Abstract
Protein splicing in trans by split inteins has increasingly become a powerful protein-engineering tool for protein ligation, both in vivo and in vitro. Over 100 naturally occurring and artificially engineered split inteins have been reported for protein ligation using protein trans-splicing. Here, we review the current status of the reported split inteins in order to delineate an empirical or rational strategy for constructing new split inteins suitable for various applications in biotechnology and chemical biology.
Keywords: protein ligation, protein splicing, protein trans-splicing, split intein
Introduction
Splitting proteins has become a routine protein engineering strategy with the goal being the development of new tools for protein complementation assay (PCA). Interaction or association of two split fragments from a protein restores the function of the original protein (Michnick et al., 2007). Protein splicing can also be used as a PCA system by splitting an intein into two fragments, which can catalyze protein splicing in trans (Shingledecker et al., 1998; Southworth et al., 1998; Yamazaki et al., 1998). Protein splicing is a posttranslational modification catalyzed by an intervening protein sequence, which is termed intein. In protein splicing the intein is auto-catalytically self-excised out, accompanied by simultaneous ligation of the flanking protein sequences (Fig. 1a) (Hirata et al., 1990; Kane et al., 1990; Paulus, 2000). Discovery of inteins opened various useful applications in biotechnology because protein complementation by split inteins in foreign contexts can ligate two foreign polypeptide chains into one (Volkmann and Iwaï, 2010; Perler and Allewell, 2014; Shah and Muir, 2014). Such protein trans-splicing (PTS) by split inteins was thought to provide a means to connect polypeptides in an aqueous solution ‘at will’ because protein splicing could occur in foreign proteins as well as in natural host proteins (Fig. 1a). In practice, it has been quickly realized that utilization of PTS as a universal protein ligation tool is not as easy as it might appear. Artificially split intein precursors were often insoluble, requiring labor- and time-consuming refolding processes (Southworth et al., 1998; Yamazaki et al., 1998; Otomo et al., 1999b). The need to optimize refolding conditions has discouraged protein chemists from using PTS. However, nature also applies the same strategy of utilizing split inteins (Wu et al., 1998a; Caspi et al., 2003; Dassa et al., 2009). Although the function of split inteins in nature is still obscure, naturally split inteins do not require any refolding. Co-expression of the two split intein precursors in the same cells is sufficient for PTS, thus opening an avenue for performing protein engineering in vivo (Paulus, 2001; Topilina and Mills, 2014). The existence of naturally split inteins inspired us to initiate a quest for robust split inteins suitable for protein engineering purposes, which resulted in the discovery of a highly efficient split DnaE intein from Nostoc punctiforme (Iwai et al., 2006; Ellilä et al., 2011). A number of artificially and naturally split inteins have been identified and characterized now by several groups. Some naturally split inteins exhibit impressive ligation kinetics (Zettler et al., 2009; Carvajal-Vallejos et al., 2012; Shah et al., 2012). Here, we review the current status of naturally and artificially split inteins, with the aim of delineating an empirical or rational strategy to design better split inteins.
Fig. 1.
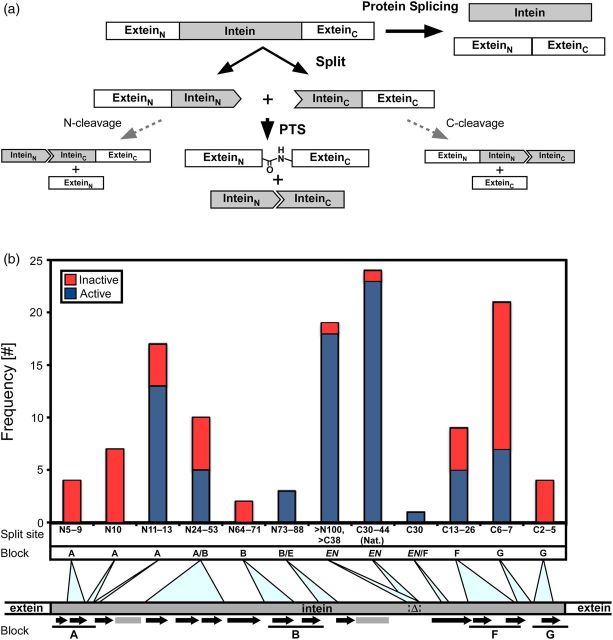
(a) Schematic representation of protein splicing in cis and trans. A cis-splicing intein is naturally or artificially split into two fragments. Upon association of the two intein fragments, protein trans-splicing (PTS) is induced by the formation of an active intein structure. This might also result in N- or/and C-cleavages. (b) Distribution of the split sites from the previously reported split inteins. The split sites are divided into ‘active’ (blue) and ‘inactive’ (red) and are classified by the new split intein nomenclature and blocks. The secondary structures are indicated by black arrows (β-sheets) and gray rectangles (helices). Lines below the secondary structures show the conserved blocks: A, B, F and G. Δ and EN represent endonuclease domain insertions found in many inteins.
How to find ‘better’ splits?
Inteins have been considered to be selfish genetic elements firstly because they do not provide any benefits to their host organisms, and secondly because many inteins contain endonuclease domains that play an essential role for their propagation by homing events (Gogarten et al., 2002; Nogami et al., 2002). The splicing and endonuclease domains are functionally and structurally independent (Hodges et al., 1992; Derbyshire et al., 1997; Duan et al., 1997). Mini-inteins without endonuclease domains occur naturally, and can also be artificially engineered (engineered mini-inteins). This suggests that canonical inteins with endonuclease insertions might have emerged by invasion of homing endonuclease domains, such as the ones encoded in introns (Derbyshire et al., 1997). Over 600 inteins have been identified only in unicellular organisms of the three kingdoms (Perler, 2002). It appears that inteins are gradually becoming extinct although they are believed to have had previous evolutionary roles in ancient times, which provided some advantages for the host organisms (Pietrokovski, 2001). Inteins are indeed more prevalent in archaea than in higher organisms (Aranko et al., 2013b). Inteins are usually inserted in the proximity of the active sites of essential proteins in which inteins reside (Gogarten et al., 2002; Swithers et al., 2009). The insertion inactivating their host proteins has been considered to be the intein's survival strategy, because inteins with other insertion points have become extinct during the evolution (Pietrokovski, 2001; Swithers et al., 2009). This hypothesis implies that ancient forms of inteins might have been more common and active in various protein contexts (thus exhibiting broader specificity) before they have been fixed into their current positions. Therefore, one strategy to hunt for ‘better’ inteins is to identify intein sequences from ancient origins. Mini-inteins, which have not yet been invaded by any homing endonuclease, might serve as prototypes of ancient inteins; even if not, naturally occurring mini-inteins have been successfully used for developing split inteins. At this time, however, we do not know if some mini-inteins are indeed prototypes of ancient inteins. Broader substrate specificity (high tolerance of insertion sites) of inteins, which is the key feature of our hypothetical ancient inteins, remains to be investigated in detail (Iwai et al., 2006; Appleby-Tagoe et al., 2011; Cheriyan et al., 2013). Directed evolution of inteins has also been employed to create ‘better’ inteins with increased specificity for a chosen non-native substrate (Lockless and Muir, 2009), or with generally broader specificities (Appleby-Tagoe et al., 2011). However, the evolved inteins have not been widely used.
