Abstract
Summary: Phenome-wide association studies (PheWAS) have been used to replicate known genetic associations and discover new phenotype associations for genetic variants. This PheWAS implementation allows users to translate ICD-9 codes to PheWAS case and control groups, perform analyses using these and/or other phenotypes with covariate adjustments and plot the results. We demonstrate the methods by replicating a PheWAS on rs3135388 (near HLA-DRB, associated with multiple sclerosis) and performing a novel PheWAS using an individual’s maximum white blood cell count (WBC) as a continuous measure. Our results for rs3135388 replicate known associations with more significant results than the original study on the same dataset. Our PheWAS of WBC found expected results, including associations with infections, myeloproliferative diseases and associated conditions, such as anemia. These results demonstrate the performance of the improved classification scheme and the flexibility of PheWAS encapsulated in this package.
Availability and implementation: This R package is freely available under the Gnu Public License (GPL-3) from http://phewascatalog.org. It is implemented in native R and is platform independent.
Contact: phewas@vanderbilt.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Genome-wide association studies (GWAS) have been an important research method for the past 10 years. These studies typically take the form of a case-control study where many genotypes are analyzed with a specific phenotype. Many software packages, such as Plink and SNPTEST, have been developed to support GWAS (Marchini et al., 2007; Purcell et al., 2007).
More recently, phenome-wide association studies (PheWAS) have been used to interrogate with which phenotypes a given genetic variant may be associated (Denny et al., 2010). PheWAS was originally described within an electronic medical record (EMR) cohort, and EMR-based PheWAS have been recently shown to replicate 66% of sufficiently powered known associations across many disease domains as well as discover new associations (Denny et al., 2013). PheWAS methods have been applied to research cohorts as well, successfully replicating many known associations and finding potential novel associations (Pendergrass et al., 2013). Visualization software has also been developed to help present and investigate PheWAS results (Pendergrass et al., 2012). Focused on evaluating many phenotypes for a given set of genotypes, PheWAS methods do not translate well to traditional GWAS software, which typically follows a one phenotype, many genotypes paradigm. Although it is possible to perform analyses using existing software packages, it requires scripting many runs of the software. To foster adoption of PheWAS, we developed an R implementation of the most common functionality needed to perform and visualize EMR-based PheWAS. This R package follows the methods detailed in Denny et al. (2013), but has been designed to use either genetic or non-genetic data as the independent variable.
2 METHODS
2.1 Data input
Data to perform a PheWAS using this package can include, for a population, the following: demographic data, International Classification of Disease codes diagnostic code data, the independent variables (e.g. genotype or laboratory data) and any other covariates, such as principal components generated to adjust for genetic ancestry. Users can pass into the phewas method any data types R supports for regression. Although the original PheWAS study used genetic data as predictors, studies using phenotypes as predictors are also feasible in this framework.
2.2 Mapping phenotypes
Investigators can perform a PheWAS using ICD-9 codes or ‘PheWAS codes’, which represent ∼1600 hierarchical phenotypes formed from grouped ICD-9 codes. Each PheWAS phenotype also includes an optional set of exclusion phenotypes for similar diagnoses to more accurately identify true controls. This step prevents patients with common ‘rule out’ codes or similar diseases from being marked as a control during the statistical analysis (e.g. a patient with an unknown arrhythmia cannot serve as a control for atrial fibrillation). As requiring multiple codes occurring on different days for a given diagnosis improves phenotype precision, users can specify count thresholds required to establish a patient as a ‘case’ for a given phenotype or simply use the code count in the regression model. This threshold is often set to a minimum of two unique code days (Denny et al., 2013).
2.3 Statistical analysis
Users can choose among different statistical tests when performing a PheWAS, including adjusted and unadjusted models. The default analyses are linear or logistic regression. χ2 and t-tests are available for fast unadjusted tests. P-values, betas, case and control counts and odds ratios (OR; if applicable) are reported. Hardy–Weinberg Equilibrium values and allele frequencies are reported if additive allele counts are supplied.
Meta-analysis of results is included using the meta package. Users can request several statistical significance thresholds: an uncorrected P-value, Bonferroni correction, false discovery rate or an adaptation of the SimpleM method. Parallelization is available with the snowfall package.
2.4 Plotting methods
The package’s phewasManhattan function allows for creation of PheWAS ‘Manhattan’ plots (Figs 1 and 2) with many options for customization, including labeling points, defining sort order and groupings, P-value threshold lines and color maps. Descriptions and groupings are provided for the PheWAS phenotype codes. The plots are generated using ggplot2 and can be altered after they are generated. Plotting functions can also plot P-values from other association studies or data not P-value based, such as ICD9 or PheWAS code counts. Users can also vary point size by OR, as in Figure 3 of Denny et al. (2011). An R Shiny interface for the phewasManhattan plotting method is included.
