Abstract
Disturbances in parvalbumin- and somatostatin-containing neurons, including deficits in the gamma-aminobutyric acid (GABA)-synthesizing enzyme GAD67 in the prefrontal cortex (PFC) in schizophrenia, may be related to disrupted pre- and/or postnatal development. Deficits in the transcription factor Lhx6, which regulates parvalbumin and somatostatin neuron development, are associated with GAD67 deficits in schizophrenia. Therefore, we investigated the potential pre- and postnatal roles of Lhx6 in GABA-related disturbances using qPCR and/or in situ hybridization to quantify PFC levels of (1) Lhx6 mRNA in a new cohort of schizophrenia subjects; (2) Lhx6 mRNA in monkeys across postnatal development; (3) GABA-related mRNAs in Lhx6 heterozygous (Lhx6+/−) mice, which model Lhx6 deficits in schizophrenia; and (4) Lhx6 mRNA in GAD67+/− mice, which model GAD67 deficits in schizophrenia. Lhx6 mRNA levels were lower (−15%) in schizophrenia and correlated with lower GAD67 mRNA levels. In addition, Lhx6 mRNA levels declined 24% from the perinatal to prepubertal periods then stabilized in monkeys. Finally, GAD67, parvalbumin, and somatostatin mRNAs were not altered in Lhx6+/− mice, and Lhx6 mRNA was not altered in GAD67+/− mice. These data suggest that PFC Lhx6 and GAD67 mRNA deficits are common components of GABA neuron pathology in schizophrenia. An excessive early postnatal decline in Lhx6 mRNA might contribute to Lhx6 mRNA deficits in schizophrenia. However, a partial loss of Lhx6 is not sufficient in isolation to produce deficits in GAD67 mRNA and vice versa, suggesting that the concurrence of Lhx6 and GAD67 mRNA deficits in schizophrenia may instead be the consequence of a common upstream factor.
Key words: Lhx6, GABA, parvalbumin, somatostatin, prefrontal cortex, schizophrenia
Introduction
Disturbances in the inhibitory circuitry of the prefrontal cortex (PFC) in schizophrenia, such as deficits in the gamma-aminobutyric acid (GABA)-synthesizing enzyme glutamate decarboxylase (GAD67),1–6 appear to be most prominent in the subpopulations of GABA neurons that express the calcium-binding protein parvalbumin or the neuropeptide somatostatin.7–11 Because prefrontal GABA neurons, in particular parvalbumin neurons, undergo anatomical, molecular, and synaptic changes across childhood and adolescence,10,12,13 incomplete postnatal development of cortical GABA neurons has been hypothesized as a potential pathogenetic process in schizophrenia.14,15 However, reports of incomplete phenotypic specification of parvalbumin neurons7 and arrested migration of somatostatin neurons16 in the PFC suggest that the disease process may begin earlier, even prenatally, and interfere with the ontogeny (ie, birth, migration, cell-type specification, and/or maturation) of these neurons. Consistent with this hypothesis, deficits in transcription factors that selectively regulate the prenatal development of parvalbumin and somatostatin neurons have also been reported in the PFC in schizophrenia.11,17
For example, we recently reported that deficits in the transcription factor Lhx6 were most prominent in the PFC of the same schizophrenia subjects who had the largest deficits in GAD67, parvalbumin, and somatostatin mRNAs.11 Lhx6 is expressed by prenatal parvalbumin and somatostatin neurons as they migrate from the medial ganglionic eminence to the cerebral cortex18–21 and continues to be expressed in this cell type–specific manner in adult human cortex.22 A complete absence of Lhx6 prenatally in mice, albeit much more severe than the Lhx6 deficit reported in schizophrenia, leads to reduced tangential migration and impeded differentiation into parvalbumin and somatostatin neurons and death of the animal shortly after birth.18,19,23 These lines of evidence suggest that altered expression of cell type–specific transcription factors, such as Lhx6, may lead to persisting deficits that predominantly affect parvalbumin and somatostatin neurons in schizophrenia.
Further investigation into the pre- and postnatal developmental roles of Lhx6 in the pathogenesis of GABA neuron dysfunction in schizophrenia requires a translational, cross-species approach that addresses the following critical questions. First, are deficits in prefrontal Lhx6 mRNA levels reliably found in schizophrenia? Replicating lower Lhx6 mRNA levels in another cohort of schizophrenia subjects would indicate that lower Lhx6 mRNA levels are a common component of GABA neuron pathology in schizophrenia. Second, are deficits in Lhx6 mRNA in schizophrenia consistent with a pattern of incomplete postnatal development similar to that reported for other GABA-related markers in the disorder? Finally, does the association between altered Lhx6 and GAD67 expression in schizophrenia reflect causality, and if so, what is the direction of the effect? That is, does a genetically induced partial decrease in Lhx6 expression that occurs prenatally and is similar in magnitude to that seen in schizophrenia result in deficits in GAD67, parvalbumin, and somatostatin mRNAs similar to those seen in schizophrenia? Alternatively, does a genetically induced partial decrease in GAD67 mRNA that occurs prenatally and is similar in magnitude to that seen in schizophrenia result in deficits in Lhx6 mRNA? To address these questions, we (1) quantified Lhx6 mRNA levels in the PFC of a new cohort of 20 schizophrenia subjects and healthy comparison subjects, (2) analyzed the postnatal developmental trajectory of Lhx6 mRNA expression in the PFC of a large cohort of monkeys, (3) examined the frontal cortex of mice with a partial reduction in Lhx6 expression for disturbances in GABA-related markers, and (4) examined the frontal cortex of mice with a partial reduction in GAD67 expression for disturbances in Lhx6 mRNA.
