Summary
Microtubule (MT) plus-end tracking proteins (+TIPs) preferentially localize to MT plus-ends. End-binding proteins (EBs) are master regulators of the +TIP complex; however, it is unknown whether EBs are regulated by other +TIPs. Here, we show that Cytoplasmic linker associated proteins (CLASPs) modulate EB localization at MTs. In CLASP-depleted cells, EBs localized along the MT lattice in addition to plus-ends. The MT-binding region of CLASP was sufficient for restoring normal EB localization, while neither EB-CLASP interactions nor EB tail-binding proteins are involved. In vitro assays revealed that CLASP directly functions to remove EB from MTs. Importantly, this effect occurs specifically during MT polymerization, but not at pre-formed MTs. Increased GTP-tubulin content within MTs in CLASP-depleted cells suggests that CLASPs facilitate GTP-hydrolysis to reduce EB lattice binding. Together, these findings suggest that CLASPs influence the MT lattice itself to regulate EB and determine exclusive plus-end localization of EBs in cells.
Introduction
MTs are inherently polar structures polymerized from GTP-tubulin heterodimers, which incorporate at the growing MT plus-end. +TIPs are a diverse group of conserved MT-associated proteins (MAPs) that specifically localize to the dynamic tips of MTs (Schuyler and Pellman, 2001). +TIPs are ideally positioned to regulate MT dynamics and many important MT-based processes. Of the +TIPs, EBs are notable in their ability to autonomously recognize a transient feature at plus-ends with great specificity (Bieling et al., 2007). In mammalian cells, there are three EB family members: EB1, EB2, and EB3. EB1 and EB3 are well studied and are similar in their structure, localization, and behavior, while EB2 is more divergent (Buey et al., 2011; Komarova et al., 2009). Recent work has revealed that EBs localize to MT tips by sensing the nucleotide-state, as well as the specific conformation, of tubulin at MT plus-ends (Maurer et al., 2011; Maurer et al., 2012; Zanic et al., 2009). The majority of +TIPs require binding to the EB tail region, via various localization signals (SxIP (Honnappa et al., 2009) or CAP-Gly (Weisbrich et al., 2007)), to achieve their specific comet-like localization (Akhmanova and Steinmetz, 2008); for this reason, EBs are considered the master regulators of the MT +TIP network.
Within this network, CLASPs are a unique class of +TIPs that exhibit two specific MT localizations. CLASPs support MT growth by binding plus-ends and promoting rescues (Akhmanova et al., 2001; Mimori-Kiyosue et al., 2005). CLASPs also stabilize MT subsets by binding along the MT lattice, for example pioneer MTs at the leading edge (Wittmann and Waterman-Storer, 2005) and Golgi-derived MTs (Efimov et al., 2007; Miller et al., 2009). In mammalian cells, there are two redundant CLASP family members, CLASP1 and CLASP2, which rely on EB interactions (via the basic/SxIP region) to localize to MT tips (Mimori-Kiyosue et al., 2005). In fission yeast, although CLASP associates with EB1, its stabilizing affect on MTs is independent of other +TIPs suggesting that CLASP directly regulates MTs (Bratman and Chang, 2007). Of note, CLASPs also independently bind tubulin heterodimers and associate with the MT lattice through their TOG (tumor overexpressed gene) domains (Al-Bassam et al., 2010; Al-Bassam et al., 2007). Here, we report evidence that CLASPs are crucial determinants of proper EB localization at MTs in cells.
Results
EB proteins localize to the MT lattice in CLASP-depleted cells
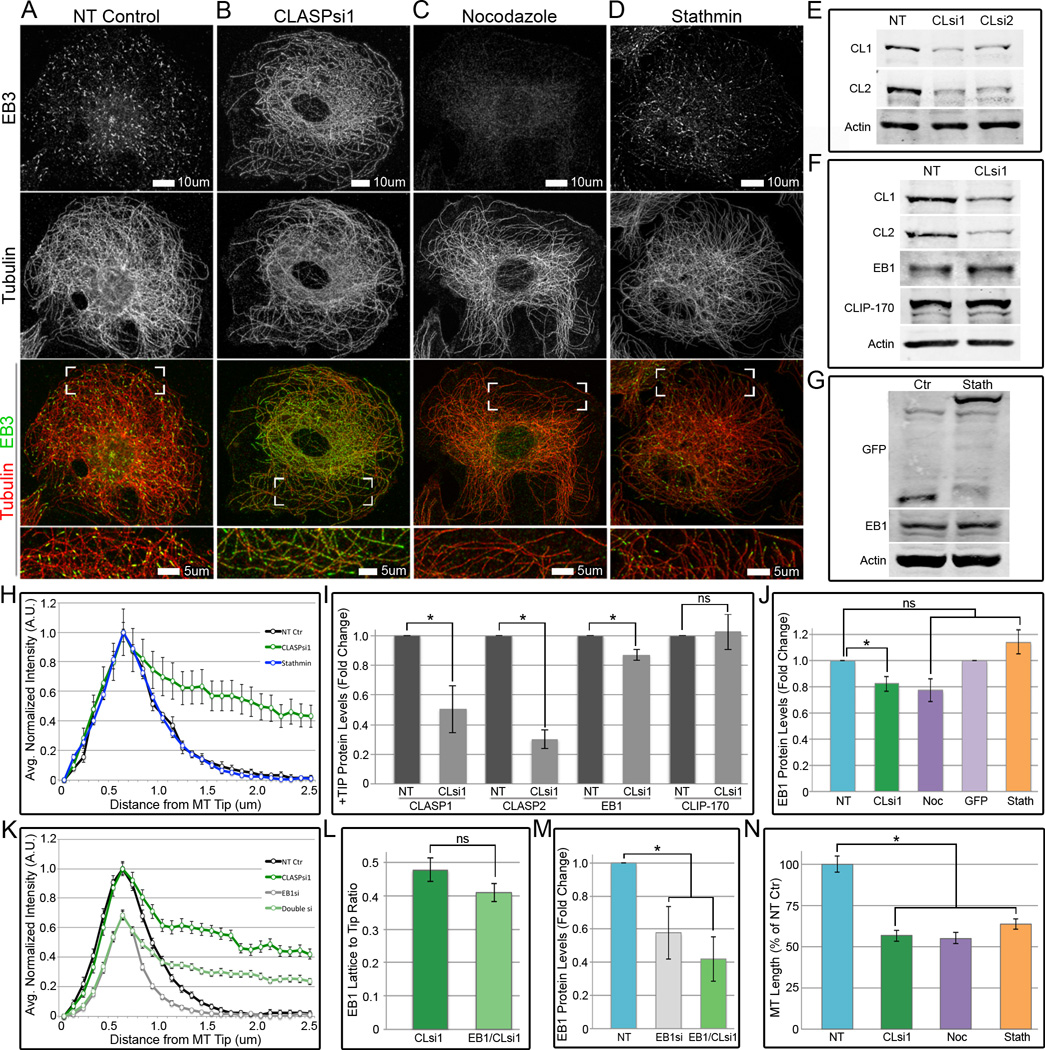
Although it is well established that EBs are the master regulators of the MT +TIP network, it is unknown whether other +TIPs regulate EB localization at MTs. Strikingly, we found that, upon depletion of CLASP1 and CLASP2 (herein referred to as CLASPs), endogenous EB1 (Figure S1A–S1B) and EB3 (Figure 1A–1B) both bind along the MT lattice in addition to their tip localizations. This dramatic change is quantified by line scan analysis of EB fluorescence intensity along MTs (Figure 1H). CLASP-dependent EB redistribution was observed in multiple cell types and is therefore not cell type specific: A7r5 (Figure 1A–1B and S1A–S1B), RPE1 (Figure 2A–2B), MEF, COS-7, HeLa, B16-F1 and Caco-2 (Figure S2) are shown in this study. Interestingly, depletion of CLASP1 or CLASP2 individually (Figure S1J) resulted in partial redistribution of EB1 and EB3 (herein referred to as EBs) along the MT lattice (Figure S1D–S1I); a combination of siRNAs against both CLASPs was used for all subsequent CLASP-depletion experiments (Figure 1E).
