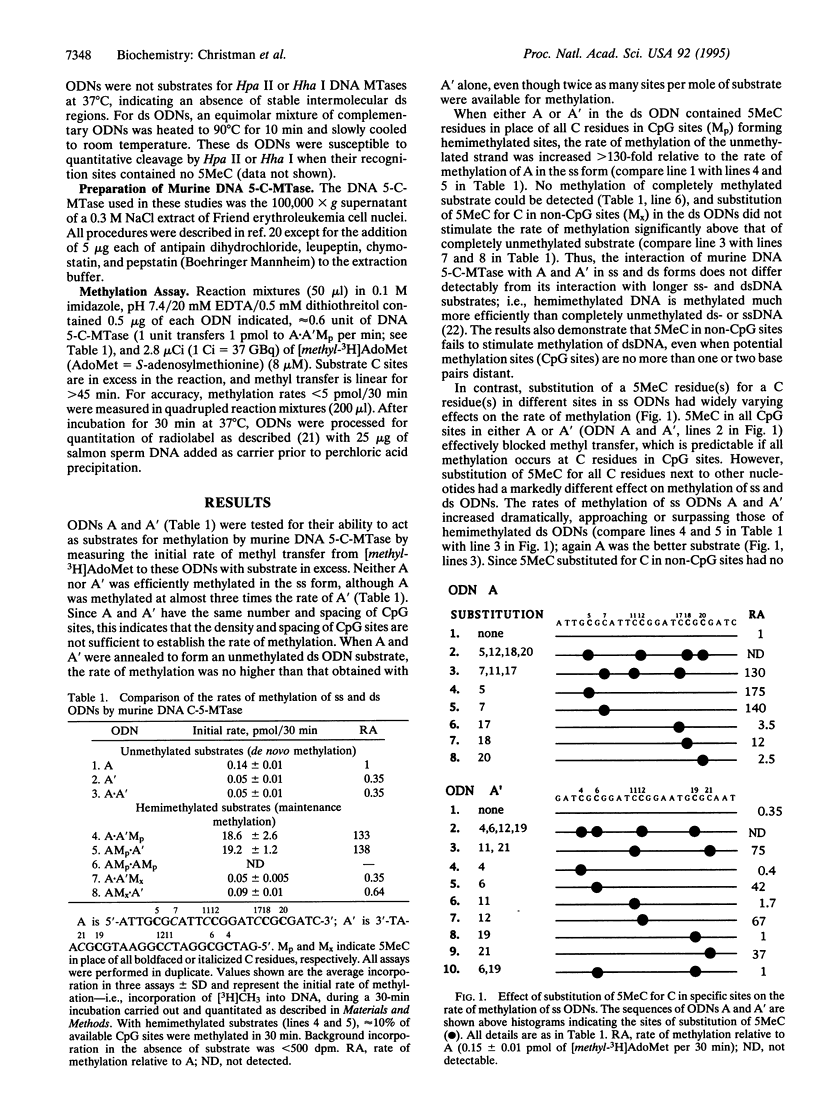
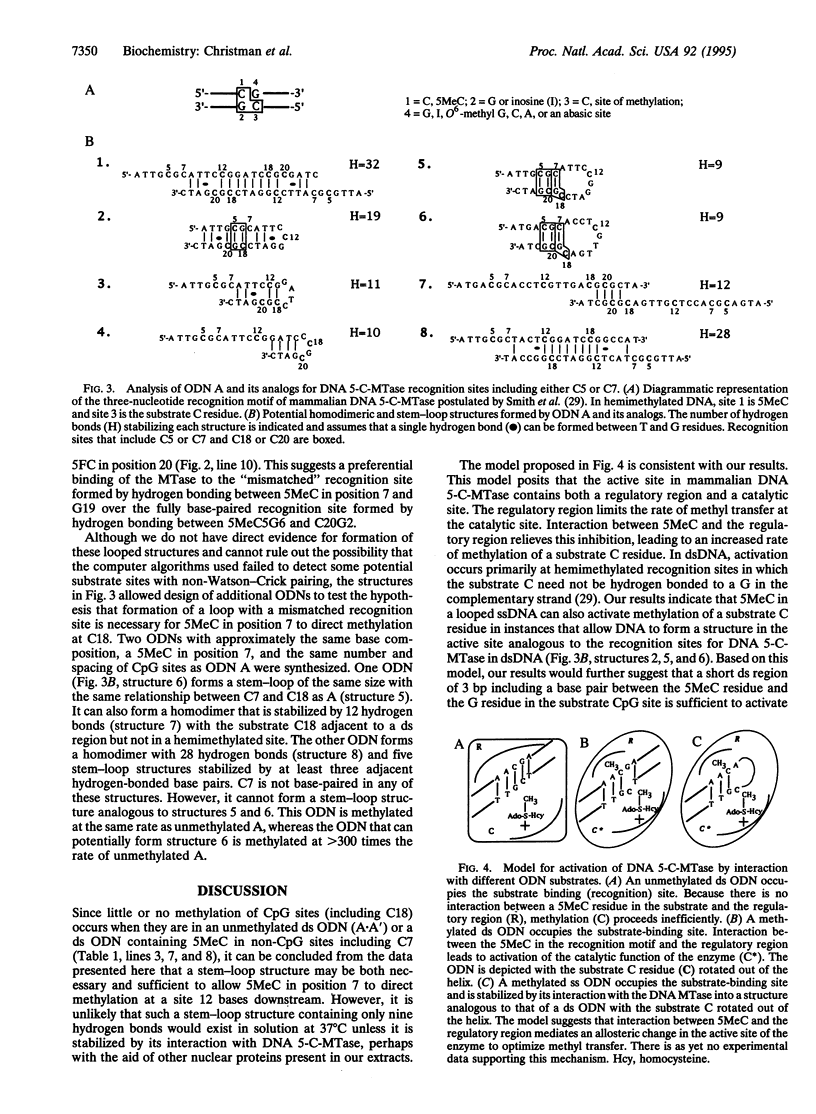
Abstract
Methylation of cytosine residues in DNA plays an important role in regulating gene expression during vertebrate embryonic development. Conversely, disruption of normal patterns of methylation is common in tumors and occurs early in progression of some human cancers. In vertebrates, it appears that the same DNA methyltransferase maintains preexisting patterns of methylation during DNA replication and carries out de novo methylation to create new methylation patterns. There are several indications that inherent signals in DNA structure can act in vivo to initiate or block de novo methylation in adjacent DNA regions. To identify sequences capable of enhancing de novo methylation of DNA in vitro, we designed a series of oligodeoxyribonucleotide substrates with substrate cytosine residues in different sequence contexts. We obtained evidence that some 5-methylcytosine residues in these single-stranded DNAs can stimulate de novo methylation of adjacent sites by murine DNA 5-cytosine methyltransferase as effectively as 5-methylcytosine residues in double-stranded DNA stimulate maintenance methylation. This suggests that double-stranded DNA may not be the primary natural substrate for de novo methylation and that looped single-stranded structures formed during the normal course of DNA replication or repair serve as "nucleation" sites for de novo methylation of adjacent DNA regions.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bestor T. H., Ingram V. M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T., Laudano A., Mattaliano R., Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988 Oct 20;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Bolden A. H., Nalin C. M., Ward C. A., Poonian M. S., Weissbach A. Primary DNA sequence determines sites of maintenance and de novo methylation by mammalian DNA methyltransferases. Mol Cell Biol. 1986 Apr;6(4):1135–1140. doi: 10.1128/mcb.6.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H., Razin A. DNA methylation and development. Biochim Biophys Acta. 1990 May 24;1049(1):1–8. doi: 10.1016/0167-4781(90)90076-e. [DOI] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Christman J. K. DNA methylation in friend erythroleukemia cells: the effects of chemically induced differentiation and of treatment with inhibitors of DNA methylation. Curr Top Microbiol Immunol. 1984;108:49–78. doi: 10.1007/978-3-642-69370-0_5. [DOI] [PubMed] [Google Scholar]