Methods
Human Subjects
Brain specimens were obtained during routine autopsies conducted at the Allegheny County Medical Examiner’s Office after consent was obtained from next-of-kin. An independent committee of experienced research clinicians made consensus Diagnostic and Statistical Manual of Mental Disorders-IV24 diagnoses for each subject using structured interviews with family members and review of medical records,25 and the absence of a psychiatric diagnosis was confirmed in healthy comparison subjects. To control for experimental variance, subjects with schizophrenia or schizoaffective disorder (n = 20) were matched individually to 1 healthy comparison subject for sex and as closely as possible for age and RNA integrity number (RIN; Agilent Bioanalyzer; supplementary table S1). Samples from subjects in a pair were processed together throughout all stages of the study, and none of these subjects were included in our previous study of Lhx6 mRNA.11 The mean age, postmortem interval, freezer storage time, brain pH, and RIN did not differ between subject groups (table 1), and each subject had a RIN ≥ 7.0 and brain pH > 6.0. All procedures were approved by the University of Pittsburgh’s Committee for the Oversight of Research Involving the Dead and Institutional Review Board.
Table 1.
Summary of Demographic and Postmortem Characteristics of Human Subjects
| Parameter | Healthy Comparison | Schizophrenia |
|---|---|---|
| N | 20 | 20 |
| Sex | 16M/4 F | 16M/4 F |
| Race | 18W/2 B | 17W/3 B |
| Age (y) | 50.0±15.1 | 49.1±12.8 |
| Postmortem interval (h) | 20.8±3.9 | 21.7±7.9 |
| Freezer storage time (mo) | 77.2±39.3 | 64.9±35.0 |
| Brain pH | 6.66±0.21 | 6.54±0.25 |
| RNA integrity number | 7.83±0.51 | 7.85±0.55 |
Note: For all, t (38) ≤ 1.7, P ≥ .09. Values are group means ± SD.
Quantitative PCR
Standardized amounts of cortical gray matter from PFC area 9 were collected in TRIzol reagent in a manner that ensured minimal white matter contamination and excellent RNA preservation26 (supplementary methods). Standardized dilutions of total RNA for each subject were used to synthesize cDNA. All primer pairs (supplementary table S2) demonstrated high amplification efficiency (>96%) across a wide range of cDNA dilutions and specific single products in dissociation curve analysis. Quantitative PCR was performed using the comparative cycle threshold (CT) method with Power SYBR Green dye and the ViiA-7 Real-Time PCR System (Applied Biosystems), as previously described27 (supplementary methods). Three reference genes (beta actin, cyclophilin A, and glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) were used to normalize target mRNA levels,28 and the geometric mean CT of the reference genes did not differ between diagnostic groups (t (38) = 0.85, P = .40). The difference in CT (dCT) for each target transcript was calculated by subtracting the geometric mean CT for the 3 reference genes from the CT of the target transcript (mean of 4 replicate measures), and differences between diagnostic groups were determined using the average dCT of each group. Because dCT represents the log2-transformed expression ratio of each target transcript to the reference genes, the relative level of the target transcript for each subject is reported as 2−dCT.25,29
Developmental Monkeys
Forty-nine rhesus macaque monkeys (Macaca mulatta) ranging in age from 1 week to 11.5 years were assigned to 4 age groups (perinatal, prepubertal, juvenile, and postpubertal) established by previous studies15 (supplementary table S3 and methods). RNA was isolated from gray matter of the frontal pole (area 10) due to the availability of existing tissue. Lhx6 mRNA levels were quantified by qPCR as described above. We previously reported that beta action and cyclophilin, but not GAPDH, mRNAs are stable through early development,27 and these 2 transcripts were used to normalize Lhx6 mRNA levels.
Lhx6 and GAD67 Heterozygous Null Mutation Mice
Fresh, frozen brains of young adult male mice (8 weeks) with an Lhx6 heterozygous null mutation (Lhx6+/−; n = 9) and wild-type littermate male mice (n = 9) were obtained from The Jackson Laboratory (supplementary methods). RNA was isolated from homogenates of frontal cortex tissue sections (12 µm) collected consecutively from Bregma +2.8 to 2.1mm (excluding sub-rhinal fissure olfactory tissue) into TRIzol. qPCR for GABA-related markers was performed as described for the human studies using the StepOnePlus Real-Time PCR System (Applied Biosystems; supplementary table S2), and the geometric mean CT of the 3 reference genes did not differ between Lhx6+/ − and wild-type mice (t (16) = 0.29, P = .78). GAD67 heterozygous null mutation (GAD67+/−) mice in which exon 2 (the first coding exon) of the Gad1 gene has been removed were previously obtained from Richard Palmiter30 and studied for other GABA-related mRNAs.31 Tissue from GAD67+/− mice was similarly processed as described in the supplementary methods.