Figure 1. Localization of EB proteins at microtubules is altered in CLASP-depleted cells.
(A–D) Immunofluorescence images of A7r5 cells stained for α-tubulin (red) and EB3 (green). (A) EB3 localizes to MT plus-ends in NT control cells. (B) EB3 coats the MT lattice, in addition to its plus-end localization, in CLASP-depleted cells. (C) EB3 does not significantly localize to the lattice in Nocodazole-treated cells. (D) EB3 localizes to MT plus-ends in GFP-Stathmin expressing cells. (A–D) Boxed corners indicate zoom region. White box=scale bar. (E) Western blot of CLASP1 and CLASP2 depletion by two different siRNA combinations. (F) Western blot showing no change in EB1 and CLIP-170 protein levels upon CLASP-depletion. (G) Western blot showing no change in EB1 protein levels upon GFP-Stathmin expression compared to GFP-control. (E–G) Actin as loading control. (H) Line scans showing distribution of EB3 at MTs in NT control (black), CLASP-depletion (green), or GFP-Stathmin (blue). Based on data similar to (A,B,D). (I) +TIP levels by western blot analysis in NT control and CLASP-depletion. (J) EB1 levels by western blot analysis in NT control, CLASP-depletion, Nocodazole treatment, and GFP-control or GFP-Stathmin expression. (K) Line scans showing distribution of EB1 at MTs in NT control (black), CLASP-depletion (green), EB1 partial depletion (grey), or EB1/CLASP double depletion (light green). (L) EB1 L:T ratio in CLASP-depletion and EB1/CLASP double depletion based on (K). (M) EB1 levels by western blot analysis in NT control, EB1 partial depletion, and EB1/CLASP double depletion. (N) Average total MT length in NT control, CLASP-depletion, Nocodazole treatment, and GFP-Stathmin expression. Based on data similar to (A–D). (H,K,L) Line scan and L:T ratio values are normalized mean intensities ± S.E.M. (n=50, 2 independent experiments). (I,J,M) Graphs are normalized means ± S.E.M in 3 independent experiments. See also Figures S1 and S2.
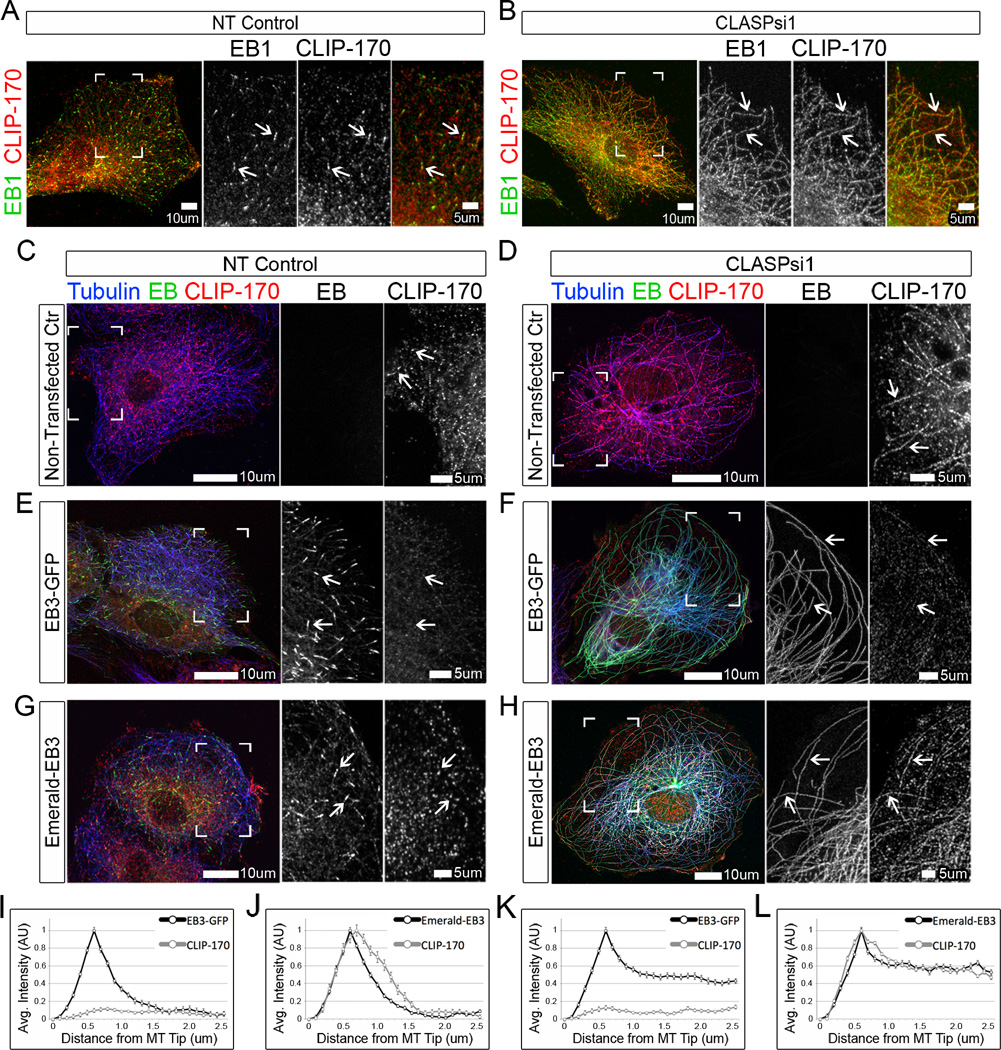
Figure 2. CAP-Gly proteins follow EBs to the microtubule lattice in CLASP-depleted cells.