Locations of experimental split sites
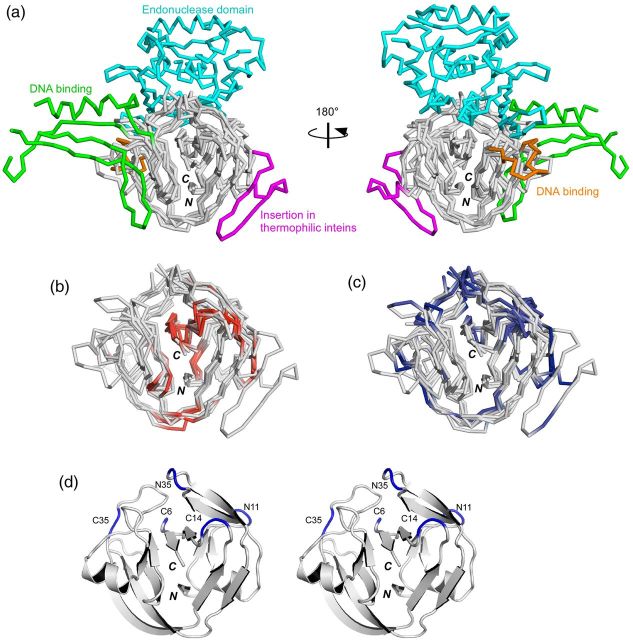
As the first step in the systematic comparison of the split inteins with various lengths, we introduced a simple nomenclature for split inteins, in which the split sites are indicated by one of the closer termini (N or C) to the split site, and the lengths from the terminus (Fig. 1b) (Aranko et al., 2014). We used this nomenclature to classify the reported split inteins in a consistent manner. This nomenclature for the split sites also allows us to conveniently compare them in terms of their locations on the three-dimensional structures. There are currently 13 unique three-dimensional structures of inteins in the Protein Data Bank. The protein splicing domain of inteins shares the HINT (Hedgehog/INTein) fold with the C-terminal domain of hedgehog proteins and bacterial intein-like (BIL) proteins (Hall et al., 1997; Aranko et al., 2013a). In Fig. 2a we show four superimposed HINT domains of the inteins: the compared molecules include a canonical intein with homing endonuclease (VMA intein from Saccharomyces cerverciae); a naturally split intein from Nostoc punctiforme (NpuDnaE intein); the engineered mini-intein from Synechocystis sp. PCC6803 (SspDnaB intein); and a mini-intein from a thermophilic organism, Thermoplasma volcanium GSS1 (TvoVMA intein) (Moure et al., 2002; Ding et al., 2003; Oeemig et al., 2009; Aranko et al., 2014).
Fig. 2.
Locations of the reported split sites on the three-dimensional structures of inteins. (a) Four intein structures superimposed with the HINT fold (SceVMA intein (1LWS), SspDnaBΔ275 intein (1MI8), NpuDnaE intein (2KEQ), TvoVMAΔ21 intein (4O1S)). The endonuclease domain (residues 183–414, colored in cyan) and the DNA binding domains (residues 54-69 and 86-156, colored respectively in orange and green) of SceVMA intein are displayed. The insertion found in thermophilic inteins is displayed in magenta (residues 28-51 in TvoVMA intein). (b) The split sites corresponding to the ‘inactive’ split inteins are colored red in the four superimposed structures, without showing the endonuclease and DNA binding domains. (c) The split sites corresponding to the ‘active’ split inteins are mapped in the same four structures in blue. (d) A stereoview of a ribbon drawing of NpuDnaE intein (2KEQ) with the five common split sites discussed in the text. N and C denote the N and C termini, respectively.
Inteins range in size from 129 to >1000 residues, with various insertions and deletions. The most typical insertions found in inteins are homing endonuclease domains. Their insertion points are highly conserved and located at block EN, a loop between blocks B and F (Fig. 1b). At least five different types of endonuclease domains are found in inteins but they are all inserted in the same loop, which is colored in cyan in Fig. 2a (Paulus, 2000; Dassa et al., 2009). Additional DNA binding regions are also observed in the HINT folds, that are colored in orange and green in Fig. 2a. In all of the six structures of inteins isolated from thermophilic organisms, an additional insertion of two β-strands, colored in magenta in Fig. 2a, was observed between blocks A and B, approximately 25 residues from the N terminus. This insertion might stabilize the HINT fold at elevated temperatures. These accessory insertions are peripherally located and it might be possible to remove them completely without loss of splicing activity (Derbyshire et al., 1997; Matsumura et al., 2006; Du et al., 2011). Tables I and II and Fig. 1b summarize the locations of the split sites of the 121 reported split inteins, which were classified into ‘active’ and ‘inactive’ categories. Although the definition of ‘active’ is subjective, the occurrence of each split site classified by motif blocks shows a general trend as depicted in Fig. 1b. The locations of the split sites are also displayed on the three-dimensional structures and are highlighted in red (inactive) in Fig. 2b and in blue (active) in Fig. 2c. Because the split sites in various naturally split inteins from cyanobacteria are highly conserved and 96% of the 24 naturally split inteins with the block EN split sites are found to be functional (Table I), the split site in block EN is the most frequent functional split site. This location, which we call C35 site (utilizing NpuDnaE intein as the numbering reference, see Fig. 2d), is also coincidently the conserved insertion site for the endonuclease domains (Fig. 2a). It seems that almost all the inteins naturally or artificially split at this site (C35) are active at least in vivo, because 95% of the 19 artificially split inteins were found to be active. Many split inteins bearing fewer than 10 residues are inactive (red in Fig. 2b). Whereas many inteins split within flexible loops are generally functional (Fig. 2c), split inteins dissected within the β-strands are almost inactive or result in very low ligation yields (Fig. 2b, Table II) (Sun et al., 2004; Aranko et al., 2009).
Table I.