In situ hybridization using an antisense S-labeled riboprobe for parvalbumin mRNA (bases 256–594 of the mouse parvalbumin gene [X59382]) was conducted as previously described31 (supplementary methods). Hybridization procedures were performed using 3 evenly spaced tissue sections (12 µm; Bregma +1.98 to +1.54mm) from each subject, which were then exposed to Biomax MR film (Kodak), coated with nuclear emulsion, developed, and counterstained with cresyl violet. Film optical density was measured in cingulate and prelimbic cortex (together termed medial PFC) and secondary motor cortex (M2)32 using a Microcomputer Imaging Device system (Imaging Research Inc). To evaluate parvalbumin mRNA expression at the cellular level, the numbers of silver grains were counted over neurons using systematically placed sampling frames through the medial PFC. Within the frames, the number of grains was counted in a circle with a fixed diameter of 16 μm placed over each Nissl-stained nucleus that had at least 3 overlying silver grains. Neurons were considered to be specifically labeled if the grain density per neuron was more than 5× background.11,31
Statistical Analysis
For the human study, the analysis of covariance (ANCOVA) model we report includes Lhx6 mRNA level as the dependent variable, diagnostic group as the main effect, subject pair as a blocking factor, and postmortem interval, brain pH, and freezer storage time as covariates. Subject pairing may be considered an attempt to account for the parallel processing of tissue samples from a pair and to balance diagnostic groups for sex, age, and RIN and not a true statistical paired design. Therefore, a second ANCOVA model without subject pair as a blocking factor and including sex, age, and RIN as covariates was also used, and both models produced similar results. Subsequent analyses of differences in mRNA levels between schizophrenia subjects grouped by substance abuse, smoking, and psychotropic medications at time of death and predictors and indicators of disease severity were conducted using the unpaired ANCOVA models with α = .05.
For the developmental monkeys, an ANOVA model with Lhx6 mRNA level as the dependent variable and age group as main effect was employed, and least significant difference was employed as a post hoc test with α = .05. For the mouse studies, measures of mRNA levels in Lhx6+/ − or GAD67+/− mice and the respective wild-type mice were compared using t tests with α = .05.
Results
Lhx6 mRNA Expression in the PFC in Schizophrenia
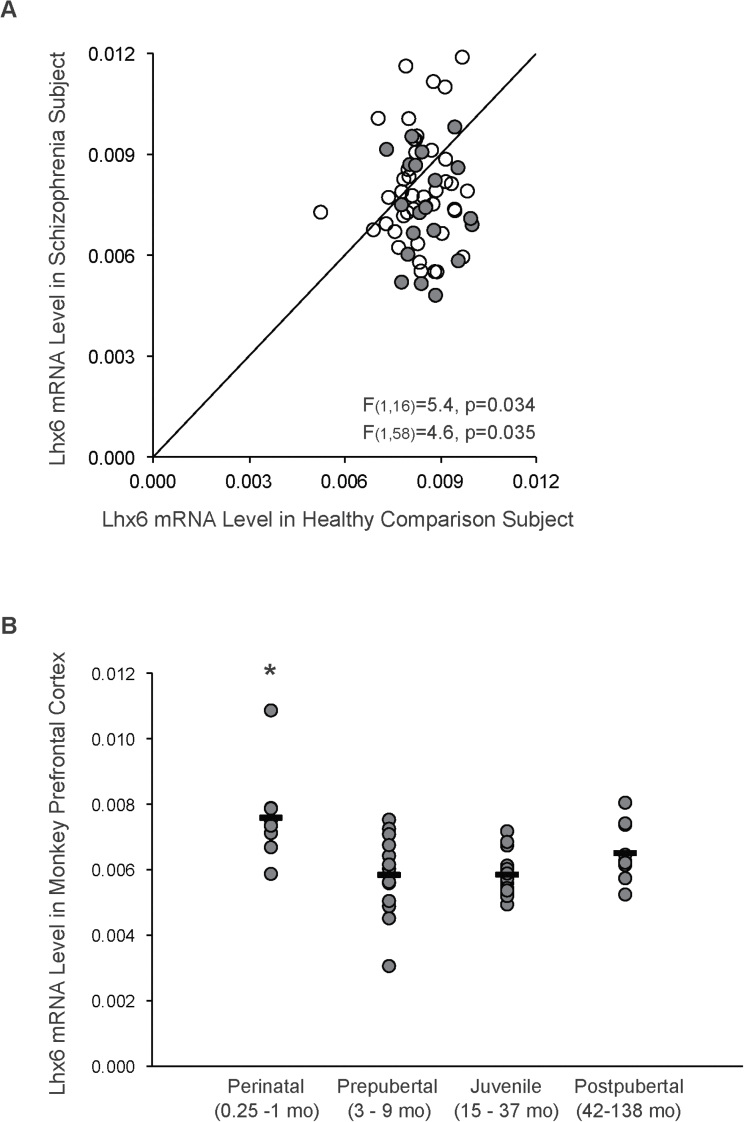
Consistent with our previous findings,11 Lhx6 mRNA levels were lower (−15%; F (1,16) = 5.4, P = .034) in the new cohort of schizophrenia subjects (mean ± SD: 0.0075±0.0015) relative to healthy comparison subjects (0.0086±0.0008; figure 1A). Because different quantification methods were employed for the present and previous studies of Lhx6 mRNA that were conducted in different subject cohorts, we quantified Lhx6 mRNA levels in the combined cohort of 62 schizophrenia subjects and 62 matched healthy subjects using the same qPCR method. As expected, Lhx6 mRNA levels were lower in the combined cohort of 62 schizophrenia subjects (0.0079±0.0016) relative to healthy subjects (0.0084±0.0008; F (1,58) = 4.6, P = .035). In addition, in the 62 schizophrenia subjects, we found no relationship between substance abuse/dependence, smoking or use of antipsychotic, antidepressant, or benzodiazepine or anticonvulsant medications at time of death and Lhx6 mRNA levels (all F ≤ 1.8, P ≥ .19; supplementary figure S1A). Furthermore, Lhx6 mRNA levels did not differ among the 62 schizophrenia subjects as a function of factors that predict a more severe illness course (early age at illness onset [≤18 years of age], first-degree relative with schizophrenia, a diagnosis of schizophrenia rather than schizoaffective disorder, male sex) or indicators of illness severity (low socioeconomic status as measured by the Hollingshead Index of Social Position, suicide, living dependently at the time of death, no history of marriage; all F ≤ 3.8, P ≥ .06; supplementary figure S1B).
