Abstract
In plants, epigenetic regulation mediates both the proper development of the plant and responses to environmental cues. Changes in epigenetic states employ DNA methylation, histone modification, and regulatory RNAs. In Arabidopsis thaliana, DNA methylation as a repressive mark is often associated with constitutively silenced loci, such as repetitive sequences, transposons, and heterochromatin. These sequences regularly give rise to small interfering RNAs, which direct DNA methylation through the RNA-directed DNA methylation (RdDM) pathway. For example, FWA locus is silenced in sporophytes and enriched with DNA methylation. Its methylated state is stable and passes to the next generation. This is an example of meiotically inherited epigenetic states. There are also epigenetic changes that can be inherited mitotically and are subsequently erased in the next generation. In this review, we use the vernalization-mediated epigenetic silencing of FLOWERING LOCUS C (FLC) as an example for this type of mitotically stable epigenetic state. Here, we discuss mechanisms of epigenetic changes that can result in meiotically or mitotically stable states with an emphasis on FWA and FLC as two examples.
Introduction
Genetic variation and environmental interactions make up most of the diversity, affecting both genotype and phenotype, within a species. Many environmental stimuli affect epigenetic variation and phenotype, although the extent of the connection between epigenetic variation and phenotype is not yet known (Nordborg and Weigel 2008). Epigenetic states refer to the stable inheritance of gene-expression states that is independent of the DNA sequence (Berger et al. 2009). These changes in gene expression have been extensively studied and they often include changes in histone modification and DNA methylation. The histone is the core unit of the nucleosome and the properties of these histones can be changed in many ways, including covalent modifications of histones (Allfrey et al. 1964; Strahl and Allis 2000), the eviction and subsequent deposition of a different variant of histone, and ATP-dependent remodeling of the nucleosome (Whitehouse et al. 1999; Teif and Rippe 2009). The modifications of histones can occur at different developmental states of plants and often can be regulated by environmental signals, such as temperature, drought, and exposure to pathogens. DNA methylation can also be influenced by environmental signals and causes the silencing of genes, transposons, and other repetitive sequences (Wagner 2003; Vanyushin 2006). The stability of DNA methylation is often linked to RNA-dependent DNA methylation (RdDM) machinery (Law and Jacobsen 2010) as well as to methylases and methyltransferases, such as CHROMOMETHYLASE 3 (CMT3) and DNA METHYLTRANSFERASE 1 (MET1). Due to the sessile nature of plants, these epigenetic changes often are necessary for modulating gene expression in response to environmental cues, and for increasing survivability and reproductive fitness of the plant.
In this review, we will focus on Arabidopsis thaliana, a model organism for both plant biology and genetics. There are several types of epigenetic states in Arabidopsis, but for the purpose of this review, we will focus on two types of epigenetically stable states, trans-generational or those epigenetic changes that persist in the next generation (meiotically stable) and intra-generational or epigenetic changes that persist for only one generation (mitotically stable). We will be using FWA as an example of a trans-generational epigenetic state and FLC as an example of an intra-generational state. Both FWA and FLC play roles in the transition from the vegetative to the reproductive state, thereby demonstrating that flowering is a useful read-out to address both categories of epigenetic states mentioned in this review.