(A–B) RPE cells stained for EB1 (green) and CLIP-170 (red). (A) EB1 and CLIP-170 localize to MT plus-ends in NT control cells. (B) EB1 and CLIP-170 relocalize to the lattice in CLASP-depleted cells. (C–H) A7r5 cells expressing EB3 (green), and stained for CLIP-170 (red) and α-tubulin (blue, pseudo-colored). (C) CLIP-170 localizes to MT plus-ends in un-transfected NT control cells, (D) CLIP-170 coats the lattice in CLASP-depleted cells. (E) In NT control cells expressing EB3-GFP, EB3-GFP localizes to MT plus-ends, while CLIP-170 localization to plus-ends is reduced. (F) In CLASP-depleted cells expressing EB3-GFP, EB3-GFP relocalizes to the MT lattice, while lattice-bound CLIP-170 is reduced. (G) In NT control cells expressing Emerald-EB3, both Emerald-EB3 and CLIP-170 localize to plus-ends. (H) In CLASP-depleted cells expressing Emerald-EB3, both Emerald-EB3 and CLIP-170 coat the lattice. (A–H) Boxed corners indicate zoom region. White box=scale bar. (C–H) White arrows=EB/CLIP-170 characteristic localization, CLIP-170 displacement, or EB/CLIP-170 lattice relocalization respectively. (I–L) Line scans showing distribution of CLIP-170 (grey) at MTs in cells expressing either EB3-GFP or Emerald-EB3 (black). Based on data similar to (E–H). (A–H) Immunofluorescence. Values are normalized mean intensities ± S.E.M. (n=50, 2 independent experiments). See also Figure S3.
As overexpression of EBs results in lattice decoration, it is in principle possible that an increased EB to MT ratio causes the enhanced EB lattice binding observed. Endogenous levels of EB1 protein did not increase upon CLASP-depletion (Figure 1F and 1I), but MT number decreased because CLASP-depletion alters MT dynamics (Akhmanova et al., 2001; Mimori-Kiyosue et al., 2005). However, in cells briefly treated with Nocodazole to decrease MT number (Figure 1C and 1N), no significant EB3 lattice relocalization was observed (Figure 1C). Also, cells expressing GFP-Stathmin, which sequesters free tubulin dimers and decreases MT number (Figure 1D and 1N), did not show significant EB3 lattice binding (Figure 1D and 1H). Total MT length did not significantly differ between CLASP-depleted, Nocodazole-treated and GFP-Stathmin-expressing cells (Figure 1N). Moreover, the number of EB3 plus-ends upon CLASP-depletion and GFP-Stathmin expression did not statistically differ (Figure S2L). Importantly, in both Nocodazole treatment and GFP-Stathmin expression, EB1 protein levels did not differ from NT control (Figure 1G, 1J, and S1K).
To further confirm that EB lattice binding in CLASP-depleted cells is not an artifact of an increased EB to MT ratio, we performed a partial depletion of EB1, either alone or in addition to CLASP-depletion (Figure 1M and S1M). For single depletion of CLASPs, the L:T ratio was 0.48 ± 0.03 (Figure 1K); similarly, upon double depletion of EB1 and CLASPs, this ratio was 0.41 ± 0.03 (Figure 1L). This difference was not statistically significant indicating that, regardless of the amount of EB protein, CLASP-depletion has the same effect on EBs. Thus, altered EB localization in cells lacking CLASPs is not a result of an increased EB to MT ratio.
+TIP proteins relocalize with EBs in CLASP-depleted cells
We next sought to determine whether CLASP influenced the localization of other +TIP families at MTs, or if CLASP regulation of EB was specific. We screened CLASP-depleted cells for localization of representative proteins from the major +TIP families: CAP-Gly, SxIP, and EB-independent. The CAP-Gly proteins, CLIP-170 and p150Glued, relocalized to the MT lattice upon CLASP-depletion and were indistinguishable from lattice-bound EBs (Figure 2 and S3A–S3B). Endogenous levels of CLIP-170 protein did not increase upon CLASP-depletion (Figure 1F and 1I). The SxIP proteins, GFP-SLAIN2 (Figure S3C–S3D) and the minimal EB-binding domain of Dystonin (GFP-Dst-EBBD, Figure S3E–S3F) relocalized to the lattice upon CLASP-depletion in a similar manner. An exception was another SxIP protein, APC, which did not change localization upon CLASP-depletion (Figure S3G–S3H), possibly due to its regulation by a large number of factors (Kita et al., 2006). Similarly, chTOG did not relocalize to the MT lattice (Figure S3I–S3J); this is likely explained by the capacity of chTOG to track MT tips independently of EBs (Brouhard et al., 2008).
To examine whether CLASPs directly regulate CAP-Gly protein localization at MTs, or if these proteins simply follow EBs to the lattice, we took advantage of a previously described approach. It has been shown that a C-Terminal GFP tag on EB occludes binding of CLIP-170 to the EEY/F tail of EBs; therefore, expression of EB3-GFP in cells displaces CLIP-170 from MT plus-ends (Lomakin et al., 2009; Skube et al., 2010). We observed that EB3-GFP, similar to endogenous protein, relocalizes to the MT lattice in CLASP-depleted cells (Figure 2F and 2K). Interestingly, upon CLASP-depletion in cells expressing EB3-GFP, CLIP-170 did not relocalize to the MT lattice (Figure 2D, 2F, and 2K). We further designed an Emerald-EB3 construct, which interacts normally with CLIP-170 and allows for correct CLIP-170 plus-end localization (Figure 2G). In CLASP-depleted cells, both Emerald-EB3 and CLIP-170 significantly relocalize to MT lattices (Figure 2H and 2L). These experiments reveal that CLIP-170 interaction with the C-terminus of EBs underlies its lattice binding in CLASP-depleted cells. Together with relocalization of GFP-Dst-EBBD, which cannot be recruited by factors other than EBs (Honnappa et al., 2009), these data suggest that both CAP-Gly and SxIP proteins likely follow EBs to the lattice rather than being independently regulated by CLASPs.