Summary of the reported active split inteins
| Blocka | Split site | Intein | Yieldb,c | Rate (s−1)c | Reference |
|---|---|---|---|---|---|
| A | N11 | PhoRadA | +++ | N.D. | Aranko et al. (2014) |
| A | N11 | RmaDnaBΔ286 | ++++ | N.D. | Lin et al. (2013) |
| A | N11 | SspDnaBΔ275 | +++ | 4.0 ± 0.2 × 10−5 | Sun et al. (2004); Appleby-Tagoe et al. (2011) |
| A | N11 | SspDnaBM86Δ275 | ++++ | 2.5 ± 0.1 × 10−3 | Appleby-Tagoe et al. (2011) |
| A | N11 | SspDnaX | ++++ | 1.7 ± 0.1 × 10−4 | Lin et al. (2013) |
| A | N11 | TvoVMA | ++++ | 2.6 ± 0.2 × 10−4 | Aranko et al. (2014) |
| A | N11 | NpuDnaE | ++ | N.D. | Aranko et al. (2014) |
| A | N12 | NpuDnaBΔ283 | ++++ | N.D. | Aranko et al. (2014) |
| A | N12 | NpuDnaE | ++ | N.D. | Lee et al. (2012) |
| A | N12 | SspDnaBΔ275 | +++ | N.D. | Ludwig et al. (2008) |
| A | N12 | SspGyrB | ++++ | N.D. | Lin et al. (2013) |
| A | N12 | TerThyXΔ132 | ++++ | 3.8 ± 0.5 × 10−4 | Lin et al. (2013) |
| A | N13 | PhoRadA | ++++ | N.D. | Aranko et al. (2014) |
| A/B | N24 | NpuDnaBΔ283 | + | N.D. | Aranko et al. (2014) |
| A/B (Nat.) | N25 | AceL-TerL | ++++ | 1.7 ± 0.2 × 10−3 | Thiel et al. (2014) |
| A/B | N35 | NpuDnaE | +++ | N.D. | Aranko et al. (2014) |
| A/B | N36 | NpuDnaE | +++ | N.D. | Lee et al. (2012) |
| A/B | N38 | PhoRadA | ++++ | N.D. | Aranko et al. (2014) |
| B/EN | N73 | PchPRP8 | ++++ | N.D. | Elleuche and Pöggeler (2007) |
| B/EN | N79 | SspDnaBΔ275 | ++++ | N.D. | Sun et al. (2004) |
| B/EN | N88 | SspDnaBΔ275 | +++ | N.D. | Sun et al. (2004) |
| EN | N100 | SspDnaBΔ275 | ++++ | N.D. | Sun et al. (2004) |
| EN | N160 | PfuRIR1-1 | ++++ | N.D. | Otomo et al. (1999b) |
| EN | N249 | Psp-GDBPol-1 | +++ | N.D. | Southworth et al. (1998) |
| EN | C107 | MtuRecAΔ228 | ++++ | N.D. | Mills et al. (1998) |
| EN | C97 | Psp-GDBPol-1 | ++ | N.D. | Southworth et al. (1998) |
| EN | C87 | PfuRIR1-2 | ++++ | N.D. | Otomo et al. (1999a) |
| EN | C64 | SceVMAΔ206 | +++ | 9.4 ± 2.0 × 10−4d | Brenzel et al. (2006) |
| EN | C51 | RmaDnaBΔ271 | ++++ | N.D. | Li et al. (2008) |
| EN | C50 | MtuRecAΔ285 | +++ | N.D. | Lew et al. (1998) |
| EN | C48 | SspDnaBΔ274 | ++++ | N.D. | Wu et al. (1998b) |
| EN | C48 | TvoVMA | ++++ | N.D. | Aranko et al. (2014) |
| EN | C47 | SspDnaBΔ275 | ++ | 9.9 ± 0.8 × 10−4 | Brenzel et al. (2006) |
| EN | C46 | PchPRP8 | ++++ | N.D. | Elleuche and Pöggeler (2007) |
| EN | C46 | PhoRadA | ++++ | N.D. | Aranko et al. (2014) |
| EN (Nat.) | C44 | gp41-8 | ++++ | 4.5 ± 0.6 × 10−2 | Carvajal-Vallejos et al. (2012) |
| EN | C43 | SceVMAΔ227 | ++++ | 1.2 ± 0.1 × 10−3d | Brenzel et al. (2006) |
| EN (Nat.) | C39 | IMPDH-1 | ++++ | 8.7 ± 3.2 × 10−2 | Carvajal-Vallejos et al. (2012) |
| EN (Nat.) | C39 | NrdJ-1 | ++++ | 9.8 ± 2.3 × 10−2 | Carvajal-Vallejos et al. (2012) |
| EN | C39 | NpuDnaBΔ283 | ++++ | N.D. | Aranko et al. (2014) |
| EN | C38 | MtuRecAΔ297 | ++++ | N.D. | Lew et al. (1999) |
| EN (Nat.) | C36 | gp41-1 | ++++ | 1.8 ± 0.5 × 10−1 | Carvajal-Vallejos et al. (2012) |
| EN (Nat.) | C35 | AovDnaE | ++++ | N.D. | Shah et al. (2012) |
| EN (Nat.) | C35 | AspDnaE | +++ | N.D. | Wei et al. (2006); Shah et al. (2012) |
| EN (Nat.) | C35 | AvaDnaE | +++ | 3.1 ± 0.2 × 10−2 | Shah et al. (2012) |
| EN (Nat.) | C35 | Cra(C5505)DnaE | ++ | 1.2 ± 0.1 × 10−2 | Shah et al. (2012) |
| EN (Nat.) | C35 | Csp(CCY0110)DnaE | ++++ | N.D. | Shah et al. (2012) |
| EN (Nat.) | C35 | Csp(PCC8801)DnaE | ++++ | 1.8 ± 0.1 × 10−2 | Shah et al. (2012) |
| EN (Nat.) | C35 | CwaDnaE | + | 5.0 ± 0.3 × 10−3 | Shah et al. (2012) |
| EN (Nat.) | C35 | Maer(NIES843)DnaE | ++++ | N.D. | Shah et al. (2012) |
| EN (Nat.) | C35 | Mcht(PCC7420)DnaE | ++++ | 2.4 ± 0.1 × 10−2 | Shah et al. (2012) |
| EN | C35 | MtuRecAΔ300 | + | N.D. | Lew et al. (1998) |
| EN (Nat.) | C35 | NpuDnaE | ++++ | 3.7 ± 0.2 × 10−2 | Iwai et al. (2006); Shah et al. (2012) |
| EN (Nat.) | C35 | NspDnaE | + | N.D. | Dassa et al. (2007) |
| EN (Nat.) | C35 | OliDnaE | ++++ | 1.6 ± 0.1 × 10−2 | Dassa et al. (2007); Shah et al. (2012) |
| EN (Nat.) | C35 | Sel(PC7942)DnaE | + | N.D. | Shah et al. (2012) |
| EN (Nat.) | C35 | SspDnaE | +++ | 1.5 ± 0.1 × 10−4 | Wu et al. (1998a); Shah et al. (2012) |
| EN (Nat.) | C35 | Ssp(PCC7002)DnaE | + | N.D. | Shah et al. (2012) |
| EN (Nat.) | C35 | TerDnaE-3 | +++ | 8.5 ± 0.5 × 10−3 | Shah et al. (2012) |
| EN (Nat.) | C34 | TelDnaE | + | N.D. | Shah et al. (2012) |
| EN (Nat.) | C34 | TvuDnaE | + | N.D. | Dassa et al. (2007) |
| EN (Nat.) | C30 | NeqPol | ++++ | N.D. | Choi et al. (2006) |
| EN/F | C30 | SspDnaE | + | N.D. | Aranko et al. (2009) |
| F | C23 | SspDnaE | + | N.D. | Aranko et al. (2009) |
| F | C16 | SspDnaE | + | N.D. | Aranko et al. (2009) |
| F | C16 | TvoVMA | ++++ | N.D. | Aranko et al. (2014) |
| F | C14 | NpuDnaE | +++ | 8.3 ± 0.7 × 10−5 | Aranko et al. (2009) |
| F | C14 | PhoRadA | ++ | N.D. | Aranko et al. (2014) |
| G | C7 | TvoVMA | + | N.D. | Aranko et al. (2014) |
| G | C6 | NpuDnaE | ++ | 5.2 ± 0.2 × 10−5 | Oeemig et al. (2009) |
| G | C6 | PhoRadA | ++++ | 4.9 ± 0.1 × 10−5 | Aranko et al. (2014) |
| G | C6 | SspDnaX | ++++ | 1.9 ± 0.3 × 10−4 | Lin et al. (2013) |
| G | C6 | SspGyrB | ++++ | 6.9 ± 2.2 × 10−5 | Appleby et al. (2009) |
| G | C6 | TerDnaE-3 | ++++ | 2.2 ± 0.2 × 10−4 | Lin et al. (2013) |
| G | C6 | TerThyXΔ132 | +++ | N.D. | Lin et al. (2013) |
aA, B, F and G stand for conserved intein motives blocks A, B, F and G; EN stands for the conserved endonuclease domain insertion site, Nat. stands for natively split intein.
