Abstract
The clinical relevance of DNA copy number alterations in chromosome 8 were investigated in oral cancers. The copy numbers of 30 selected genes in 33 OSCC patients were detected using the multiplex ligation-dependent probe amplification (MLPA) technique. Amplifications of the EIF3E gene were found in 27.3% of the patients, MYC in 18.2%, RECQL4 in 15.2% and MYBL1 in 12.1% of patients. The most frequent gene losses found were the GATA4 gene (24.2%), FGFR1 gene (24.2%), MSRA (21.2) and CSGALNACT1 (12.1%). The co-amplification of EIF3E and RECQL4 was found in 9% of patients and showed significant association with alcohol drinkers. There was a significant association between the amplification of EIF3E gene with non-betel quid chewers and the negative lymph node status. EIF3E amplifications did not show prognostic significance on survival. Our results suggest that EIF3E may have a role in the carcinogenesis of OSCC in non-betel quid chewers.
In 2008, there are about 263,900 new cases of oral cancer and 128,000 deaths reported worldwide1. Oral cancer incidence rates vary extensively across the world mainly due to the types of oral cancer-associated lifestyles practiced by different groups of people2. The lifestyle behaviours associated with oral cancers include tobacco smoking, alcohol drinking and betel quid chewing3.
The understanding of the genetic basis of oral squamous cell carcinoma (OSCC) carcinogenesis has progressed significantly in recent years. It is generally known that OSCC arises through a multistep process in which the accumulation of genetic alterations in proto-oncogenes and tumour-suppressor genes sets the foundation for carcinogenesis4. Moreover, a progression model for oral cancer has been proposed where mucosal fields with these genetic alterations can replace the normal epithelium in the oral cavity. Clonal evolution of the mucosal fields leads to invasive carcinoma5,6.
Chromosomal alterations in chromosome 8 have been frequently discovered in oral cancers7,8,9. Gains in chromosome 8q and losses in 8p are the common alterations found7,8,9,10 and have been shown to be involved in the lymph node metastasis of OSCCs7,9. Located in chromosome 8 is the well-known gene, c-MYC (8q24), which is frequently amplified in various cancers11,12,13. The common deleted regions found in oral cancers are 8p21, 8p22 and 8p2314,15. At 8p22, the FEZ1 (also known as LZTS1) gene has been shown to be frequently deleted in oral cancers16. Although many genetic aberrations have been found, the clinical relevance of many of these genes has not been characterized in OSCCs.
To get a better understanding of these genetic alterations in chromosome 8, we used a high resolution PCR-based method called the multiplex ligation-dependent probe amplification (MLPA) as it is able to measure chromosomal alterations of up to 40 target locations using only small amounts of DNA17. MLPA allows the concurrent analysis of a large set of oncogenes and tumour suppressor genes which are found in chromosomal regions that have shown copy number changes.
In this study, we aim to examine the genetic alterations of various genes in chromosome 8 using MLPA and find the associations between gene copy number changes in oral cancer with sociodemographic and clinicopathological parameters.