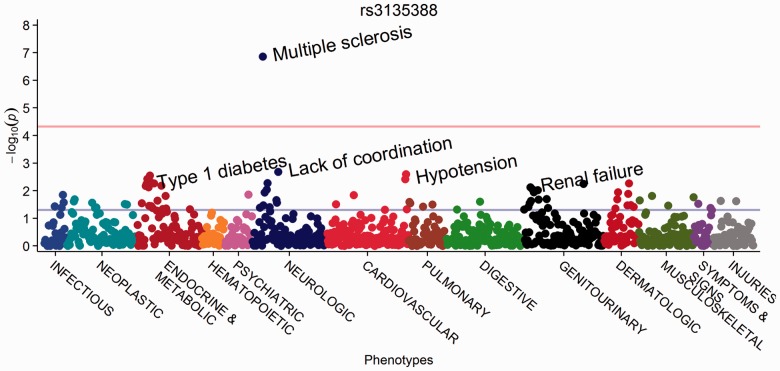
Fig. 1.
PheWAS Manhattan plot for rs3135388, with phenotypes ordered by PheWAS code
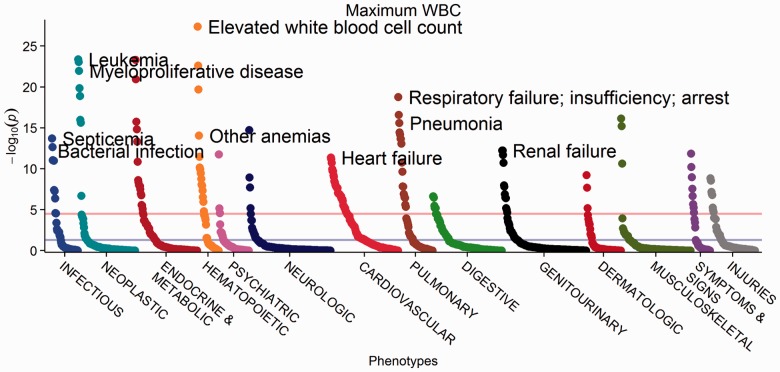
Fig. 2.
PheWAS Manhattan plot for maximum WBC, with phenotypes ordered within each phenotype category by P-value
2.5 Example applications of the PheWAS package
To show the functionality of this package, we repeated one of the original PheWAS analyses for rs3135388, which is highly correlated with HLA-DRB1*1501 and associated with multiple sclerosis with an OR of 2.24 and a P-value of 2.8 × 10−6. For this example, we analyzed the same 6005 individuals and data included in the original analysis (Denny et al., 2010). We performed logistic regression, adjusting for age and gender as covariates, and used the current PheWAS hierarchy (Denny et al., 2013) with all PheWAS codes occurring in ≥20 individuals (n = 1127 PheWAS codes). This is an improvement over the previous analysis that included 733 phenotypes and used a χ2 test. The second example applied linear regression to PheWAS codes adjusted for age and gender to predict maximum white blood cell count (WBC) in the same population.
3 RESULTS AND DISCUSSION
We found an association between multiple sclerosis and rs3135388 with an OR of 2.56 and P-value of 1.4 × 10−7 (Fig. 1). The improved methods, including a covariate-adjusted analysis and revised phenotypes, yielded an OR more consistent with the largest published study on this association, which reported an OR of 2.75 (De Jager et al., 2009). Maximum WBC was associated with infections, leukemias and other expected conditions (Fig. 2).
These analyses were performed from ICD-9 codes and demographic data using the createPhewasTable, phewas, and phewasManhattan methods of the R PheWAS package. The top lines represent Bonferroni significance, and differing output options were used to showcase functionality. Supplementary Tables S1 and S2 include the top 25 hits by P-value for each analysis.
As more investigators leverage EMR data for clinical and genomic analyses, available validated methods will become more valuable. These methods should permit easier adoption of EMR-based PheWAS by more researchers. As shown in the WBC analysis, the PheWAS methodology can also be applied to non-genetic data, providing new avenues of investigation for PheWAS.
Funding: The project was supported by National Library of Medicine (5T15LM007450 and R01 LM010685).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- De Jager PL, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am. J. Hum. Genet. 2011;89:529–542. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 2013;31:1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Pendergrass SA, et al. Visually integrating and exploring high throughput phenome-wide association study (PheWAS) results using PheWAS-view. BioData Min. 2012;5:5. doi: 10.1186/1756-0381-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrass SA, et al. Phenome-wide association study (PheWAS) for detection of pleiotropy within the population architecture using genomics and epidemiology (PAGE) network. PLoS Genet. 2013;9:e1003087. doi: 10.1371/journal.pgen.1003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.