Fig. 1.
qPCR determination of prefrontal cortex (PFC) Lhx6 mRNA levels in schizophrenia subjects and across postnatal development in monkeys. (A) Lhx6 mRNA levels for a new (n = 20) and a previous studied (n = 42) cohort of schizophrenia subjects relative to matched healthy subjects are indicated by gray and open circles, respectively. Data points to the right of the unity line indicate lower levels in the schizophrenia subject relative to the healthy subject and vice versa. (B) Lhx6 mRNA levels in PFC area 10 from 49 monkeys at different stages of postnatal development are plotted in 4 age-defined groups.15
Postnatal Development of Lhx6 mRNA Levels in Monkey PFC
We next examined the developmental trajectory of Lhx6 mRNA levels from the neonatal period to adulthood in the PFC of macaque monkeys, which recapitulates the cytoarchitecture and protracted development of human PFC. Statistical analysis revealed a significant effect of age group on Lhx6 mRNA levels in monkey PFC (F (3,45) = 6.9, P = .001; figure 1B), which remained significant after excluding a potential outlier (>2 SDs from the mean) in the perinatal age group (F (3,44) = 5.3, P = .003). Post hoc comparisons revealed that Lhx6 mRNA levels declined (−24%) from the perinatal to prepubertal time period (P = .0002) then remained stable across the prepubertal, juvenile, and postpubertal time periods (P ≥ .13).
Expression of GABA-Related Markers in Lhx6+/− Mice
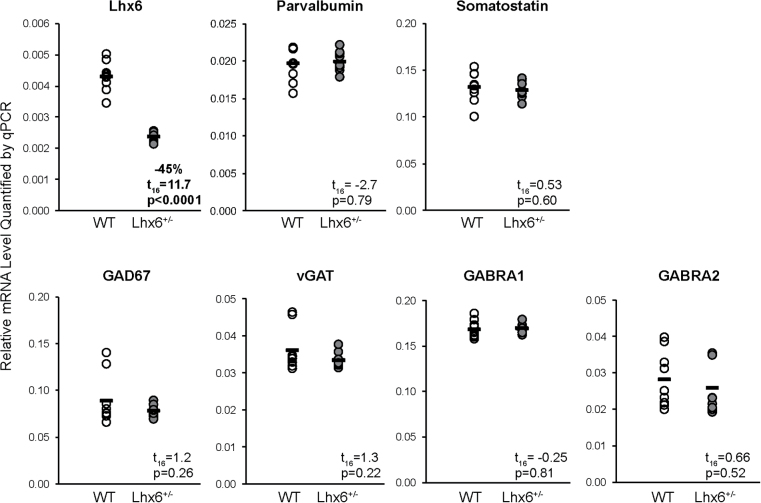
Mean Lhx6 mRNA levels were 45% lower in frontal cortex homogenates of Lhx6+/− mice relative to wild-type littermate mice (t (16) = 11.7, P < .0001; figure 2), which is consistent with a loss of function in 1 Lhx6 allele. This reduction in frontal cortex Lhx6 mRNA expression is highly similar in magnitude to the largest deficit in Lhx6 mRNA observed in a schizophrenia subject relative to the matched healthy comparison subject (−45%; figure 1), which suggests that the Lhx6+/− mouse model has construct validity to determine whether a partial loss of Lhx6 is sufficient to produce deficits in GABA-related markers similar to those seen in schizophrenia.
Fig. 2.
qPCR determination of gamma-aminobutyric acid -related mRNA levels in Lhx6+/ − mice. Transcript levels (black bar indicates group mean) for Lhx6, GAD67, parvalbumin, somatostatin, the vesicular gamma-aminobutyric acid transporter (vGAT), and the α1 (GABRA1) and α2 (GABRA2) subunits of the GABAA receptor were quantified in the frontal cortex of Lhx6+/− mice and wild-type (WT) littermate mice.
We next determined whether a partial loss of Lhx6 in the Lhx6+/− mice results in disturbances in GAD67, parvalbumin, and somatostatin mRNA levels similar to those reported in schizophrenia. However, qPCR analysis of frontal cortex revealed that transcript levels for GAD67 (t (16) = 1.2, P = .26), parvalbumin (t (16) = .27, P = .79), and somatostatin (t (16) = .53, P = .60) did not differ between Lhx6+/− and wild-type littermate mice (figure 2). We then determined whether a partial loss of Lhx6 might result in alterations in other GABA-related markers that have previously been reported to be abnormally expressed in the PFC in schizophrenia.15,33,34 However, qPCR analysis also found no differences in mRNA levels for the vesicular GABA transporter and the α1 and α2 subunits of the GABAA receptor in the Lhx6+/− mice relative to wild-type littermate mice (t (16) ≤ |1.3|, P ≥ .22; figure 2).