Meiotically stable epigenetic inheritance
The context of DNA methylation in Arabidopsis
To understand how DNA methylation can remain meiotically stable in plants, it is important to highlight some underlying mechanisms both for RdDM-mediated silencing and for activity of methylase/methyltransferase. DNA methylation occurs at genomic regions that are often transcriptionally inactive, such as pericentric heterochromatin, repetitive sequences associated with transposable elements, and regions that produce siRNA (Zhang et al. 2006). DNA methylation occurs in three different contexts on cytosine residues at CG, CNG (where N is any nucleotide), and CHH (where H is either C, T, or A) sequences in Arabidopsis (Zhang et al. 2006; Popova et al. 2013). Deposition of the methylation on the cytosine bases requires different methylases for each of the different contexts. CG-methylation requires MET1, homolog of the mammalian de novo methyltransferase (DNMT1), using a pre-methylated parent strand as the template for the deposition of new methyl groups on the adjacent strand (Bartee et al. 2001; Mathieu et al. 2007). In a study of the DNA methylation at genome-wide scale, met1 mutants showed an increase in the expression of pseudogenes found within pericentric heterochromatin, indicating the importance of CG methylation in the silencing of not only transposable elements but also of constitutive heterochromatin (Zhang et al. 2006). CNG methylation, like CG methylation, is also symmetric and facilitated by DOMAINS REARRANGED METHYLASE1/2 (DRM1/2), orthologs of mammalian DNMT3a/b, and CMT3, a plant-specific methyltransferase (Chan et al. 2006a). Methylation within the context of CNG occurs through a different mechanism compared with CG methylation (Gruenbaum et al. 1981; Chan et al. 2006a). Much like MET1, CMT3 also uses the methylated strand as a template for methylation of the other strand. Unlike methylation of CG and CNG, methylation of CHH is asymmetric and not directly copied onto a newly replicated DNA strand. Although the methylation-context is different, these sites are redundantly methylated by the same de novo DNA methyltransferases as CNG sequences, namely DRM1/2 and CMT3 (Zilberman et al. 2004; Law and Jacobsen 2010). CHH methylation is also mediated via the Snf2 family remodeler DDM1 (Lippman et al. 2004) and can do so independently of the RdDM pathway (Zemach et al. 2013). These CHH methylation sites are found throughout the life cycle of plants. In humans, non-CG methylation (including CHH methylation) is abundant in the embryonic stem cells but disappear upon the induced-differentiation of the embryonic stem cells (Lister et al. 2009). In the Arabidopsis triple mutant drm1/drm2/cmt3, euchromatic genes are mostly up-regulated, indicating an important role for non-CG methylation in the proper expression of functional genes in plants (Zhang et al. 2006). These mutants also exhibited pleiotropic developmental defects, suggesting that non-CG methylation serves as a controlling factor for the expression of developmental genes (Cao and Jacobsen 2002).
RdDM: a mechanism for DNA methylation
One mechanism for DNA methylation in the CNG or CHH context is RdDM, which functions to silence areas of the chromatin with either repetitive sequences, such as those located within centromeric heterochromatin, or transposon sequences, by generating 24-nt siRNA that target DNA for methylation (Law and Jacobsen 2010). Transgenes can also be the target of RdDM due to the likelihood of formation of double-stranded RNA (dsRNA) due to high levels of expression or by repetitive sequences. The dsRNA precursors are produced by a combination of RNA polymerase IV (Pol IV) and RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) activities. These precursors can then be processed into siRNA by DICER-LIKE 3 (DCL3) (Liu et al. 2006). When combined with ARGONAUTE4 (AGO4), these siRNA can bind to transcripts produced by RNA polymerase V (Pol V) (Wierzbicki et al. 2009; Zheng et al. 2009). These siRNA acts as scaffolds that promote the recruitment of RdDM machinery, including DNA methylases as well as histone-modifying proteins, to the target loci (Wierzbicki et al. 2009). The maintenance of this silencing mechanism requires the accumulation of siRNA for the associated target sequence, suggesting that there is basal transcription of these repeat sequences (Lister et al. 2008). RdDM has also been implicated in various responses of plants to stress, such as heat tolerance in response to high temperatures (Popova et al. 2013). In addition, Arabidopsis has many well-studied epialleles with different patterns of DNA methylation without DNA sequence variations (Bender and Fink 1995; Kakutani et al. 1996; Jacobsen and Meyerowitz 1997; Soppe et al. 2000; Liu et al. 2004; Rangwala et al. 2006; Mathieu et al. 2007; Saze and Kakutani 2007; Johannes et al. 2009; Reinders et al. 2009; Foerster et al. 2011). These epialleles generally require RdDM for their maintenance (Law and Jacobsen 2010). Thus, they have been used in genetic screens to elucidate the mechanisms and machinery of RdDM pathways (Bender and Fink 1995; Jacobsen and Meyerowitz 1997; Saze and Kakutani 2007; Reinders et al. 2009; Cao and Jacobsen 2002; Jackson et al. 2002; Zilberman et al. 2003; Riddle and Richard 2005; Chan et al. 2006a, 2006b; Johnson et al. 2008; Woo et al. 2008; Greenberg et al. 2011).