TOG2 region of CLASP, but not EB binding, is required to restore normal EB plus-end localization
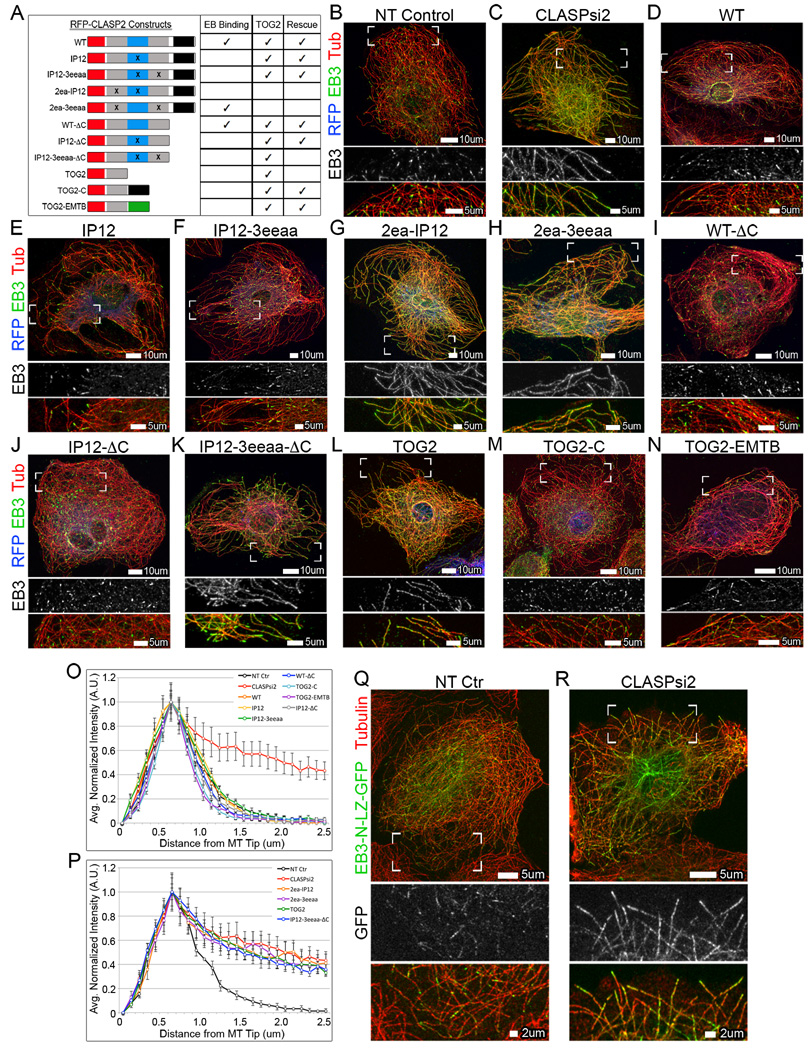
To determine which CLASP domains are instrumental for the mechanism of CLASP-dependent EB regulation, we tested the capacity of various CLASP mutants (Figure 3 and S4) to rescue EB localization. CLASPs have three main protein-interacting regions: The TOG/TOG-like domains confer MT binding (Al-Bassam et al., 2007; Patel et al., 2012), while the basic/SxIP region dictates EB binding (Mimori-Kiyosue et al., 2005; Patel et al., 2012), and the C-terminus mediates CLASP dimerization and interactions with protein partners (Akhmanova et al., 2001; Efimov et al., 2007; Lansbergen et al., 2006). For this study, the TOG-like domains of CLASP2 will be referred to as TOG2 and TOG3, as in (Leano et al., 2013). As expected, re-expression of CLASP2 in CLASP-depleted cells rescued EB3 plus-end localization (Figure 3D and 3O).
Figure 3. TOG2 domain of CLASP is necessary to restore normal EB plus-end localization.
(A) Schematic representation of CLASP2 rescue constructs. Red=RFP tag, grey=MT-binding domains (TOG2 and TOG3), blue=EB-binding region (basic/SxIP), black=C-Terminus, green=MT-binding domain of Ensconsin (EMTB). Construct capacity to bind EB and rescue EB localization is shown. (B) EB3 localizes to MT tips in NT control cells. (C) EB3 extensively coats the lattice, in addition to plus-end localization, in CLASP-depleted cells. (D–N) A7r5 cells expressing various RFP-CLASP2 rescue constructs (pseudo-colored blue) stained for endogenous EB3 (green) and α-tubulin (pseudo-colored red). (D,E,F,I,J,M,N) CLASP-depleted cells expressing WT (D), IP12 (E), IP12-3eeaa (F), WT-ΔC (I), IP12-ΔC (J), TOG2-C (M), or TOG2-EMTB (N) rescue constructs all restore normal EB3 plus-end localization. (G,H,K,L) CLASP-depleted cells expressing 2ea-IP12 (G), 2ea-3eeaa (H), IP12-3eeaa-ΔC (K), or TOG2 (L) rescue constructs were not sufficient to restore normal EB3 localization. EB3 binds along the lattice. Line scan analysis for constructs that rescue EB3 plus-end localization (O) or constructs unable to rescue (P). (O–P) Line scan values are normalized mean intensities ± S.E.M. (n=50, 2 independent experiments). (Q–R) A7r5 cells expressing minimal MT-binding region of EB3, EB3-N-LZ-GFP (green) and stained for α-tubulin (red). EB3-N-LZ-GFP localizes to MT tips in NT control cells (Q) and relocalizes to the lattice in CLASP-depleted cells (R). (B–N,Q–R) Immunofluorescence. Boxed corners indicate zoom region. White box=scale bar. See also Figure S4.
Intriguingly, a CLASP construct with mutated EB-binding motifs readily rescued EB plus-end localization (Figure 3E and 3O), indicating that CLASP-EB interactions are not involved in the observed phenomenon. To confirm this conclusion, we utilized an artificial dimer of the EB3 MT binding domain, EB3-N-LZ-GFP (Komarova et al., 2009), which lacks the CLASP-interacting EB tail region. EB3-N-LZ-GFP localized along the MT lattice in CLASP-depleted cells (Figure 3R) indicating that CLASPs influence EB localization without a contribution from: 1) CLASP-EB interaction or 2) binding of other proteins to the EB tail domain, such as another MAP that might recruit EB to the lattice when CLASPs are not present. Together, these data support a model in which CLASPs alter MTs themselves to restrict EB-MT interactions to MT tips.