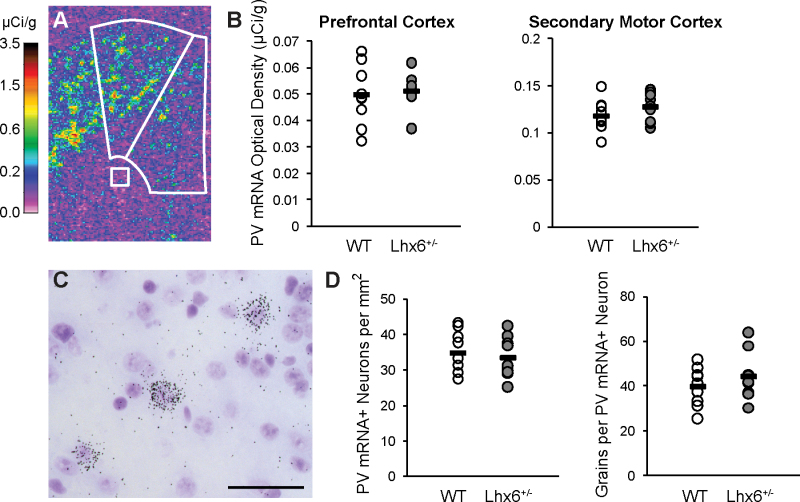
In schizophrenia, in situ hybridization grain-counting studies have found that PFC GABA neurons underexpress parvalbumin mRNA but that the number of neurons expressing detectable parvalbumin mRNA levels appears unchanged.7 Therefore, we used in situ hybridization to determine whether the partial loss of Lhx6 mRNA in Lhx6+/− mice produces the same pattern of deficits in parvalbumin mRNA expression present in the PFC in schizophrenia that might not be detectable by quantitative PCR. Optical density analysis revealed that parvalbumin mRNA levels did not differ between Lhx6+/− and wild-type mice in medial PFC (t (16) = 0.37, P = .72; figure 3A and B). We also analyzed optical density in the secondary motor cortex, which is adjacent to the medial PFC and contains a higher density of parvalbumin neurons than the medial PFC,31 and did not find a difference in parvalbumin mRNA levels between Lhx6+/− and wild-type mice (t (16) = 1.3, P = .21; figure 3A and B). Finally, cellular grain–counting analyses revealed no differences in the number of parvalbumin mRNA-labeled neurons per tissue area (t (16) = 0.54, P = .60) or in the number of grains per parvalbumin mRNA-labeled neuron (t (16) = 1.1, P = .31) in the medial PFC of Lhx6+/− mice (figure 3C and D).
Fig. 3.
In situ hybridization for parvalbumin mRNA in Lhx6+/− mice. (A) Pseudocolored film autoradiograph of a frontal cortex processed by in situ hybridization for parvalbumin (PV) mRNA with calibration scale (µCi/g). White lines denote medial PFC and secondary motor cortex and white matter background measures. (B) Film optical density analysis found that mean parvalbumin mRNA levels did not differ between wild-type (WT) and Lhx6+/− mice in medial PFC or secondary motor cortex. (C) Emulsion-dipped tissue section processed by in situ hybridization for parvalbumin mRNA reveals the presence of silver grain clusters over a minority of lightly Nissl-stained neurons but not over smaller, intensely Nissl-stained glia, which is consistent with specific labeling of parvalbumin neurons. Calibration bar = 50 µm. (D) Cellular grain–counting analyses in the medial PFC found no differences between WT and Lhx6+/− mice in parvalbumin mRNA-labeled neurons/mm2 or mean number of grains per parvalbumin mRNA-labeled neuron.
Relationship Between Lhx6 and GAD67 in Schizophrenia and in GAD67 Heterozygous Null Mutation Mice
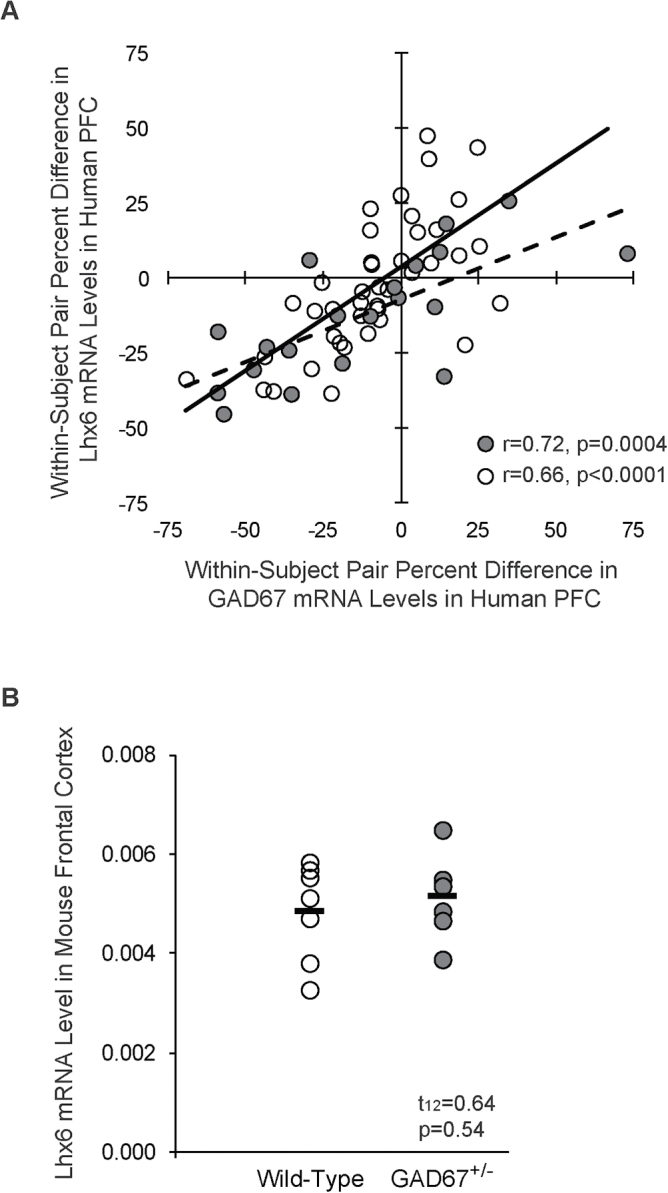
Because we found that a partial reduction of Lhx6 similar to that reported in schizophrenia did not result in alterations in frontal cortex GAD67 mRNA levels, we further examined whether a relationship may exist between lower Lhx6 and GAD67 mRNA levels in schizophrenia subjects. We found that within-subject pair differences in Lhx6 and GAD67 mRNA levels were strongly correlated in the new cohort of 20 schizophrenia and matched healthy comparison subjects (r = .72; P = .0004; figure 4A) and in the combined cohort of 62 schizophrenia subject and matched healthy comparison subjects (r = .66, P < .0001), which suggests that a relationship between Lhx6 and GAD67 is indeed present in the disease. Consequently, in order to determine if the deficits in Lhx6 could be downstream of deficits in GAD67, we quantified Lhx6 mRNA levels in frontal cortex homogenates of GAD67 heterozygous null mutation (GAD67+/−) mice in which we previously demonstrated lower GAD67 mRNA levels (−37%).31 However, Lhx6 mRNA levels did not differ between GAD67+/− and wild-type mice (t (12) = .64, P = .54; figure 4B).