H3K9 dimethylation correlates with DNA methylation
H3K9 dimethylation (H3K9me2) often is correlated with genome-wide CNG methylation, suggesting that these two epigenetic marks could be connected by some mechanism (Bernatavichute et al. 2008). The histone H3K9 methyltransferases are also recruited by AGO4 to the targeted sequence as part of the mechanism for RdDM-mediated silencing (Zilberman et al. 2003). This mechanism is instrumental in the maintenance of DNA methylation because the loss of functional AGO4 can result in the suppression of both DNA methylation and H3K9 methylation, in turn resulting in re-activation of heterochromatin (Zilberman et al. 2003; Xie et al. 2004). In plants, H3K9me2 is deposited by multiple histone methyltransferases (Liu et al. 2007). H3K9 histone methyltransferases in Arabidopsis include SU(VAR)3-9 HOMOLOGUE4 (SUV4 or KRYPTONITE), SUV5, and SUV6 (Jackson et al. 2002). As both DNA methylation in the context of CHG and H3K9me2 often are correlated, these proteins may be involved in a positive feedback loop (Lindroth et al. 2004; Johnson et al. 2007). The H3K9 methyltransferase KYP has an SRA domain that can recognize methylation either in the context of a CNG or a CHH (Lindroth et al. 2004). Similarly, CMT3-mediated DNA methylation can occur because CMT3 contains a chromodomain that recognizes H3K9me2, suggesting that DNA methylation and H3K9me2 can perpetuate each other.
FWA: a trans-generationally stable epiallele
“Epiallelic” variations exist for the FWA gene in Arabidopsis. In wild-type, FWA is inactive due to the high level of DNA methylation that can be stably inherited (Koornneef et al. 1991; Soppe et al. 2000). In a dominant epiallele, fwa-1, DNA hypomethylation results in ectopic expression of FWA which results in late-flowering (Soppe et al. 2000). The maintenance of DNA methylation at FWA locus depends on the presence of transposon-derived sequence at its promoter. It was also noted that a late-flowering phenotype observed in the ddm1 mutant background, which causes hypomethylation of the DNA and a wide variety of phenotypic defects, was genetically linked to the FWA chromatin (Kakutani et al. 1996), suggesting a role for methylation involved in flowering. FWA is demethylated after gametogenesis in the maternal allele and imprinted in the endosperm, whereas the paternal allele remains methylated (Kinoshita et al. 2004). When the plant is in the vegetative state, both the maternal and the paternal alleles are methylated and their silencing is maintained throughout the life cycle by MET1 (Kinoshita et al. 2004). Ectopic expression of FWA influences the transition from the vegetative to the floral state in Arabidopsis. In a dominant fwa-1 mutant, increased expression of FWA in vegetative tissues led to late flowering (Soppe et al. 2000); wild-type FWA is not expressed in vegetative tissues but only in the endosperm (Kinoshita et al. 2004) (Fig. 1A). fwa-1 mutant does not have a mutated nucleotide sequence, rather exhibits a decreased level of DNA methylation within its promoter region, thus making this gene transcriptionally active. DNA methylation at FWA locus is caused by a tandemly repeated transposon-like sequence within the promoter region. In the un-methylated form, ectopic FWA expression in vegetative tissues can result in late-flowering (Fujimoto et al. 2008). The methylation of the repeat sequence is directed by the RdDM pathway (Lippman et al. 2004; Chan et al. 2006b). It is interesting to note that the structures of this transposon-like sequence are variable among different ecotypes of Arabidopsis (Fujimoto et al. 2008), suggesting the variation on the presence of epialleles in population.
Fig. 1.