Further rescue experiments with CLASP mutant constructs revealed that the TOG2 domain, but not the TOG3 domain of CLASP, was required for regulation of EB (Figure 3F–3H and 3O–3P). Importantly, the TOG2 domain of CLASP has been reported to mediate strong MT interactions (Patel et al., 2012). The monomeric TOG2 domain alone was not sufficient to restore EB plus-end localization (Figure 3L and 3P); although, truncated CLASP mutants containing TOG2 plus an additional MT-interacting region (TOG3 or the Ensconsin MT-binding domain (EMTB)) were sufficient for CLASP-dependent regulation of EBs (Figure 3I–3J and 3N–3O). Also, TOG2 directly linked to the C-terminus of CLASP rescued EB localization (Figure 3M and 3O), possibly because the C-terminal domain weakly interacts with MTs (Wittmann and Waterman-Storer, 2005) or promotes MT-binding via dimerization (Al-Bassam et al., 2010; Patel et al., 2012). Nevertheless, the C-terminal domain of CLASP was not strictly required to rescue EB localization (Figure 3I and 3O), which reveals that CLASP does not bind its cellular partners for regulation of EB at MTs. Together, these mutant studies demonstrate that the TOG2 domain of CLASP is necessary to restore EB localization. Accordingly, we suggest that CLASP influences the MT lattice itself to promote normal EB plus-end distribution.
CLASP modulates microtubule-affinity and EB localization in vitro
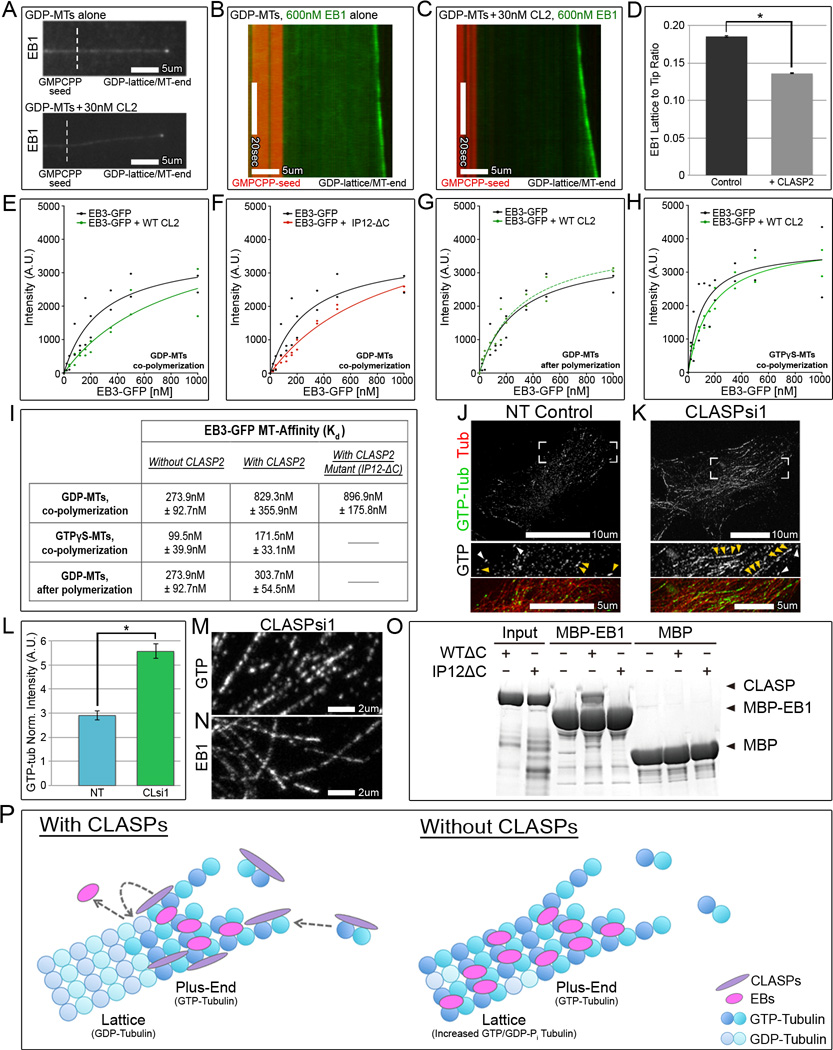
To address whether CLASPs independently regulate EB localization at MTs, we performed in vitro MT plus-end tracking assays. For these experiments, we analyzed the capacity for CLASP2 to remove a minimal EB1 construct, EB1-N-LZ-GFP, from the lattice. In the absence of CLASP, this minimal EB1 construct clearly decorated MT lattices in addition to its plus-end recruitment (Figure 4A and 4B) with a lattice to tip (L:T) ratio of 0.185 ± 0.001 (Figure 4D). Copolymerization with CLASP2 resulted in significant reduction of lattice binding (Figure 4A and 4C) resulting in a ratio of 0.136 ± 0.001 (Figure 4D). Thus, CLASP independently modulated EB1-N-LZ-GFP lattice localization in vitro. Confirming our results in cells, these data indicate that the observed effects are due to CLASP interactions with MTs, rather than a CLASP-EB interaction, because the minimal EB1 construct lacks the CLASP-interacting region.
Figure 4. CLASP modulates microtubule-affinity of EB and promotes GTP-hydrolysis at microtubules in vitro.
