are done in the same 3T GE SIGNA scanner and the acquisition parameters can be found in [3]. We applied the proposed persistent homological method in obtaining the filtrations for sparse correlations and inverse covariances in the same 548 nodes (Figure 1). The resulting filtration patterns also show similar pattern of rapid increase in disconnected components (Figure 2 and 4) for sparse correlations. The Jackknife-based one-sample T-test also shows significant group difference for correlations (p-value < 0.001). This results are due to consistent abnormality observed in both MRI and DTI modalities. PI exhibited stronger white matter homogeneity and less spatial variability compared to normal controls in both MRI and DTI measurements. The inverse covariance was not able to discriminate the groups.
5 Conclusions and Discussions
Using the persistent homological framework, we have shown that PI group shows less anatomic variation in MRI compared to the normal controls. This result is consistent with DTI, which shows similar patterns. The reason we did not detect the group difference in the inverse covariances might be that, as shown in Figure 4, the changes in the first Betti number are occurring in a really narrow window and losing the discrimination power. On the other hand the sparse correlations exhibit more slow changes in the Betti number over the wide window making it easier to discriminate the groups. The proposed method is general enough to run on any type of volumetric imaging data that is spatially normalized.
Acknowledgments
This work was supported by NIH Research Grants MH61285 and MH68858 to S.D.P., NIMH Grant MH84051 to R.J.D. and US National Institute of Drug Abuse (Fellowship DA028087) to J.L.H. The authors like to thank Matthew Arnold of University of Bristol for the discussion on the KS-test procedure and pointing out the reference [6].
References
- 1.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friston KJ, Holmes AP, Worsley KJ, Poline JP, Frith CD, Frackowiak RSJ. Statistical parametric maps in functional imaging: a general linear approach. Human Brain Mapping. 1995;2:189–210. [Google Scholar]
- 3.Hanson JL, Adluru N, Chung MK, Alexander AL, Davidson RJ, Pollak SD. Early neglect is associated with alterations in white matter integrity and cognitive functioning. Child Development. 2013 doi: 10.1111/cdev.12069. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang S, Li J, Sun L, Ye J, Fleisher A, Wu T, Chen K, Reiman E. Learning brain connectivity of Alzheimer’s disease by sparse inverse covariance estimation. Neuro Image. 2010;50:935–949. doi: 10.1016/j.neuroimage.2009.12.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee H, Chung MK, Kang H, Kim BN, Lee DS. Computing the shape of brain networks using graph filtration and Gromov-Hausdorff metric. MICCAI, Lecture Notes in Computer Science. 2011;6892:302–309. doi: 10.1007/978-3-642-23629-7_37. [DOI] [PubMed] [Google Scholar]
- 6.Mazumder R, Hastie T. Exact covariance thresholding into connected components for large-scale graphical lasso. The Journal of Machine Learning Research. 2012;13:781–794. [PMC free article] [PubMed] [Google Scholar]