Fig. 4.
Relationship between Lhx6 and GAD67 in schizophrenia and GAD67+/− mice. (A) Within-subject pair differences in Lhx6 and GAD67 mRNA levels were strongly correlated in the PFC of a new cohort of 20 schizophrenia and healthy comparison subjects (gray circles and dashed trendline) and in the combined cohort of 62 schizophrenia and healthy comparison subjects, which included 42 previously studied subject pairs (open circles and solid trendline). (B) Mean Lhx6 mRNA levels did not differ in the frontal cortex of wild-type and GAD67+/− mice.
Discussion
We utilized a cross-species approach to investigate the possible pre- and postnatal roles of Lhx6 in the pathogenesis of cortical parvalbumin and somatostatin neuron dysfunction in schizophrenia. First, we replicated our prior finding of deficits in Lhx6 mRNA levels in a new cohort of schizophrenia subjects, suggesting that deficits in Lhx6 mRNA levels, along with deficits in GAD67 mRNA levels, are a common component of cortical GABA circuitry dysfunction in schizophrenia. Second, we found that Lhx6 mRNA levels normally decline from the perinatal period until the prepubertal period, then remain stably expressed through adulthood in monkey PFC. Third, using a mouse model, we found that a partial loss of Lhx6 expression similar in magnitude to that seen in schizophrenia is not sufficient to produce the pattern of disturbances in GABA-related markers reported in schizophrenia. Finally, we found that while deficits in GAD67 mRNA also do not drive deficits in Lhx6 in another mouse model, deficits in Lhx6 and GAD67 mRNA levels are strongly correlated in schizophrenia. These findings help clarify and constrain the potential pre- and postnatal roles of deficits in Lhx6 mRNA levels in cortical parvalbumin and somatostatin neuron dysfunction in schizophrenia and may indicate that other upstream factors contribute to coordinated disturbances in the expression of Lhx6 and GAD67 in the disorder.
We found that the postnatal expression of prefrontal Lhx6 mRNA levels was highest during the perinatal time period and declined until the prepubertal period in monkeys, which is consistent with reports of a primary role of Lhx6 in the prenatal development of cortical parvalbumin and somatostatin neurons.18,19,23 Tissue from prenatal monkeys was not available for this study, and consequently, it is unclear whether Lhx6 mRNA levels are even higher at late prenatal stages. While speculative, a decline in Lhx6 mRNA levels during this late prenatal/early postnatal period that occurs to an excessive degree might produce the Lhx6 mRNA deficits seen in schizophrenia. In contrast, the stability of Lhx6 mRNA levels across the prepubertal and peripubertal periods suggests that deficits in Lhx6 mRNA expression in schizophrenia are unlikely to be attributable to arrested development during adolescence and may not be involved in the anatomical and molecular changes in parvalbumin neurons that occur during adolescence.10,12,13,15 However, the strong and stable expression of Lhx6 throughout life that is restricted to parvalbumin and somatostatin neurons22 may indicate other important roles for Lhx6 in adulthood such as maintenance of cellular phenotype.
The absence of an effect of a partial loss of Lhx6 on GABA-related markers, including GAD67, parvalbumin, and somatostatin mRNAs, in the frontal cortex of Lhx6+/ − mice highlights the importance of understanding the relevance of gene dose effects to hypothesized pathogenetic contributions of underexpressed molecular markers in schizophrenia. For example, a complete loss of Lhx6 (homozygous null mutation) has profound effects on the development of cortical parvalbumin and somatostatin neurons, including reduced tangential migration and impeded differentiation into parvalbumin and somatostatin neurons followed by death of the animal.18,19,23 However, we found that a partial reduction in Lhx6 levels (45% less than wild-type mice) that more closely modeled the largest reduction in Lhx6 mRNA levels seen in a schizophrenia subject was not sufficient to impair the migration or differentiation of parvalbumin neurons or to alter frontal cortex levels of GABA-related markers. Interestingly, while this study was being conducted, another recent study utilized a mouse model of a severe but not total loss of Lhx6 (ie, mice with an Lhx6 null mutation allele and a hypomorphic approximately 40% functional Lhx6 allele)23 that was not lethal. The severe loss of Lhx6 did not affect parvalbumin or somatostatin neuron migration but substantially impaired differentiation into cortical somatostatin, but not parvalbumin, neurons. Thus, available evidence indicates that a complete loss of Lhx6 prevents parvalbumin and somatostatin neurons from differentiating, a major loss of Lhx6 (ie, more than half) reduces the terminal differentiation of somatostatin but not parvalbumin neurons, and a moderate loss of Lhx6 (ie, less than half) does not have a discernable effect on parvalbumin and somatostatin neuron development. Going forward, these data suggest that (1) gene dose is an important factor to consider in future studies applying mouse models of loss of gene function to the understanding of pathogenetic processes in schizophrenia and (2) differing degrees of loss of gene function can differentially affect the development of different neuronal subpopulations that normally express that gene. In the case of Lhx6, parvalbumin neurons appear to require only a small amount of Lhx6 (ie, 1 partially functioning Lhx6 allele) to successfully complete migration, differentiation, and maturation, while somatostatin neurons require at least 1 completely functional Lhx6 allele for normal development.