(A–C) in vitro MT plus-end tracking assays. (A) Localization of 600nM EB1-N-LZ-GFP to GDP-MTs grown from GMPCPP seeds. (B,C) Kymographs of MTs (shown in A). EB1-N-LZ-GFP alone (B) or in the presence of 30nM CLASP2 (C). Red=GMPCPP-seed, Green=EB1, CLASP2 and tubulin are unlabeled. (D) EB1-N-LZ-GFP L:T ratio based on kymographs similar to (B,C). n=87 for control, n=93 for CLASP, 2 independent experiments. Values are normalized mean intensities ± S.E.M. (E–H) EB3-GFP binding curves for MTs: GDP-MTs copolymerized with 150nM CLASP2 (E), GDP-MTs copolymerized with 150nM IP12-ΔC (EB-binding mutant) (F), GDP-MTs with 150nM CLASP2 added after polymerization (G), or GTPγS-MTs copolymerized with 150nM CLASP2 (H). (E–H) Intensities of MT-bound EB3-GFP as a function of EB concentration with corresponding hyperbolic fit. (E–G) Intensities and binding curves (solid black line) for EB3-GFP bound to GDP-MTs same for each graph. (I) Derived dissociation constants (Kd). Analysis performed over 2 independent experiments. Kd are ± standard error. (J,K) RPE cells stained for GTP-tubulin (hMB11, grey/green) expressing mCherry-tubulin (red). (J) In NT control cells, hMB11 localizes to MT tips and at GTP-tubulin remnants. (K) In CLASP-depleted cells, lattice decoration by hMB11 is increased compared to NT control. (J,K) Boxed corners indicate zoom region. White arrowheads=MT tips, yellow arrowheads=GTP-tubulin remnants. (L) GTP-tubulin intensity in NT control and CLASP-depleted cells. Based on data similar to (J,K). Values are normalized mean intensities ± S.E.M. (n=20, 2 independent experiments). (M,N) Zoomed images highlight speckled nature of GTP-tubulin (M) and EB1 (N). (J,K,M,N) Immunofluorescence. (B,C,J,K,M,N) White box=scale bar. (O) Coomasie showing CLASP (WTΔC and IP12ΔC) pull-down by maltose-binding protein (MBP)-EB1. MBP as negative control. (P) Model for CLASP-dependent regulation of EB localization at MTs. EB nucleotide-state sensing and EB-CLASP interactions are not shown. Purple=CLASP, pink=EB, dark blue=GTP-tubulin, light blue=GDP-tubulin. Grey arrows indicate possible mechanisms of CLASP function.
To quantitatively analyze the effect of CLASP on EB binding, we determined EB affinity for MTs in vitro. GDP-MTs were polymerized from GTP-tubulin in the presence or absence of CLASP2 then incubated with varying concentrations of EB3-GFP prior to cosedimentation. Hyperbolic fit shows EB3-GFP has lower affinity for GDP-MTs copolymerized with CLASP2 compared to GDP-MTs alone (Figure 4E and 4I). Consistent with our data in cells, a CLASP2 mutant unable to bind to EBs (IP12-ΔC) similarly reduced EB3-GFP affinity (Figure 4F, 4I, and 4O).
In order to test whether CLASPs function during MT polymerization, we examined EB3-GFP affinity for GDP-MTs that were first assembled then incubated with CLASP2; importantly, EB3-GFP affinity for these MTs did not significantly differ from GDP-MTs alone (Figure 4G and 4I). This reveals that CLASPs must be present during MT polymerization in order to regulate EB affinity for MTs.
Interestingly, the use of GTPγS, a slowly hydrolysable GTP analog (experimental model for the natural GDP-Pi state EBs recognize) eliminated the difference between MTs polymerized with and without CLASP. As in previously published reports (Maurer et al., 2011), EB3-GFP displayed higher affinity for GTPγS-MTs than GDP-MTs (Figure 4H and 4I). Co-polymerization of GTPγS-MTs with CLASP2 did not significantly reduce EB3-GFP affinity (Figure 4H and 4I). This result indicates that CLASPs recognize or modify a particular tubulin and/or MT conformation to regulate MT-affinity and localization of EBs, possibly via altering the GTP-state.
GTP-tubulin content at the MT lattice is increased in CLASP-depleted cells
Because EBs have higher affinity for MTs formed from GTP-tubulin analogs compared to GDP-tubulin (Maurer et al., 2011; Maurer et al., 2012; Zanic et al., 2009), redistribution of EBs to the MT lattice could be explained by a change in GTP-tubulin content after CLASP-depletion. To explore this hypothesis, we took advantage of the recently described hMB11 antibody, which is thought to recognize GTP-tubulin in cells (Dimitrov et al., 2008). Currently, it is not clear whether hMB11 recognizes GTP-tubulin conformation (or GDP-Pi) or a more general GTP-tubulin-like structural feature. In NT control cells, hMB11 highlights MT tips and occasional lattice patches, known as GTP-tubulin remnants (Figure 4J). In CLASP-depleted cells, GTP-tubulin content was significantly increased: hMB11 lattice patches were more abundant and extended along whole MTs in regions (Figure 4K and 4L).
Consistent with previous findings (Dimitrov et al., 2008), the pattern of GTP-tubulin lattice remnants in CLASP-depleted cells was non-homogenous (Figure 4M); Interestingly, it closely resembled the speckle-like lattice distribution of EBs upon CLASP-depletion (Figure 4N). Unfortunately, due to hMB11 antibody restrictions, we were unable to co-stain for EB and GTP-tubulin at MTs; as an alternative, we used computational modeling to determine whether EB1 lattice speckles and GTP-tubulin remnants are similar in size and distribution along MTs. Image processing was applied to compensate for differences in contrast between EB1 and GTP-tubulin (see Supplemental Experimental Procedures). EB1 and GTP-tubulin speckles, though not identical, had similar distribution characteristics (Figure S4N–S4O); thus, these patterns could, in principle, indicate the same lattice regions. Therefore, we propose that EB lattice binding may be the result of increased GTP-tubulin content along MTs in CLASP-depleted cells, and that the speckle-like appearance of EB staining reflects uneven distribution of GTP-tubulin remnants at the lattice. These experiments reveal a possible mechanism for relocalization of EBs to the MT lattice in CLASP-depleted cells, via recognition of increased GTP-tubulin content, or another hMB11-recognized lattice feature.
Discussion
Our study has uncovered a novel regulatory mechanism for the localization and MT affinity of EBs. Our data indicate that, when CLASPs bind a MT, they influence the lattice itself to reduce EB binding, thereby promoting EB plus-end distribution. This regulation likely occurs during MT polymerization, setting up the nucleotide state sensed by EB for tip localization (Maurer et al., 2012; Zanic et al., 2009). Specifically, we show that GTP-tubulin content is increased at the lattice of CLASP-depleted cells and thus propose that high-affinity EB binding sites within the lattice are likely GTP-tubulin remnants. Because it is unclear what the hMB11 antibody recognizes, our findings in CLASP-depleted cells may indicate that MT lattice structure is altered to more closely resemble MT tips.
These findings add another level of complexity to existing evidence on the regulation of EB interactions with MTs. While CLASPs clearly act as prominent regulators of EB localization, EBs are still enriched at plus-ends, in addition to their lattice localization, in CLASP-depleted cells. Also, in vitro, EBs autonomously track MT plus-ends and are not enriched at the lattice at physiological concentrations, despite CLASPs not being present (Bieling et al., 2007 and others). In our assays, a higher concentration of EB, which binds the lattice, displayed decreased lattice localization in the presence of CLASP leading to restricted EB binding to MT tips. It is likely that while GTP hydrolysis does not strictly require CLASPs, this important process is hindered in their absence both in cells and in vitro.