b(++++) = 80–100%, (+++) = 60–79%, (++) = 40–59%, (+) = 1–39%, N.D. = not determined.
cThe highest yield/fastest rate reported is shown.
dWith rapamycin.
Abbreviations of inteins: AceL-TerL, Ace lake terminase large subunit intein from unknown host; AovDnaE, DnaE intein from Aphanizomenon ovalisporum; AspDnaE, DnaE intein from Anabaena species; AvaDnaE, DnaE intein from Anabaena variabilis; Cra(CS505)DnaE, DnaE intein from Cylindrospermopsis raciborskii CS505; Csp(CCY00110)DnaE, DnaE intein from Cyanothece sp CCY00110; Csp(PCC8801)DnaE, DnaE intein from Cyanothece sp PCC8801; CwaDnaE, DnaE intein from Crocosphaera watsonii; gp41-1 and gp41-8, gp41 DNA helicase inteins from unknown host; IMPDH-1, IMPDH intein from unknown host; Maer(NIES843)DnaE, DnaE intein from Microcystis aerigunosa NIES843; Mcht(PCC7420)-2DnaE, DnaE intein from Microcoleus chthonoplastes sp PCC7420; MtuRecAΔ228/285/300, minimized RecA inteins from Mycobacterium tuberculosis; NeqPol, DNA polymerase intein from Nanoarchaeum equitans; NpuDnaBΔ283, minimized DnaB intein from Nostoc punctiforme; NpuDnaE, DnaE intein from Nostoc punctiforme; NrdJ, NrdJ intein from unknown host; NspDnaE, DnaE intein from Nostoc sp PCC7120; OliDnaE, DnaE intein from Oscillatoria limnetica; PchPRP8, PRP8 intein from Penicillium chrysogenum; PfuRIR1-1, RIR1 intein from Pyrococcus furiosus; PfuRIR1-2, RIR1 intein from Pyrococcus furiosus; PhoRadA, RadA intein from Pyrococcus horikoshii; Psp-GDBPol-1 DNA polymerase intein from Pyrococcus sp GB-D; RmaDnaBΔ271/Δ286, minimized DnaB inteins from Rhodothermus marinus; SceVMAΔ206/227, minimized VMA inteins from Saccharomyces cerevisiae; Sel(PC7942)DnaE, DnaE intein from Synechococcus elongatus PC7942; SspDnaBΔ274/275, minimized DnaB inteins from Synechocystis sp PCC6008; SspDnaBM86Δ275, M86 mutant of minimized DnaB intein from Synechocystis sp PCC6008; SspDnaE, DnaE intein from Synechocystis sp PCC6008; Ssp(PCC7002)DnaE, DnaE intein from Synechococcus sp PCC7002; SspDnaX, DnaX intein from Synechocystis sp PCC6008; SspGyrB, GyrB intein from Synechocystis sp PCC6008; TelDnaE, DnaE intein from Thermosynechococcus elongatus; TerDnaE-3, DnaE intein from Trichodesmium erythraeum; TerThyXΔ132, ThyX intein from Trichodesmium erythraeum; TvoVMA, VMA intein from Thermoplasma volcanium; TvuDnaE, DnaE intein from Thermosynechococcus vulcanus.
Table II.
Summary of the reported inactive split inteins
| Blocka | Split site | Intein | Reference |
|---|---|---|---|
| A | N5 | SspDnaBΔ275 | Ludwig et al. (2008) |
| A | N5 | PhoRadA | Aranko et al. (2014) |
| A | N6 | PhoRadA | Aranko et al. (2014) |
| A | N9 | SspDnaBΔ275 | Ludwig et al. (2008) |
| A | N10 | SspDnaBΔ275 | Ludwig et al. (2008) |
| A | N10 | TerRIR-4Δ244 | Lin et al. (2013) |
| A | N10 | TerDnaE-1Δ1226 | Lin et al. (2013) |
| A | N10 | TerRIR-1Δ257 | Lin et al. (2013) |
| A | N10 | CnePrp8 | Lin et al. (2013) |
| A | N10b | TerRIR-2Δ238 | Lin et al. (2013) |
| A | N10b | TthRIR1-1Δ287 | Lin et al. (2013) |
| A | N11b | TerDnaB-1Δ1843 | Lin et al. (2013) |
| A | N11 | TerDnaE-2Δ288 | Lin et al. (2013) |
| A | N11 | TerDnaE-3 | Lin et al. (2013) |
| A | N11b | TerRIR-3Δ188 | Lin et al. (2013) |
| A/B | N25 | SspDnaBΔ275 | Sun et al. (2004) |
| A/B | N34 | NpuDnaBΔ283 | Aranko et al. (2014) |
| A/B | N35 | SspDnaBΔ275 | Sun et al. (2004) |
| A/B | N44 | SspDnaBΔ275 | Sun et al. (2004) |
| A/B | N53 | SspDnaBΔ275 | Sun et al. (2004) |
| B | N64 | SspDnaBΔ275 | Sun et al. (2004) |
| B | N71 | SspDnaBΔ275 | Sun et al. (2004) |
| EN | N108 | Psp-GDBPol-1 | Southworth et al. (1998) |
| EN (Nat.) | C35 | AhaDnaE | Shah et al. (2012) |
| F | C26 | PchPRP8 | Elleuche and Pöggeler (2007) |
| F | C22 | SspDnaBΔ275 | Sun et al. (2004) |
| F | C14 | SspDnaBΔ275 | Sun et al. (2004) |
| F | C13 | NpuDnaBΔ283 | Aranko et al. (2014) |
| G | C9 | SspDnaE | Aranko et al. (2009) |
| G | C7 | CnePrp8 | Lin et al. (2013) |
| G | C6 | RmaDnaBΔ286 | Lin et al. (2013) |
| G | C6 | SspDnaBΔ275 | Sun et al. (2004) |
| G | C6b | TerDnaB-1Δ1843 | Lin et al. (2013) |
| G | C6 | TerDnaE-2Δ288 | Lin et al. (2013) |
| G | C6 | TerDnaE-1Δ1226 | Lin et al. (2013) |
| G | C6 | TerRIR-1Δ257 | Lin et al. (2013) |
| G | C6b | TerRIR-2Δ238 | Lin et al. (2013) |
| G | C6b | TerRIR-3Δ188 | Lin et al. (2013) |
| G | C6 | TerRIR-4Δ244 | Lin et al. (2013) |
| G | C6 | TthDnaE-1 | Appleby et al. (2009) |
| G | C6 | TthDnaE-2 | Appleby et al. (2009) |
| G | C6b | TthRIR1-1Δ287 | Lin et al. (2013) |
| G | C5 | SspDnaE | Aranko et al. (2009) |
| G | C3 | SspDnaE | Aranko et al. (2009) |
| G | C3 | NpuDnaE | Aranko et al. (2014) |
| G | C2 | NpuDnaE | Aranko et al. (2014) |
aA, B, F and G stand for conserved intein motives blocks A, B, F and G; EN stands for the conserved endonuclease domain insertion site; Nat. stands for naturally occurring split intein.