Results
Analysis of genetic alterations using MLPA
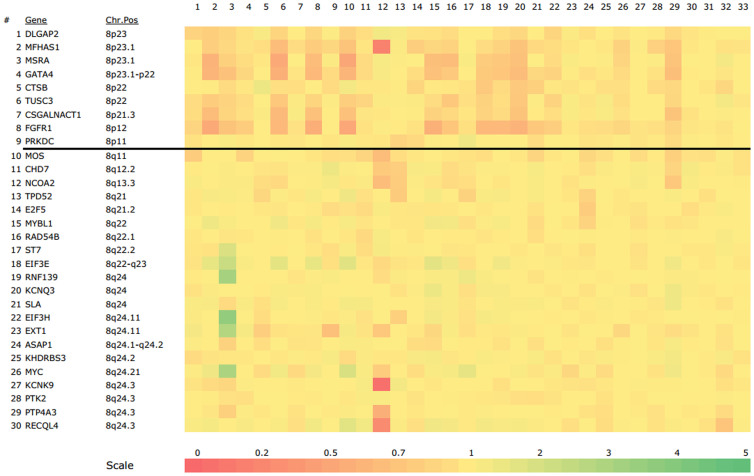
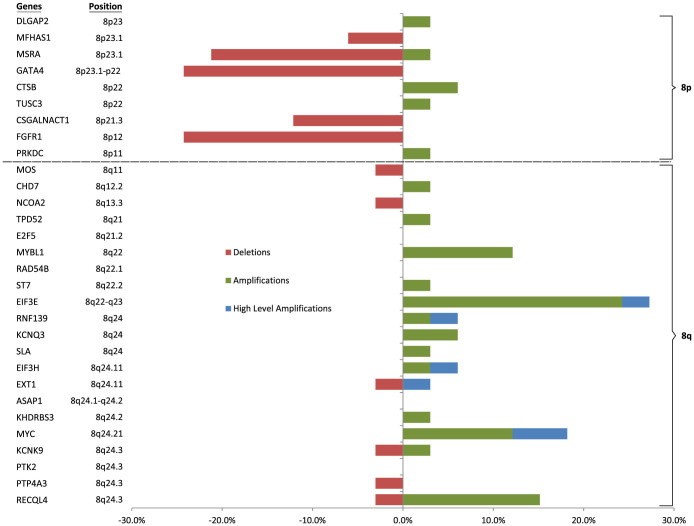
An overview of the copy number ratios of all 33 OSCC patients is presented in a heatmap (Fig. 1). The percentage of gene alterations for all 30 genes is shown in Figure 2. The most frequent losses were found in the 8p region while most of the frequent gains were found in the 8q. Eighteen patients (54.5%) showed at least a loss or a gain of a gene. Twelve patients out of these 18 patients (66.7%) had both gains and losses, five (27.8%) patients showed only gains and 1 patient (5.6%) had only gene losses. Fifteen patients (45.5%) did not show any gene alterations.
Figure 1. Heat map of copy number ratios of 30 genes on chromosome 8 for 33 OSCC patients.
Greener squares indicate higher copy number ratio while redder squares indicated lower copy number ratios. Yellow squares indicate normal copy number ratios. The dark line shows the border between the short arm and long arm of chromosome 8. Each column represents one OSCC patient.
Figure 2. Percentage of amplifications (green), deletions (red) and high-level amplifications (blue) for 30 genes in chromosome 8 of 33 OSCC patients.
The black dashed line shows the position of the centromere.
The genes with the most frequent gains were EIF3E in 27.3% of the patients, MYC (18.2%), RECQL4 (15.2%) and MYBL1 (12.1%). Two out 33 (6.1%) patients had high level amplifications (copy number > 2) of MYC. The rest of the genes with high level amplifications were ST7, EIF3E, EIF3H, EXT1 and RNF139 which were found in one of the patients with high level amplification of MYC.
The most frequent gene losses were GATA4 gene (24.2% of patients), FGFR1 gene (24.2%), MSRA (21.2) and CSGALNACT1 (12.1%).
The most frequent co-amplifications were EIF3E and MYC in 15% of patients, EIF3E and MYBL1 in 12% of patients, and EIF3E and RECQL4 in 9% of patients. All MYBL1 amplified (n = 4) OSCCs were co-amplified with EIF3E. Of 6 MYC amplifications, 83% (n = 5) were EIF3E co-amplified. Out of 5 RECQL4 amplifications, 3 were co-amplified with EIF3E.
Associations with sociodemographic parameters
There was a significant association (p = 0.047) between the gain of EIF3E gene and non-betel quid chewers. The co-amplification of EIF3E and RECQL4 showed significant association (p = 0.022) with alcohol drinkers. A detailed description of this co-amplification is shown in Table 1. No associations were found for sex, ethnicity and tobacco smoking habit.