There are unknown differences in the lattices of MTs polymerized in cells and in vitro (McEwen and Edelstein, 1980; Wade and Chretien, 1993). Indeed, a recent study demonstrates that MT properties in vitro only resemble those in cells when additional cellular proteins are present; Interestingly, they found that another TOG-containing MAP, XMAP215, influences EB behavior via an allosteric interaction though MTs (Zanic et al., 2013). In our study, the effect of CLASP on EB localization is TOG-domain dependent. Thus, we suggest a similar model in which CLASP allosterically regulates EB localization: as a MT polymerizes, CLASPs likely alter the MT lattice, behind the tip, thus restricting high-affinity EB-binding sites to the plus-end, for example by promoting GTP-hydrolysis. Alternatively, because CLASP binds tubulin dimers (Al-Bassam et al., 2010), CLASPs may prime GTP-tubulin dimers for efficient hydrolysis upon lattice incorporation. Either mechanism would lead to increased GTP-tubulin content at MTs and enhanced EB lattice binding if mis-regulated.
Another potential mechanism of CLASP-dependent regulation of EB involves steric hindrance. In this scenario, under physiological conditions, CLASPs would bind the lattice during MT polymerization and remain associated with GTP-tubulin remnants, thereby blocking potential EB recruitment to these sites (and antibody recognition). In this model, CLASPs would bind MTs in a way that prevents EB-lattice interactions. Moreover, this type of MT association should be different from CLASP association with already polymerized MTs observed under overexpression conditions, in which EB is artificially recruited to MTs (Mimori-Kiyosue et al., 2005). The presence of mini-bundles along the MT lattice in CLASP-depleted cells could also explain EB lattice localization; however, MT bundling in CLASP-depleted cells was not detected (Figure S4P).
Because GTP-tubulin remnants are proposed to be sites of MT rescue, it is interesting that in CLASP-depleted cells MTs undergo low rescue frequency. If our model of increased GTP-tubulin content is correct, this may reflect that the absence of a strong rescue factor like CLASP leads to inefficient rescues even in the presence of multiple GTP-tubulin remnants. In the steric hindrance model, the number of GTP-tubulin remnants would not differ in CLASP-depleted cells, and low rescue activity would be explained by the absence of CLASP at these sites.
However, existing evidence leans toward the model of allosteric EB regulation by CLASPs rather than the steric hindrance model, because typically only low amounts of CLASPs are observed at the MT lattice. Furthermore, a recent study on the effects of γ-tubulin depletion reported an increase in GTP-tubulin remnants and a similar EB distribution at MT lattices (Bouissou et al., 2014). Because γ-tubulin is a major factor defining consistency in MT structure, we hypothesize that the lattice structure in γ-tubulin depleted cells is changed, and as a result, EB lattice affinity is altered. This finding supports the idea that a structural change in MT lattices would result in EB recruitment.
In conclusion, our findings have revealed novel functions for CLASPs, have uncovered an additional regulatory mechanism for EB tip tracking, and have major implications for understanding establishment of the +TIP network at MT plus-ends.
Experimental Procedures
Cells
A7r5 cells (ATCC) were maintained in low glucose (1g/L) Dulbecco’s Modified Eagle’s Medium (DMEM), without Phenol Red. hTert-RPE1 cells (Clontech) were maintained in DMEM/F12, which was supplemented with 500ug/mL G418 for an mCherry-tubulin RPE stable line (R. Ohi, Vanderbilt University, TN). HeLa (R. Ohi, Vanderbilt University, TN), MEF (A. Kenworthy, Vanderbilt University, TN), COS-7 (A. Kenworthy), and B16-F1 cells (M. Tyska, Vanderbilt University, TN) were maintained in DMEM. Caco-2 cells (M. Tyska) were maintained in DMEM supplemented with 20% fetal bovine serum (FBS). All cells were grown with 10%FBS (unless indicated) and in 5%CO2 at 37°C. Cells were plated on fibronectin-coated glass coverslips 72hrs prior to experiments.
Antibodies
Mouse monoclonal primary antibodies: anti-α-tubulin (DM1A, Sigma); anti-EB1 (BD Transduction), anti-p150Glued (BD Transduction). Rabbit polyclonal primary antibodies: anti-α-tubulin (Abcam); anti-EB3 (A. Akhmanova); anti-CLIP-170 (A. Akhmanova); anti-chTOG (L. Cassimeris); anti-APC (R. Coffey). A recombinant human primary antibody, hMB11, against GTP-MTs (F. Perez) was also used. Secondary antibodies: Alexa488, Alexa568, or Alexa647-conjugated highly cross-absorbed goat anti-mouse, anti-rabbit, or anti-Human (Invitrogen/Molecular Probes). For +TIPs/MT staining, cells were fixed in methanol (5min, −20°C). hMB11 staining was done according to Dimitrov et al., 2008.
in vitro MT-affinity and MT plus-end tracking assays
For in vitro MT-affinity assays, 20uM tubulin was polymerized alone or with 150nM CLASP2 or IP12ΔC in BRB80 buffer (80mM K-Pipes pH 6.8, 2mM MgCl2, 1 mM EGTA) supplemented with 1mM GTP and 140mM KCl. Samples were incubated at 37°C for 30min, stabilized with Taxol (30uM). For GTPγS-MTs, 1mM GTPγS was used to polymerize MTs for 2hrs. After centrifugation at 35°C for 20min at 60,000rpm, supernatant was removed and MT pellets were incubated with EB3-GFP (0–1000nM) for 5min at room temperature (RT). For experiments where CLASP was added after polymerization, 150nM CLASP2 was incubated with MT pellet for 5min at RT prior to EB3-GFP addition for another 5min. MTs were pelleted again then supernatants and pellets were analyzed by Coomassie.
For in vitro MT plus-end tracking assays, preformed Hilyte647-labeled GMPCPP-MT seeds were attached to PLL-PEG-50%biotin-passivated coverslips via biotin-streptavidin linkers. Using TIRF, dynamic MTs and 600nM EB1-N-LZ-GFP were observed in the presence of 13uM tubulin in assay buffer: 50mM KCl, 1mM GTP, 0.6mg/ml κ-casein, 0.2% methyl cellulose, 4mM DTT, 0.2mg/ml catalase, 0.4mg/ml glucose oxidase, and 50mM glucose in MRB80 (80mM PIPES, pH 6.8 with KOH, 1mM EGTA, 4mM MgCl2), with 30nM CLASP2 or equal volume buffer control.