binactive cis-splicing mini-intein.
Abbreviations of inteins: AhaDnaE, DnaE intein from Aphanothece halophytica; CnePRP8, PRP8 intein from Cryptococcus neoformans; NpuDnaBΔ283, minimized DnaB intein from Nostoc punctiforme; NpuDnaE, DnaE intein from Nostoc punctiforme; PchPRP8, PRP8 intein from Penicillium chrysogenum; PhoRadA, RadA intein from Pyrococcus horikoshii; Psp-GDBPol-1 DNA polymerase intein from Pyrococcus sp GB-D; RmaDnaBΔ286, minimized DnaB intein from Rhodothermus marinus; SspDnaBΔ275, minimized DnaB intein from Synechocystis sp PCC6008; SspDnaE, DnaE intein from Synechocystis sp PCC6008; TerDnaB-1Δ1843, minimized DnaB intein from Trichodesmium erythraeum; TerDnaE-1Δ1226 and TerDnaE-2Δ288, minimized DnaE inteins from Trichodesmium erythraeum; TerDnaE-3, DnaE intein from Trichodesmium erythraeum; TerRIR-1Δ257, TerRIR-2Δ238, TerRIR-3Δ188, and TerRIR-4Δ244, minimized RIR inteins from Trichodesmium erythraeum; TthDnaE-1 and TthDnaE-2, DnaE inteins from Thermus thermophilus; TthRIR1-1Δ287, minimized RIR intein from Thermus thermophilus.
Where to split?
The HINT fold can be divided into two subdomains related by pseudo two-fold symmetry. It was proposed to be the result of gene duplication (Fig. 3a) (Hall et al., 1997; Liu, 2000). These two subdomains can be related to each other very well, with the exception of the last C-terminal β-strand (Fig. 3b). Each subdomain contains only a few loops. Judging from the position of the loops present in the subdomains of various intein structures, possible split sites for engineering can be reduced to four symmetry-related sites (N11/C59, N24/C49, N35/C35 and N63/C14), as well as the C6 site that is located outside of the region related by pseudo-symmetry (the numbering is based on NpuDnaE intein) (Fig. 3c). In other words, there are only nine sites in the HINT domain that are potentially suitable for splitting inteins. The best split site among them is the aforementioned C35 site, which most reliably produces functional split inteins (Tables I and II) (Sun et al., 2004; Elleuche and Pöggeler, 2007; Aranko et al., 2014). The symmetry-related N35 site in the other subdomain is also a relatively successful split site. However, due to insertions such as the ones observed in the inteins from thermophilic organisms (Fig. 2a), it might not be easy to identify the corresponding N35 site without comparing the three-dimensional structures. Generally speaking, split inteins with their split sites located in the C-terminal subdomain seem to be more productive than those split within the N-terminal subdomain. We speculate that these differences stem from dissimilarities in the protein folding process of each subdomain. The C6 site has also been successfully used for protein ligation (Appleby et al., 2009; Oeemig et al., 2009; Lin et al., 2013; Aranko et al., 2014). However, it often ends up mostly in the cleavage reaction because these six residues might not be required for the N-S acyl shift, which is the first step of the protein splicing reaction (Appleby et al., 2009; Oeemig et al., 2009; Aranko et al., 2014). The shorter split inteins bearing fewer than six residues are probably not suitable for protein trans-splicing without further optimization, even though their shorter lengths make them attractive for semi-synthesis.
Fig. 3.
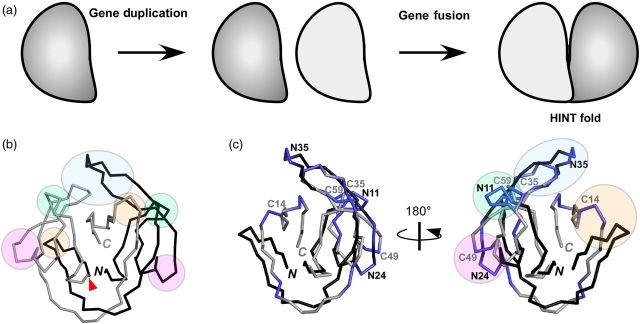
(a) A gene duplication model for the evolution of the HINT fold. (b) The pseudo C2-symmetry relationship in NpuDnaE intein. The backbones of the two subdomains (residues 1-67 and 68-137) are colored in black and gray, respectively. Green, magenta, blue and orange circles in the structure represent the C2-symmetry-related split sites (N11/C59, N24/C49, N35/C35 and N63/C14, respectively) in NpuDnaE intein. (c) Superposition of the main chains of the two subdomains of NpuDnaE intein. The four main split sites in the subdomain are highlighted as in (b), together with the split sites according to the numbering in NpuDnaE intein. N and C denote the N and C termini, respectively.
Faster is better?