The lack of downstream effects from an early partial reduction in Lhx6 mRNA levels on GABA-related mRNAs may indicate that lower Lhx6 levels in adults with schizophrenia are a consequence, rather than a cause, of parvalbumin and somatostatin neuron pathology. That is, perhaps, Lhx6 mRNA expression becomes downregulated in schizophrenia when some other upstream disease-related factors disrupt the phenotype of parvalbumin and somatostatin neurons. This interpretation may, in part, explain why (1) the deficit in Lhx6 mRNA levels are commonly seen across different cohorts of schizophrenia subjects similar to the replicated findings of deficits in GAD67, parvalbumin, and somatostatin mRNAs and (2) the deficit in Lhx6 mRNA levels has not been reported to be larger than the deficits in GAD67, parvalbumin, and somatostatin mRNA levels in schizophrenia. Consistent with this interpretation, we also found that while deficits in Lhx6 mRNA levels are strongly correlated with deficits in GAD67 mRNA levels in schizophrenia subjects, Lhx6 mRNA levels are not affected by a partial loss of GAD67 mRNA in GAD67+/− mice.
Other upstream disease-related factors may potentially disrupt the prenatal ontogeny of cortical GABA neurons in schizophrenia. For example, deficits in cortical levels of the transcription factor Dlx1, which regulates the prenatal ontogeny of cortical GABA neurons,35 have also been reported in schizophrenia.17 Furthermore, maternal infection during the first and second trimesters, when cortical parvalbumin and somatostatin neurons are born and begin to migrate in humans,21 is associated with an increased risk of developing schizophrenia in offspring.36 Murine models of maternal immune activation have been reported to have deficits in parvalbumin immunoreactivity and in GAD67 mRNA and protein levels in the PFC of offspring37,38 and lower Lhx6 levels in fetal brain.39 Consequently, perhaps prenatal insults that are both maternal (ie, immune activation) and fetal (ie, deficits in multiple transcription factors such as Lhx6 and Dlx1) in origin may be required to interact and disrupt the development of cortical parvalbumin and somatostatin neurons in schizophrenia. Thus, future studies that employ gene × gene and/or gene × environment interactions in animal models are needed to help direct and constrain pathogenetic hypotheses involving disrupted prenatal ontogeny of cortical parvalbumin and somatostatin neurons in schizophrenia.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
The National Institutes of Health (MH084016, MH100066 to D.W.V.; MH043784, MH051234 to D.A.L.); the Hamilton Family Award for Basic Neuroscience Research in Psychiatry (to D.W.V.).
Supplementary Material
Acknowledgments
Dr Lewis currently receives investigator-initiated research support from Bristol-Myers Squibb and Pfizer and serves as a consultant in the areas of target identification and validation and new compound development to Bristol-Myers Squibb, Concert Pharmaceuticals and Autifony. All other authors have nothing to disclose.