Table 1. Sociodemographic and clinicopathological parameters of EIF3E-RECQL4 co-amplifications with OSCC cases.
| EIF3E-RECQL4 | ||||
|---|---|---|---|---|
| Parameters | N | Amplified (%) | Not Amplified (%) | P value (Fisher's exact test) |
| Sex | ||||
| Male | 13 | 3 (23.1) | 10 (76.9) | 0.052 |
| Female | 20 | 0 (0.0) | 20 (100.0) | |
| Ethnicity | ||||
| Indian | 16 | 0 (0.0) | 16 (100.0) | 0.227 |
| Others | 17 | 3 (17.6) | 14 (82.4) | |
| Alcohol Drinking | ||||
| Drinkers | 10 | 3 (30.0) | 7 (70.0) | 0.022 |
| Non-drinkers | 23 | 0 (0.0) | 23 (100.0) | |
| Smoking | ||||
| Smokers | 7 | 2 (28.6) | 5 (71.4) | 0.106 |
| Non-smokers | 26 | 1 (3.8) | 25 (96.2) | |
| Betel Quid Chewing | ||||
| Chewers | 18 | 0 (0.0) | 18 (100.0) | 0.083 |
| Non-chewers | 15 | 3 (20.0) | 12 (80.0) | |
| Habits | ||||
| Yes | 26 | 3 (11.5) | 23 (88.5) | 1.00 |
| No | 7 | 0 (0.0) | 7 (100.0) | |
| Tumour Sub-sites | ||||
| Tongue | 18 | 3 (16.7) | 15 (83.3) | 0.233 |
| Others (Buccal) | 15 | 0 (0.0) | 15 (100.0) | |
| Tumour Size | ||||
| T1 & T2 | 18 | 2 (11.1) | 16 (88.9) | 1.00 |
| T3 & T4 | 12 | 1 (8.3) | 11 (91.7) | |
| Lymph Node Status | ||||
| Positive | 11 | 0 (0.0) | 11 (100.0) | 0.535 |
| Negative | 20 | 3 (15.0) | 17 (85.0) | |
| TNM Staging | ||||
| Stage I/II | 12 | 2 (16.7) | 10 (83.3) | 0.540 |
| Stage III/IV | 20 | 1 (5.0) | 19 (95.0) | |
| Histological Grading | ||||
| Well differentiated | 14 | 1 (7.1) | 13 (92.9) | 1.00 |
| Moderate differentiated | 17 | 2 (11.8) | 15 (88.2) | |
Associations with clinicopathological parameters
The gain of EIF3E showed significant association with negative lymph node status (p = 0.01). MYC gains showed a trend towards association with negative lymph node status (p = 0.066). The detailed analysis of EIF3E and MYC are shown in Table 2 and 3 respectively. There were no associations found for primary tumour size, tumour site, tumour stage, and histological grading.
Table 2. Sociodemographic and clinicopathological parameters of EIF3E amplifications with OSCC cases.