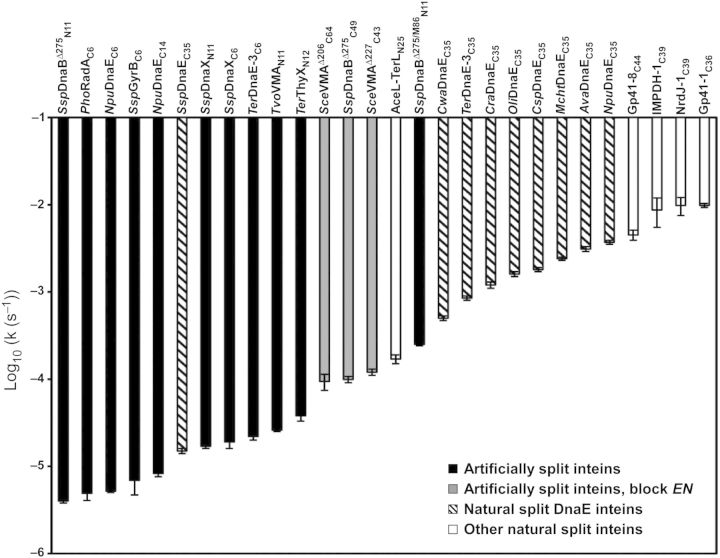
Ligation kinetics is one of the important factors for the application of split inteins as protein ligation tools. There are already many reports that provide the kinetic parameters of protein trans-splicing (as summarized in Table I). The ligation reaction rates observed for individual inteins vary greatly (easily up to a 104-fold difference). However, one has to pay special attention when directly comparing these numbers: inteins are single-turnover enzymes with attached substrates, which differ substantially from the classical enzymes (Paulus, 2001), and protein splicing is not only dependent on reaction conditions which are crucial for classical enzymes (Martin et al., 2001), such as pH and temperature, but also depends considerably on the junction sequences (Iwai et al., 2006; Lockless and Muir, 2009; Appleby-Tagoe et al., 2011; Cheriyan et al., 2013) and on the exteins (Aranko et al., 2009). Precise comparison of enzyme kinetics requires them to have the same reaction conditions as well as identical substrates. Most enzymes have evolved to process specific substrates and that is also true for inteins (their substrates are exteins, including their junctions) (Iwai et al., 2006; Lockless and Muir, 2009; Appleby-Tagoe et al., 2011; Cheriyan et al., 2013). However, it is almost impossible to determine the enzyme kinetics of inteins without applying conditional splicing approaches because an intein auto-catalytically accomplishes self-excision and ligation immediately after protein translation (folding), which typically takes place inside cells. Split inteins provide an opportunity to analyze the kinetic parameters of protein splicing reactions that can be initiated by mixing the two split fragments. The ligation reaction rates of split inteins vary from 10−1 to 10−5 s−1 (Table I, Fig. 4). Naturally split DnaE inteins have reaction rates of k≈ 10−2–10−3 s−1, the exception being SspDnaE intein, which exhibits a slightly slower reaction rate (k≈ 10−4 s−1) (Fig. 4). The fastest reaction rates reported so far are, however, those from a recently discovered group of naturally split inteins that do not belong to the DnaE intein family (k≈ 10−1–10−2 s−1) (Dassa et al., 2009; Carvajal-Vallejos et al., 2012). Generally speaking, naturally split inteins have faster ligation rates. However, the differences in the substrates (the extein sequences) used for the analysis have to be taken into consideration, as they could exert profound effects on the kinetic parameters. The artificially split inteins split at block EN are usually the fastest among artificially split inteins (k≈ 10−3 s−1), while artificially split inteins split at the other sites are a ten- to a hundred-fold slower (k≈ 10−4–10−5 s−1). It was feasible to improve the ligation rate for a split intein derived from a cis-splicing variant of SspDnaB intein (k≈ 10−3 s−1) to a value similar to that of naturally split inteins by sequential directed evolution (Appleby-Tagoe et al., 2011). This result indicated that protein engineering approaches could improve artificial split inteins to the level of naturally split inteins (Appleby-Tagoe et al., 2011). The ligation kinetics of split inteins derived from a single intein also vary greatly depending on the split sites, emphasizing the importance of the association and folding of the split halves (Aranko et al., 2009; Oeemig et al., 2009). This observation also explains why the junction sequences, as well as the entire extein sequence, influence the ligation kinetics. Net charges and molecular sizes are expected to affect the association rates, particularly when the ligation rate is closer to the diffusion limits of the precursor fragments. The association constants of the naturally split inteins have been reported to be in the low nanomolar range (Shi and Muir, 2005), although some reports used mutants bereft of splicing abilities (Shah et al., 2011; Zheng et al., 2012). The association of the two split fragments is considered to be guided by local charge differences in the interaction interfaces (Shi and Muir, 2005; Dassa et al., 2007). Since it is necessary for the two precursors to associate to initiate protein trans-splicing, the affinity between the two split intein fragments is likely to correlate with the ligation kinetics.
Fig. 4.
A comparison of the ligation rates from the reported split inteins, sorted by the kinetic parameters. The names of the split inteins follow the previously introduced split intein nomenclature (Aranko et al., 2014). Gray and filled bars represent artificially engineered inteins split at the endonuclease insertion site (EN) or at other split sites, respectively. Hashed and white bars represent naturally occurring split inteins found in DnaE polymerase and other host proteins, respectively. The errors are taken from the references.
Being fast is not enough
What are the ‘better’ split inteins? The answer probably depends on what they are going to be applied. We consider inteins to be ‘better’ particularly for their in vitro applications. In vitro protein ligation is very demanding because it requires high solubility of the two precursors, faster ligation kinetics, and lower side reactions (such as cleavages). In addition, ‘better’ inteins should ideally have broad substrate specificity (high tolerance of the splicing junctions) and high specificity of the split intein fragments (orthogonality). Faster ligation is only one of the features required for broader application of split inteins. We found that inteins with an excellent in vivo splicing ability are not necessarily excellent protein ligation tools in vitro (Ellilä et al., 2011; Aranko et al., 2014). For example, split inteins split within the endonuclease domain are generally superior in ligation kinetics but they are often poorly soluble when the precursors are individually expressed and purified, because the split intein fragments are prone to aggregation and misfolding due to the presence of long, unfolded polypeptide chains (Mills et al., 1998; Southworth et al., 1998; Otomo et al., 1999b; Brenzel et al., 2006). Even some of the naturally split inteins are poorly soluble in specific cases (Shah et al., 2012; Zheng et al., 2012), but inteins with shorter chain lengths from the N or C terminus might be less likely to disturb the solubility of the target (extein) and are advantageous for semi-synthesis (Aranko et al., 2009; Mootz, 2009; Oeemig et al., 2009). However, split inteins with shorter chain lengths have other problems, such as the aforementioned premature cleavages. In addition, shorter intein fragments also suffer from lower specificity (non-orthogonality), making them cross-reactive to homologous sequences (Aranko et al., 2014). Such cross-reactivity is typically observed among allelic naturally split DnaE inteins (Iwai et al., 2006; Dassa et al., 2007; Sorci et al., 2013; Aranko et al., 2014). High specificity (orthogonality) of split intein fragments is required for specific applications, such as ‘one-pot’ multiple-fragment ligation (Otomo et al., 1999b; Shi and Muir, 2005; Busche et al., 2009; Shah et al., 2011). Ultimately, we would like to have ample amounts of ligated products for, e.g. segmental isotopic labeling. To improve the yield of protein ligation, ‘better’ split inteins must have no side reactions, because the yield probably matters more in some applications than the ligation kinetics do. Suppression of premature and undesired cleavages is thus very critical. Small differences in the structure and folding process could easily direct the reaction to non-productive pathways, as different split inteins derived from an identical intein could induce more cleavages than ligation. While fine-tuning of the coordination of the catalytic residues in inteins is thus crucial for productive protein ligation, BIL proteins have evolved specifically for cleavages (Amitai et al., 2003; Aranko et al., 2013a). The comparison of the mechanisms and structures of the members of the HINT superfamily might provide better insight into how to prevent undesired side reactions, or how to enhance the more productive use of inteins in protein purification as self-cleaving tags (Chong et al., 1997).