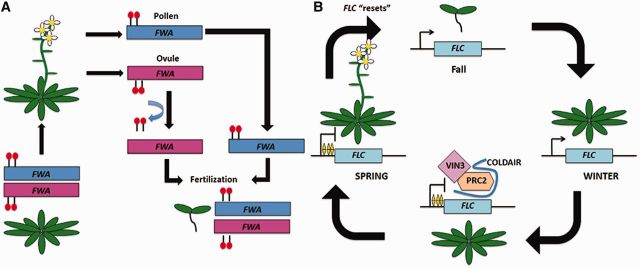
Two examples of epigenetic regulation in Arabidopsis. (A) Meiotically stable epigenetic change at the FWA locus. In the vegetative state, both the maternal allele (pink) and paternal allele (blue) of FWA are DNA-methylated at its promoter (red), thereby promoting the transition to a flowering state. During gametogenesis, the paternal allele maintains methylation, whereas the maternal allele is demethylated. After fertilization, the maternal allele is re-methylated. (B) Mitotically stable epigenetic change at the FLC locus. In fall and winter, FLC is actively transcribed, thereby preventing flowering. During the winter, COLDAIR transcripts increase and PRC2 deposits H3K27me3 (yellow diamond). After the return to higher temperatures in the spring, the repression of FLC is stably maintained and the plant transitions to the reproductive state. In the following generation, H3K27me3 mark is erased and FLC is “reset” and the cycle continues.
Mitotically stable epigenetic inheritance: vernalization
Histone modifications are reversible epigenetic marks that often are tied to changes in gene expression by environmental signals and developmental cues. These epigenetic marks can either have repressive or activating behaviors, depending on the amino-acid residue and type of modification otherwise known as the “Histone Code” (Jenuwein and Allis 2001). Acetylation of histone (Grunstein 1997), as well as some phosphorylation (Wei et al. 1999) and ubiquitination (Pham and Sauer 2000), often is correlated with loose chromatin and active transcription, whereas biotinylation (Kothapalli et al. 2005) and sumolyation (Shiio and Eisenman 2003) often are associated with tightly wound chromatin and repressed transcription. Histone methylations can be associated both with active and repressive histone states depending on amino-acid resides (Jenuwein and Allis 2001). Long noncoding RNA (lncRNA) has recently emerged to cause transcriptionally active or repressive states of genes through modification of histone residues (Weinberg and Morris 2013). In the following sections, we describe how repressive histone modification and lncRNA can confer a mitotically repressive state of the floral repressor (FLOWERING LOCUS C) FLC and thereby accelerate flowering.
The role of histone modification in the vernalization-mediated repression of FLC
Perhaps, one of the most well-studied environmental epigenetic effects in plants is vernalization, a response to the prolonged cold of winter. In many ecotypes of Arabidopsis, vernalization is required for promoting the floral transition. Vernalization promotes flowering after a prolonged exposure to cold, as in winter, and is an adaptation that allows greater reproductive success. Vernalization provides a plant “memory” of winter that remains even after a return to higher temperatures. In the vernalization pathway of Arabidopsis, FLC is a major component, acting as a primary floral repressor. FLC is a MADS-domain transcription factor that prevents the expression of the floral integrators SUPPRESSOR OF CONSTANS 1 (SOC1) and FLOWERING LOCUS T (FT) (Hepworth et al. 2002), thereby preventing the transition to the reproductive state. During prolonged exposure to cold, levels of VERNALIZATION INSENSITIVE 3 (VIN3) increase and form a complex with POLYCOMB REPRESSIVE COMPLEX 2 (PRC2) to deposit histone H3 lysine 27 tri-methylation (H3K27me3) at FLC chromatin (Wood et al. 2006; De Lucia et al. 2008). These modifications of histone persist after a return to warm conditions, thereby creating cellular “memory.” However, the epigenetic marks are only stable throughout mitosis and are removed the following generation, allowing for the reactivation of FLC (Trevaskis et al. 2007) (Fig. 1B). As Arabidopsis follows the annual habit, the resetting of FLC occurs with the setting of seed, but in plants with a perennial habit, such as Arabis alpina, there cannot be a “hard” memory of winter. PERPETUAL FLOWERING 1 (PEP1) is an ortholog to Arabidopsis FLC and has similar expression patterns to FLC (Wang et al. 2009; Albani et al. 2012). Unlike FLC, after vernalization and the return to warmer conditions, the expression levels of PEP1 return to pre-vernalization levels and able to respond to vernalization again (Wang et al. 2009), implicating the fundamental difference in the mechanisms underlying the repression of FLC by vernalization.