Statistics
All quantitative data were collected from experiments performed in at least duplicate and are expressed as mean ± S.E.M generated in Excel. Student’s t-test (two-tailed, unpaired) was performed to determine statistical difference between groups. A p-value of p<0.05 was considered statistically significant.
Supplementary Material
Highlights.
In CLASP-depleted cells, EB localizes along the MT lattice in addition to MT tips.
Proper EB localization requires the TOG2 region of CLASP, but not EB binding.
CLASP reduces GTP-tubulin lattice content possibly promoting GTP-hydrolysis at MTs.
In vitro, CLASP modulates MT-affinity and localization of EB without other factors.
Acknowledgements
We thank the Kaverina, Tyska, Ohi, and Lee labs for insightful discussions as well as Nadia Efimova and Changsong Yang for help with the platinum replica experiments. We are grateful to A. Akhmanova, L. Cassimeris, F. Perez, and R. Coffey for reagents. This study was supported by NIH grant GM078373 (I.K.), AHA grant-in-aid 13GRNT16980096 (I.K.), AHA pre-doctoral fellowship 12PRE12040153 (A.D.G.), and Scientific Research grant-in-aid 22570190 (I.H.). B.P.F. is supported by EPSRC-funded MOAC Doctoral Training Centre. A.S. is a Lister Institute Research Prize Fellow. This collaboration has benefited from a Strategic Partnership Grant from the University of Warwick (A.S.) and a Marie Curie Cancer Care programme grant (A.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, et al. Clasps are CLIP-115 and-170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nature reviews Molecular cell biology. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Kim H, Brouhard G, van Oijen A, Harrison SC, Chang F. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Developmental cell. 2010;19:245–258. doi: 10.1016/j.devcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Larsen NA, Hyman AA, Harrison SC. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 2007;15:355–362. doi: 10.1016/j.str.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, Surrey T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- Bouissou A, Verollet C, de Forges H, Haren L, Bellaiche Y, Perez F, Merdes A, Raynaud-Messina B. gamma-Tubulin Ring Complexes and EB1 play antagonistic roles in microtubule dynamics and spindle positioning. The EMBO journal. 2014;33:114–128. doi: 10.1002/embj.201385967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratman SV, Chang F. Stabilization of overlapping microtubules by fission yeast CLASP. Developmental cell. 2007;13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey RM, Mohan R, Leslie K, Walzthoeni T, Missimer JH, Menzel A, Bjelic S, Bargsten K, Grigoriev I, Smal I, et al. Insights into EB1 structure and the role of its C-terminal domain for discriminating microtubule tips from the lattice. Molecular biology of the cell. 2011;22:2912–2923. doi: 10.1091/mbc.E11-01-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey RM, Sen I, Kortt O, Mohan R, Gfeller D, Veprintsev D, Kretzschmar I, Scheuermann J, Neri D, Zoete V, et al. Sequence determinants of a microtubule tip localization signal (MtLS) The Journal of biological chemistry. 2012;287:28227–28242. doi: 10.1074/jbc.M112.373928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov A, Quesnoit M, Moutel S, Cantaloube I, Pous C, Perez F. Detection of GTP-tubulin conformation in vivo reveals a role for GTP remnants in microtubule rescues. Science. 2008;322:1353–1356. doi: 10.1126/science.1165401. [DOI] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Developmental cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Kita K, Wittmann T, Nathke IS, Waterman-Storer CM. Adenomatous polyposis coli on microtubule plus ends in cell extensions can promote microtubule net growth with or without EB1. Molecular biology of the cell. 2006;17:2331–2345. doi: 10.1091/mbc.E05-06-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, et al. Mammalian end binding proteins control persistent microtubule growth. The Journal of cell biology. 2009;184:691–706. doi: 10.1083/jcb.200807179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G, Grigoriev I, Mimori-Kiyosue Y, Ohtsuka T, Higa S, Kitajima I, Demmers J, Galjart N, Houtsmuller AB, Grosveld F, et al. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Developmental cell. 2006;11:21–32. doi: 10.1016/j.devcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Leano JB, Rogers SL, Slep KC. A cryptic TOG domain with a distinct architecture underlies CLASP-dependent bipolar spindle formation. Structure. 2013;21:939–950. doi: 10.1016/j.str.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin AJ, Semenova I, Zaliapin I, Kraikivski P, Nadezhdina E, Slepchenko BM, Akhmanova A, Rodionov V. CLIP-170-dependent capture of membrane organelles by microtubules initiates minus-end directed transport. Developmental cell. 2009;17:323–333. doi: 10.1016/j.devcel.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Bieling P, Cope J, Hoenger A, Surrey T. GTPgammaS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs) Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3988–3993. doi: 10.1073/pnas.1014758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B, Edelstein SJ. Evidence for a mixed lattice in microtubules reassembled in vitro. Journal of molecular biology. 1980;139:123–145. doi: 10.1016/0022-2836(80)90300-9. [DOI] [PubMed] [Google Scholar]
- Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nature cell biology. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. The Journal of cell biology. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K, Nogales E, Heald R. Multiple domains of human CLASP contribute to microtubule dynamics and organization in vitro and in Xenopus egg extracts. Cytoskeleton. 2012;69:155–165. doi: 10.1002/cm.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler SC, Pellman D. Microtubule"plus-end-tracking proteins": The end is just the beginning. Cell. 2001;105:421–424. doi: 10.1016/s0092-8674(01)00364-6. [DOI] [PubMed] [Google Scholar]
- Skube SB, Chaverri JM, Goodson HV. Effect of GFP tags on the localization of EB1 and EB1 fragments in vivo. Cytoskeleton. 2010;67:1–12. doi: 10.1002/cm.20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade RH, Chretien D. Cryoelectron microscopy of microtubules. Journal of structural biology. 1993;110:1–27. doi: 10.1006/jsbi.1993.1001. [DOI] [PubMed] [Google Scholar]
- Weisbrich A, Honnappa S, Jaussi R, Okhrimenko O, Frey D, Jelesarov I, Akhmanova A, Steinmetz MO. Structure-function relationship of CAP-Gly domains. Nature structural & molecular biology. 2007;14:959–967. doi: 10.1038/nsmb1291. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. The Journal of cell biology. 2005;169:929–939. doi: 10.1083/jcb.200412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M, Stear JH, Hyman AA, Howard J. EB1 recognizes the nucleotide state of tubulin in the microtubule lattice. PloS one. 2009;4:e7585. doi: 10.1371/journal.pone.0007585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M, Widlund PO, Hyman AA, Howard J. Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nature cell biology. 2013;15:688–693. doi: 10.1038/ncb2744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.