References
- 1. Akbarian S, Kim JJ, Potkin SG, et al. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52:258–266 [DOI] [PubMed] [Google Scholar]
- 2. Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry. 2000;57:237–245 [DOI] [PubMed] [Google Scholar]
- 3. Guidotti A, Auta J, Davis JM, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57:1061–1069 [DOI] [PubMed] [Google Scholar]
- 4. Straub RE, Lipska BK, Egan MF, et al. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Mol Psychiatry. 2007;12:854–869 [DOI] [PubMed] [Google Scholar]
- 5. Duncan CE, Webster MJ, Rothmond DA, Bahn S, Elashoff M, Shannon Weickert C. Prefrontal GABA(A) receptor alpha-subunit expression in normal postnatal human development and schizophrenia. J Psychiatr Res. 2010;44:673–681 [DOI] [PubMed] [Google Scholar]
- 6. Curley AA, Arion D, Volk DW, et al. Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: clinical, protein, and cell type-specific features. Am J Psychiatry. 2011;168:921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hashimoto T, Volk DW, Eggan SM, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23:6315–6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morris HM, Hashimoto T, Lewis DA. Alterations in somatostatin mRNA expression in the dorsolateral prefrontal cortex of subjects with schizophrenia or schizoaffective disorder. Cereb Cortex. 2008;18:1575–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry. 2009;65:1006–1014 [DOI] [PubMed] [Google Scholar]
- 10. Fung SJ, Webster MJ, Sivagnanasundaram S, Duncan C, Elashoff M, Weickert CS. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am J Psychiatry. 2010;167:1479–1488 [DOI] [PubMed] [Google Scholar]
- 11. Volk DW, Matsubara T, Li S, et al. Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. Am J Psychiatry. 2012;169:1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Erickson SL, Lewis DA. Postnatal development of parvalbumin- and GABA transporter-immunoreactive axon terminals in monkey prefrontal cortex. J Comp Neurol. 2002;448:186–202 [DOI] [PubMed] [Google Scholar]
- 13. Fish KN, Hoftman GD, Sheikh W, Kitchens M, Lewis DA. Parvalbumin-containing chandelier and basket cell boutons have distinctive modes of maturation in monkey prefrontal cortex. J Neurosci. 2013;33:8352–8358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hyde TM, Lipska BK, Ali T, et al. Expression of GABA signaling molecules KCC2, NKCC1, and GAD1 in cortical development and schizophrenia. J Neurosci. 2011;31:11088–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoftman GD, Volk DW, Bazmi HH, et al. Altered cortical expression of GABA-related genes in schizophrenia: illness progression vs developmental disturbance [published online ahead of print 2013]. Schizophr Bull. 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang Y, Fung SJ, Rothwell A, Tianmei S, Weickert CS. Increased interstitial white matter neuron density in the dorsolateral prefrontal cortex of people with schizophrenia. Biol Psychiatry. 2011;69:63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Joshi D, Fung SJ, Rothwell A, Weickert CS. Higher gamma-aminobutyric acid neuron density in the white matter of orbital frontal cortex in schizophrenia. Biol Psychiatry. 2012;72:725–733 [DOI] [PubMed] [Google Scholar]
- 18. Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27:3078–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JL. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J Comp Neurol. 2008;510:79–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fertuzinhos S, Krsnik Z, Kawasawa YI, et al. Selective depletion of molecularly defined cortical interneurons in human holoprosencephaly with severe striatal hypoplasia. Cereb Cortex. 2009;19:2196–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jakovcevski I, Mayer N, Zecevic N. Multiple origins of human neocortical interneurons are supported by distinct expression of transcription factors. Cereb Cortex. 2011;21:1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Georgiev D, González-Burgos G, Kikuchi M, Minabe Y, Lewis DA, Hashimoto T. Selective expression of KCNS3 potassium channel α-subunit in parvalbumin-containing GABA neurons in the human prefrontal cortex. PLoS One. 2012;7:e43904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neves G, Shah MM, Liodis P, et al. The LIM homeodomain protein Lhx6 regulates maturation of interneurons and network excitability in the mammalian cortex. Cereb Cortex. 2013;23:1811–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. American Psychiatric Association. DSM-IV. Diagnostic and Statistical Manual of Mental Disorders. 4th ed. Washington, DC: American Psychiatric Association; 1994 [Google Scholar]
- 25. Volk DW, Eggan SM, Lewis DA. Alterations in metabotropic glutamate receptor 1α and regulator of G protein signaling 4 in the prefrontal cortex in schizophrenia. Am J Psychiatry. 2010;167:1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Volk DW, Siegel BI, Verrico CD, Lewis DA. Endocannabinoid metabolism in the prefrontal cortex in schizophrenia. Schizophr Res. 2013;147:53–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Volk DW, Radchenkova PV, Walker EM, Sengupta EJ, Lewis DA. Cortical opioid markers in schizophrenia and across postnatal development. Cereb Cortex. 2012;22:1215–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hashimoto T, Bazmi HH, Mirnics K, Wu Q, Sampson AR, Lewis DA. Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Am J Psychiatry. 2008;165:479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chattopadhyaya B, Di Cristo G, Wu CZ, et al. GAD67-mediated GABA synthesis and signaling regulate inhibitory synaptic innervation in the visual cortex. Neuron. 2007;54:889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Curley AA, Eggan SM, Lazarus MS, Huang ZJ, Volk DW, Lewis DA. Role of glutamic acid decarboxylase 67 in regulating cortical parvalbumin and GABA membrane transporter 1 expression: implications for schizophrenia. Neurobiol Dis. 2013;50:179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paxinos G, Franklin K. The Mouse Brain in Stereotaxic Coordinates. 2nd ed. San Diego, CA: Academic Press; 2001 [Google Scholar]
- 33. Beneyto M, Abbott A, Hashimoto T, Lewis DA. Lamina-specific alterations in cortical GABA(A) receptor subunit expression in schizophrenia. Cereb Cortex. 2011;21:999–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Glausier JR, Lewis DA. Selective pyramidal cell reduction of GABA(A) receptor α1 subunit messenger RNA expression in schizophrenia. Neuropsychopharmacology. 2011;36:2103–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476 [DOI] [PubMed] [Google Scholar]
- 36. Brown AS, Derkits EJ. Prenatal infection and schizophrenia: a review of epidemiologic and translational studies. Am J Psychiatry. 2010;167:261–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meyer U, Nyffeler M, Yee BK, Knuesel I, Feldon J. Adult brain and behavioral pathological markers of prenatal immune challenge during early/middle and late fetal development in mice. Brain Behav Immun. 2008;22:469–486 [DOI] [PubMed] [Google Scholar]
- 38. Richetto J, Calabrese F, Riva MA, Meyer U. Prenatal immune activation induces maturation-dependent alterations in the prefrontal GABAergic transcriptome. Schizophr Bull. 2014;40:351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oskvig DB, Elkahloun AG, Johnson KR, Phillips TM, Herkenham M. Maternal immune activation by LPS selectively alters specific gene expression profiles of interneuron migration and oxidative stress in the fetus without triggering a fetal immune response. Brain Behav Immun. 2012;26:623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.