Long noncoding RNAs
In addition to modification of histone lncRNAs, such as COLDAIR and COOLAIR, play a role in the intra-generational repression of FLC. COLDAIR is an lncRNA that is expressed in the sense direction from the first intron of FLC. COLDAIR is induced during cold exposure and associates with PRC2. Its knockdown leads to a decrease in epigenetic silencing, less H3K27me3, implicating it as a required component in the recruitment of PRC2 (Heo and Sung 2011) (Fig. 1B). Conversely, COOLAIR is a collection of FLC antisense transcripts with differential polyadenylations and alternative splicings (Swiezewski et al. 2009). COOLAIR is also induced by cold exposure but does not physically interact with PRC2 (Heo and Sung 2011). It has been suggested that COOLAIR may have a role in an early step in the vernalization-response by regulating the transcription of sense-strands in a stage-dependent manner (Swiezewski et al. 2009). Taken together, the repression of FLC by vernalization is an epigenetic phenomenon in which both histone modification and lncRNAs operate in response to an environmental cue, low temperature. It should be noted that DNA methylation does play a role in the repression of FLC (Finnegan et al. 2005), suggesting that various routes to maintain the epigenetic states in plants.
Concluding remarks
In this review, we highlighted different mechanistic approaches for the persistence of epigenetic changes in both meiotically stable and mitotically stable states. The meiotically stable, or trans-generational, state uses DNA methylation, histone modification (particularly H3K9me2), and the RdDM pathway to silence repetitive sequences, transposable elements, and heterochromatic regions. This type of epigenetic change is well represented by the case of FWA. Conversely, mitotically stable or intra-generational states, at least in the case of FLC, do not involve DNA methylation and instead rely on histone-modification and lncRNAs. These examples, particularly FLC, are influenced by environmental signals creating a potential connection between the environment and inheritance. Additionally, the epigenome unlocks the potential for changes in gene expression that are independent of the genome, possibly increasing the capacity for evolution and adaptation.
Acknowledgments
Sung Lab is supported by grants from NIH (R01GM100108) and NSF (IOS0950785).
References
- Albani MC, Castaings L, Wötzel S, Mateos J L, Wunder J, Wang R, Coupland G. PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genet. 2012;8:1–14. doi: 10.1371/journal.pgen.1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey G, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci. 1964;315:786–94. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Gen Dev. 2001;15:1753–8. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J, Fink GR. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell. 1995;83:725–34. doi: 10.1016/0092-8674(95)90185-x. [DOI] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Gen Dev. 2009;23:781–3. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatavichute Y, Zhang X, Cokus S, Pellegrini M, Jacobsen S. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One. 2008;3:1–11. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen S. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–44. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Chan S, Henderson I, Zhang X, Shah G, Chien J, Jacobsen S. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet. 2006a;2:791–7. doi: 10.1371/journal.pgen.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S, Zhang X, Bernatavichute Y, Jacobsen S. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006b;4:1923–33. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones A, Greb T, Dean C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad. 2008;105:16831–6. doi: 10.1073/pnas.0808687105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA, Jaligot E, Sheldon CC, Peacock WJ, Dennis ES. The downregulation of FLOWERING LOCUS C (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms. Plant J. 2005;44:420–32. doi: 10.1111/j.1365-313X.2005.02541.x. [DOI] [PubMed] [Google Scholar]
- Foerster A, Dinh H, Sedman L, Wohlrab B, Mittelsten Scheid O. Genetic rearrangements can modify chromatin features at epialleles. PLoS Genet. 2011;7:1–13. doi: 10.1371/journal.pgen.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto R, Kinoshita Y, Kawabe A, Kinoshita T, Takashima K, Nordborg M, Kakutani T. Evolution and control of imprinted FWA genes in the genus Arabidopsis. PLoS Genet. 2008;4:1–11. doi: 10.1371/journal.pgen.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M, Ausin I, Chan S, Cokus S, Cuperus J, Feng S, Jacobsen S. Identification of genes required for de novo DNA methylation in Arabidopsis. Epigenetics. 2011;6:344–54. doi: 10.4161/epi.6.3.14242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Naveh-Many T, Cedar H, Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981;292:860–2. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–52. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–9. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- Hepworth S, Valverde F, Ravenscroft D, Mouradov A, Coupland G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 2002;21:4327–37. doi: 10.1093/emboj/cdf432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–60. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Meyerowitz EM. Hypermethylated SUPERMAN epigenetic alleles in arabidopsis. Science. 1997;277:1100–3. doi: 10.1126/science.277.5329.1100. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johannes F, Porcher E, Teixeira FK, Saliba-Colombani V, Simon M, Agier N, Colot V. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5:1–11. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17: 379–84. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Law J, Khattar A, Henderson IR, Jacobsen SE. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 2008;4:1–13. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh J, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci USA. 1996;93:12406–11. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Kakutani T. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–3. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Kothapalli N, Camporeale G, Kueh A, Chew YC, Oommen AM, Griffin JB, Zempleni J. Biological functions of biotinylated histones. J Nutr Biochem. 2005;16: 446–8. doi: 10.1016/j.jnutbio.2005.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establish patterns in plants and animals. Nat Rev Genet. 2010;11: 204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Jacobsen SE. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4286–96. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–6. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar H, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Nery JR, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, He Y, Amasino R, Chen X. siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Gen Dev. 2004;18:2873–8. doi: 10.1101/gad.1217304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yu Y, Ruan Y, Meyer D, Wolff M, Xu L, Shen WH. Plant SET- and RING-associated domain proteins in heterochromatinization. Plant J. 2007;52:914–26. doi: 10.1111/j.1365-313X.2007.03286.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514–20. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–62. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Weigel D. Next-generation genetics in plants. Nature. 2008;456:720–3. doi: 10.1038/nature07629. [DOI] [PubMed] [Google Scholar]
- Pham A, Sauer F. Ubiquitin-activating/conjugating activity of TAF250, a mediator of activation of gene expression in Drosophila. Science. 2000;289:2357–60. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- Popova OV, Dinh HQ, Aufsatz W, Jonak C. The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol Plant. 2013;6:396–410. doi: 10.1093/mp/sst023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala SH, Elumalai R, Vanier C, Ozkan H, Galbraith DW, Richards EJ. Meiotically stable natural epialleles of Sadhu, a novel Arabidopsis retroposon. PLoS Genet. 2006;2:270–81. doi: 10.1371/journal.pgen.0020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Wulff BB, Mirouze M, Marı A. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Gen Dev. 2009;23:939–50. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Richards EJ. Genetic variation in epigenetic inheritance of ribosomal RNA gene methylation in Arabidopsis. Plant J. 2005;41:524–32. doi: 10.1111/j.1365-313X.2004.02317.x. [DOI] [PubMed] [Google Scholar]
- Saze H, Kakutani T. Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 2007;26: 3641–52. doi: 10.1038/sj.emboj.7601788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA. 2003;100:13225–30. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters J. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- Teif VB, Rippe K. Predicting nucleosome positions on the DNA: combining intrinsic sequence preferences and remodeler activities. Nucleic Acids Res. 2009;37:5641–55. doi: 10.1093/nar/gkp610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007;12:352–7. doi: 10.1016/j.tplants.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Vanyushin BF. DNA methylation and epigenetics. Russ J Genet. 2006;42:985–97. [Google Scholar]
- Wagner D. Chromatin regulation of plant development. Curr Opin Plant Biol. 2003;6:20–8. doi: 10.1016/s1369526602000079. [DOI] [PubMed] [Google Scholar]
- Wang R, Farrona S, Vincent C, Joecker A, Schoof H, Turck F, Albani MC. PEP1 regulates perennial flowering in Arabis alpina. Nature. 2009;459:423–7. doi: 10.1038/nature07988. [DOI] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Morris KV. Long non-coding RNA targeting and transcriptional de-repression. Nucleic Acid Ther. 2013;23:9–14. doi: 10.1089/nat.2012.0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–7. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- Wierzbicki A, Ream T, Jeremy H, Pikaard C. RNA Polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–34. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Dittmer T, Richards EJ. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 2008;4:1–13. doi: 10.1371/journal.pgen.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell C. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA. 2006;103:14631–6. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:642–52. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Ecker JR. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Gen Dev. 2009;23:2850–60. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–9. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol. 2004;14:1214–20. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]