Conclusions and future perspectives
After many attempts at creating inteins with various artificial split sites, it seems that we came back to the starting point: Nature's recipe for splitting inteins still seems to be the best. Naturally split inteins dominate the podium in the competition for ‘the best’ split intein in terms of ligation kinetics. However, none of the naturally split inteins fully fulfil our wishes for ‘better’ split inteins. Splitting experiments with various inteins have clearly highlighted the bottlenecks in artificially engineered split inteins, such as poor solubility and dominant side reactions. The recent discoveries of naturally split inteins with faster ligation kinetics are clearly a step forwards toward ‘better’ split inteins. The remaining challenge is to develop split inteins of various lengths with optimal combinations of solubility, substrate specificity, orthogonality, and robustness. It remains to be seen if it will be at all possible to develop ideal split inteins for biotechnological applications. However, with the detailed structural information in hand, it should be feasible for protein engineers to develop clever strategies to create ‘better’ split inteins by design or by evolutional approach. We still have to develop our own ‘better’ recipe to overcome the bottlenecks in the existing split inteins until it is beyond the nature's golden recipe, by increasing our knowledge of the protein trans-splicing mechanism at the atomic level.
Acknowledgements
A.S.A. acknowledges Viikki Doctoral Programme in Molecular Biosciences for financial support. This work was supported by the Academy of Finland (137995), Sigrid Jusélius Foundation, Biocenter Finland, and in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Amitai G., Belenkiy O., Dassa B., Shainskaya A., Pietrokovski S. Mol. Microbiol. 2003;47:61–73. doi: 10.1046/j.1365-2958.2003.03283.x. [DOI] [PubMed] [Google Scholar]
- Appleby J.H., Zhou K., Volkmann G., Liu X.Q. J. Biol. Chem. 2009;284:6194–6199. doi: 10.1074/jbc.M805474200. [DOI] [PubMed] [Google Scholar]
- Appleby-Tagoe J.H., Thiel I.V., Wang Y., Wang Y., Mootz H.D., Liu X.Q. J. Biol. Chem. 2011;286:34440–34447. doi: 10.1074/jbc.M111.277350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranko A.S., Züger S., Buchinger E., Iwaï H. PloS ONE. 2009;4:e5185. doi: 10.1371/journal.pone.0005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranko A.S., Oeemig J.S., Iwaï H. FEBS J. 2013a;280:3256–3269. doi: 10.1111/febs.12307. [DOI] [PubMed] [Google Scholar]
- Aranko A.S., Oeemig J.S., Kajander T., Iwaï H. Nat. Chem. Biol. 2013b;9:616–622. doi: 10.1038/nchembio.1320. [DOI] [PubMed] [Google Scholar]
- Aranko A.S., Oeemig J.S., Zhou D., Kajander T., Wlodawer A., Iwaï H. Mol. Biosyst. 2014;10:1023–1034. doi: 10.1039/c4mb00021h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenzel S., Kurpiers T., Mootz H.D. Biochemistry. 2006;45:1571–1578. doi: 10.1021/bi051697+. [DOI] [PubMed] [Google Scholar]
- Busche A.E., Aranko A.S., Talebzadeh-Farooji M., Bernhard F., Dotsch V., Iwaï H. Angew Chem. Int. Ed. 2009;48:6128–6131. doi: 10.1002/anie.200901488. [DOI] [PubMed] [Google Scholar]
- Carvajal-Vallejos P., Pallisse R., Mootz H.D., Schmidt S.R. J. Biol. Chem. 2012;287:28686–28696. doi: 10.1074/jbc.M112.372680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi J., Amitai G., Belenkiy O., Pietrokovski S. Mol. Microbiol. 2003;50:1569–1577. doi: 10.1046/j.1365-2958.2003.03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheriyan M., Pedamallu C.S., Tori K., Perler F. J. Biol. Chem. 2013;288:6202–6211. doi: 10.1074/jbc.M112.433094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.J., Nam K.H., Min B., Kim S.J., Soll D., Kwon S.T. J. Mol. Biol. 2006;356:1093–1106. doi: 10.1016/j.jmb.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Chong S., Mersha F.B., Comb D.G., et al. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- Dassa B., Amitai G., Caspi J., Schueler-Furman O., Pietrokovski S. Biochemistry. 2007;46:322–330. doi: 10.1021/bi0611762. [DOI] [PubMed] [Google Scholar]
- Dassa B., London N., Stoddard B.L., Schueler-Furman O., Pietrokovski S. Nucleic Acids Res. 2009;37:2560–2573. doi: 10.1093/nar/gkp095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire V., Wood D.W., Wu W., Dansereau J.T., Dalgaard J.Z., Belfort M. Proc. Natl. Acad. Sci. USA. 1997;94:11466–11471. doi: 10.1073/pnas.94.21.11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Xu M.Q., Ghosh I., Chen X., Ferrandon S., Lesage G., Rao Z. J. Biol. Chem. 2003;278:39133–39142. doi: 10.1074/jbc.M306197200. [DOI] [PubMed] [Google Scholar]
- Du Z., Liu J., Albracht C.D., et al. J. Biol. Chem. 2011;286:38638–38648. doi: 10.1074/jbc.M111.290569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X., Gimble F.S., Quiocho F.A. Cell. 1997;89:555–564. doi: 10.1016/s0092-8674(00)80237-8. [DOI] [PubMed] [Google Scholar]
- Elleuche S., Pöggeler S. Biochem. Biophys. Res. Commun. 2007;355:830–834. doi: 10.1016/j.bbrc.2007.02.035. [DOI] [PubMed] [Google Scholar]
- Ellilä S., Jurvansuu J.M., Iwaï H. FEBS Lett. 2011;585:3471–3477. doi: 10.1016/j.febslet.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Gogarten J.P., Senejani A.G., Zhaxybayeva O., Olendzenski L., Hilario E. Annu. Rev. Microbiol. 2002;56:263–287. doi: 10.1146/annurev.micro.56.012302.160741. [DOI] [PubMed] [Google Scholar]
- Hall T.M., Porter J.A., Young K.E., Koonin E.V., Beachy P.A., Leahy D.J. Cell. 1997;91:85–97. doi: 10.1016/s0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]
- Hirata R., Ohsumk Y., Nakano A., Kawasaki H., Suzuki K., Anraku Y. J. Biol. Chem. 1990;265:6726–6733. [PubMed] [Google Scholar]