- Creusot F., Acs G., Christman J. K. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2'-deoxycytidine. J Biol Chem. 1982 Feb 25;257(4):2041–2048. [PubMed] [Google Scholar]
- DePamphilis M. L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Midgett R. M., Slagel V. A., Githens S., Kuo K. C., Gehrke C. W., Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983 Jun 24;740(2):212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- Grant S. G., Chapman V. M. Mechanisms of X-chromosome regulation. Annu Rev Genet. 1988;22:199–233. doi: 10.1146/annurev.ge.22.120188.001215. [DOI] [PubMed] [Google Scholar]
- Groudine M., Conkin K. F. Chromatin structure and de novo methylation of sperm DNA: implications for activation of the paternal genome. Science. 1985 May 31;228(4703):1061–1068. doi: 10.1126/science.2986289. [DOI] [PubMed] [Google Scholar]
- Hasse A., Schulz W. A. Enhancement of reporter gene de novo methylation by DNA fragments from the alpha-fetoprotein control region. J Biol Chem. 1994 Jan 21;269(3):1821–1826. [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992 Jun 12;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Makos M., Nelkin B. D., Lerman M. I., Latif F., Zbar B., Baylin S. B. Distinct hypermethylation patterns occur at altered chromosome loci in human lung and colon cancer. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1929–1933. doi: 10.1073/pnas.89.5.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummaneni P., Bishop P. L., Turker M. S. A cis-acting element accounts for a conserved methylation pattern upstream of the mouse adenine phosphoribosyltransferase gene. J Biol Chem. 1993 Jan 5;268(1):552–558. [PubMed] [Google Scholar]
- Osterman D. G., DePillis G. D., Wu J. C., Matsuda A., Santi D. V. 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry. 1988 Jul 12;27(14):5204–5210. doi: 10.1021/bi00414a039. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Jones P. A. 5-methylcytosine, gene regulation, and cancer. Adv Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Allen N. D., Surani M. A. DNA methylation and genomic imprinting in mammals. EXS. 1993;64:469–486. doi: 10.1007/978-3-0348-9118-9_21. [DOI] [PubMed] [Google Scholar]
- Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- Smith S. S., Kan J. L., Baker D. J., Kaplan B. E., Dembek P. Recognition of unusual DNA structures by human DNA (cytosine-5)methyltransferase. J Mol Biol. 1991 Jan 5;217(1):39–51. doi: 10.1016/0022-2836(91)90609-a. [DOI] [PubMed] [Google Scholar]
- Smith S. S., Kaplan B. E., Sowers L. C., Newman E. M. Mechanism of human methyl-directed DNA methyltransferase and the fidelity of cytosine methylation. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4744–4748. doi: 10.1073/pnas.89.10.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. S., Lingeman R. G., Kaplan B. E. Recognition of foldback DNA by the human DNA (cytosine-5-)-methyltransferase. Biochemistry. 1992 Jan 28;31(3):850–854. doi: 10.1021/bi00118a030. [DOI] [PubMed] [Google Scholar]
- Tate P. H., Bird A. P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993 Apr;3(2):226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- Toth M., Lichtenberg U., Doerfler W. Genomic sequencing reveals a 5-methylcytosine-free domain in active promoters and the spreading of preimposed methylation patterns. Proc Natl Acad Sci U S A. 1989 May;86(10):3728–3732. doi: 10.1073/pnas.86.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M., Müller U., Doerfler W. Establishment of de novo DNA methylation patterns. Transcription factor binding and deoxycytidine methylation at CpG and non-CpG sequences in an integrated adenovirus promoter. J Mol Biol. 1990 Aug 5;214(3):673–683. doi: 10.1016/0022-2836(90)90285-T. [DOI] [PubMed] [Google Scholar]
- Wainfan E., Dizik M., Stender M., Christman J. K. Rapid appearance of hypomethylated DNA in livers of rats fed cancer-promoting, methyl-deficient diets. Cancer Res. 1989 Aug 1;49(15):4094–4097. [PubMed] [Google Scholar]
- Wyszynski M. W., Gabbara S., Kubareva E. A., Romanova E. A., Oretskaya T. S., Gromova E. S., Shabarova Z. A., Bhagwat A. S. The cysteine conserved among DNA cytosine methylases is required for methyl transfer, but not for specific DNA binding. Nucleic Acids Res. 1993 Jan 25;21(2):295–301. doi: 10.1093/nar/21.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde K., Zlatanova J. Unusual DNA structures, chromatin and transcription. Bioessays. 1994 Jan;16(1):59–68. doi: 10.1002/bies.950160110. [DOI] [PubMed] [Google Scholar]