- Hodges R.A., Perler F.B., Noren C.J., Jack W.E. Nucleic Acids Res. 1992;20:6153–6157. doi: 10.1093/nar/20.23.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai H., Züger S., Jin J., Tam P.H. FEBS Lett. 2006;580:1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- Kane P.M., Yamashiro C.T., Wolczyk D.F., Neff N., Goebl M., Stevens T.H. Science. 1990;250:651–657. doi: 10.1126/science.2146742. [DOI] [PubMed] [Google Scholar]
- Lee Y.T., Su T.H., Lo W.C., Lyu P.C., Sue S.C. PLoS One. 2012;7:e43820. doi: 10.1371/journal.pone.0043820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew B.M., Mills K.V., Paulus H. J. Biol. Chem. 1998;273:15887–15890. doi: 10.1074/jbc.273.26.15887. [DOI] [PubMed] [Google Scholar]
- Lew B.M., Mills K.V., Paulus H. Biopolymers. 1999;51:355–362. doi: 10.1002/(SICI)1097-0282(1999)51:5<355::AID-BIP5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Li J., Sun W., Wang B., Xiao X., Liu X.Q. Hum. Gene Ther. 2008;19:958–964. doi: 10.1089/hum.2008.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Li M., Song H., Xu L., Meng Q., Liu X.Q. PLoS One. 2013;8:e59516. doi: 10.1371/journal.pone.0059516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.Q. Annu. Rev. Genet. 2000;34:61–76. doi: 10.1146/annurev.genet.34.1.61. [DOI] [PubMed] [Google Scholar]
- Lockless S.W., Muir T.W. Proc. Natl. Acad. Sci. USA. 2009;106:10999–11004. doi: 10.1073/pnas.0902964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C., Schwarzer D., Mootz H.D. J. Biol. Chem. 2008;283:25264–25272. doi: 10.1074/jbc.M802972200. [DOI] [PubMed] [Google Scholar]
- Martin D.D., Xu M.Q., Evans T.C., Jr. Biochemistry. 2001;40:1393–1402. doi: 10.1021/bi001786g. [DOI] [PubMed] [Google Scholar]
- Matsumura H., Takahashi H., Inoue T., et al. Proteins. 2006;63:711–715. doi: 10.1002/prot.20858. [DOI] [PubMed] [Google Scholar]
- Michnick S.W., Ear P.H., Manderson E.N., Remy I., Stefan E. Nat. Rev. Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- Mills K.V., Lew B.M., Jiang S., Paulus H. Proc. Natl. Acad. Sci. USA. 1998;95:3543–3548. doi: 10.1073/pnas.95.7.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz H.D. ChemBioChem. 2009;10:2579–2589. doi: 10.1002/cbic.200900370. [DOI] [PubMed] [Google Scholar]
- Moure C.M., Gimble F.S., Quiocho F.A. Nat. Struct. Biol. 2002;9:764–770. doi: 10.1038/nsb840. [DOI] [PubMed] [Google Scholar]
- Nogami S., Fukuda T., Nagai Y., Yabe S., Sugiura M., Mizutani R., Satow Y., Anraku Y., Ohya Y. Yeast. 2002;19:773–782. doi: 10.1002/yea.872. [DOI] [PubMed] [Google Scholar]
- Oeemig J.S., Aranko A.S., Djupsjöbacka J., Heinämäki K., Iwaï H. FEBS Lett. 2009;583:1451–1456. doi: 10.1016/j.febslet.2009.03.058. [DOI] [PubMed] [Google Scholar]
- Otomo T., Ito N., Kyogoku Y., Yamazaki T. Biochemistry. 1999a;38:16040–16044. doi: 10.1021/bi991902j. [DOI] [PubMed] [Google Scholar]
- Otomo T., Teruya K., Uegaki K., Yamazaki T., Kyogoku Y. J. Biomol. NMR. 1999b;14:105–114. doi: 10.1023/a:1008308128050. [DOI] [PubMed] [Google Scholar]
- Paulus H. Annu. Rev. Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- Paulus H. Bioorg. Chem. 2001;29:119–129. doi: 10.1006/bioo.2001.1203. [DOI] [PubMed] [Google Scholar]
- Perler F.B. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perler F.B., Allewell N.M. J. Biol. Chem. 2014;289:14488–14489. doi: 10.1074/jbc.R114.570531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrokovski S. Trends Genet. 2001;17:465–472. doi: 10.1016/s0168-9525(01)02365-4. [DOI] [PubMed] [Google Scholar]
- Shah N.H., Muir T.W. Chem. Sci. 2014;5:446–461. doi: 10.1039/C3SC52951G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N.H., Vila-Perello M., Muir T.W. Angew Chem. Int. Ed. 2011;50:6511–6515. doi: 10.1002/anie.201102909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N.H., Dann G.P., Vila-Perello M., Liu Z., Muir T.W. J. Am. Chem. Soc. 2012;134:11338–11341. doi: 10.1021/ja303226x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Muir T.W. J. Am. Chem. Soc. 2005;127:6198–6206. doi: 10.1021/ja042287w. [DOI] [PubMed] [Google Scholar]
- Shingledecker K., Jiang S.Q., Paulus H. Gene. 1998;207:187–195. doi: 10.1016/s0378-1119(97)00624-0. [DOI] [PubMed] [Google Scholar]
- Sorci M., Dassa B., Liu H., Anand G., Dutta A.K., Pietrokovski S., Belfort M., Belfort G. Anal. Chem. 2013;85:6080–6088. doi: 10.1021/ac400949t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth M.W., Adam E., Panne D., Byer R., Kautz R., Perler F.B. EMBO J. 1998;17:918–926. doi: 10.1093/emboj/17.4.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Yang J., Liu X.Q. J. Biol. Chem. 2004;279:35281–35286. doi: 10.1074/jbc.M405491200. [DOI] [PubMed] [Google Scholar]
- Swithers K.S., Senejani A.G., Fournier G.P., Gogarten J.P. BMC Evol. Biol. 2009;9:303. doi: 10.1186/1471-2148-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel I.V., Volkmann G., Pietrokovski S., Mootz H.D. Angew Chem. Int. Ed. Engl. 2014;53:1306–1310. doi: 10.1002/anie.201307969. [DOI] [PubMed] [Google Scholar]
- Topilina N.I., Mills K.V. Mob DNA. 2014;5:5. doi: 10.1186/1759-8753-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann G., Iwaï H. Mol. Bio. Syst. 2010;6:2110–2121. doi: 10.1039/c0mb00034e. [DOI] [PubMed] [Google Scholar]
- Wei X.Y., Sakr S., Li J.H., Wang L., Chen W.L., Zhang C.C. Res. Microbiol. 2006;157:227–234. doi: 10.1016/j.resmic.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Wu H., Hu Z., Liu X.Q. Proc. Natl. Acad. Sci. USA. 1998a;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Xu M.Q., Liu X.Q. Biochim. Biophys. Acta. 1998b;1387:422–432. doi: 10.1016/s0167-4838(98)00157-5. [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Otomo T., Oda N., Kyogoku Y., Uegaki K., Ito N., Ishino Y., Nakamura H. J. Am. Chem. Soc. 1998;120:5591–5592. [Google Scholar]
- Zettler J., Schutz V., Mootz H.D. FEBS Lett. 2009;583:909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wu Q., Wang C., Xu M.Q., Liu Y. Biosci. Rep. 2012;32:433–442. doi: 10.1042/BSR20120049. [DOI] [PMC free article] [PubMed] [Google Scholar]