| EIF3E | ||||
|---|---|---|---|---|
| Parameters | N | Amplified (%) | Not Amplified (%) | P value (Fisher's exact test) |
| Sex | ||||
| Male | 13 | 6 (46.2) | 7 (53.8) | 0.107 |
| Female | 20 | 3 (15.0) | 17 (85.0) | |
| Ethnicity | ||||
| Indian | 16 | 2 (12.5) | 14 (87.5) | 0.118 |
| Others | 17 | 7 (41.2) | 10 (58.8) | |
| Alcohol Drinking | ||||
| Drinkers | 10 | 4 (40.0) | 6 (60.0) | 0.40 |
| Non-drinkers | 23 | 5 (21.7) | 18 (78.3) | |
| Smoking | ||||
| Smokers | 7 | 4 (57.1) | 3 (42.9) | 0.068 |
| Non-smokers | 26 | 5 (19.2) | 21 (80.8) | |
| Betel Quid Chewing | ||||
| Chewers | 18 | 2 (11.1) | 16 (88.9) | 0.047 |
| Non-chewers | 15 | 7 (46.7) | 8 (53.3) | |
| Habits | ||||
| Yes | 26 | 6 (23.1) | 20 (76.9) | 0.358 |
| No | 7 | 3 (42.9) | 4 (57.1) | |
| Tumour Sub-sites | ||||
| Tongue | 18 | 6 (33.3) | 12 (66.7) | 0.458 |
| Others (Buccal) | 15 | 3 (20.0) | 12 (80.0) | |
| Tumour Size | ||||
| T1 & T2 | 18 | 5 (27.8) | 13 (72.2) | 1.00 |
| T3 & T4 | 12 | 4 (33.3) | 8 (66.7) | |
| Lymph Node Status | ||||
| Positive | 11 | 0 (0.0) | 11 (100.0) | 0.012 |
| Negative | 20 | 9 (45.0) | 11 (55.0) | |
| TNM Staging | ||||
| Stage I/II | 12 | 5 (41.7) | 7 (58.3) | 0.240 |
| Stage III/IV | 20 | 4 (20.0) | 16 (80.0) | |
| Histological Grading | ||||
| Well differentiated | 14 | 4 (28.6) | 10 (71.4) | 1.00 |
| Moderate differentiated | 17 | 5 (29.4) | 12 (70.6) | |
Table 3. Sociodemographic and clinicopathological parameters of MYC amplifications with OSCC cases.
| MYC | ||||
|---|---|---|---|---|
| Parameters | N | Amplified (%) | Not Amplified (%) | P value (Fisher's exact test) |
| Sex | ||||
| Male | 13 | 3 (23.1) | 10 (76.9) | 0.659 |
| Female | 20 | 3 (15.0) | 17 (85.0) | |
| Ethnicity | ||||
| Indian | 16 | 1 (6.3) | 15 (93.8) | 0.175 |
| Others | 17 | 5 (29.4) | 12 (70.6) | |
| Alcohol Drinking | ||||
| Drinkers | 10 | 2 (20.0) | 8 (80.0) | 1.00 |
| Non-drinkers | 23 | 4 (17.4) | 19 (82.6) | |
| Smoking | ||||
| Smokers | 7 | 3 (42.9) | 4 (57.1) | 0.093 |
| Non-smokers | 26 | 3 (11.5) | 23 (88.5) | |
| Betel Quid Chewing | ||||
| Chewers | 18 | 1 (5.6) | 17 (94.4) | 0.070 |
| Non-chewers | 15 | 5 (33.3) | 10 (66.7) | |
| Habits | ||||
| Yes | 26 | 4 (15.4) | 22 (84.6) | 0.584 |
| No | 7 | 2 (28.6) | 5 (71.4) | |
| Tumour Sub-sites | ||||
| Tongue | 18 | 4 (22.2) | 14 (77.8) | 0.665 |
| Others (Buccal) | 15 | 2 (13.3) | 13 (86.7) | |
| Tumour Size | ||||
| T1 & T2 | 18 | 3 (16.7) | 15 (83.3) | 0.660 |
| T3 & T4 | 12 | 3 (25.0) | 9 (75.0) | |
| Lymph Node Status | ||||
| Positive | 11 | 0 (0.0) | 11 (100.0) | 0.066 |
| Negative | 20 | 6 (30.0) | 14 (70.0) | |
| TNM Staging | ||||
| Stage I/II | 12 | 3 (25.0) | 9 (75.0) | 0.647 |
| Stage III/IV | 20 | 3 (15.0) | 17 (85.0) | |
| Histological Grading | ||||
| Well differentiated | 14 | 3 (21.4) | 11 (78.6) | 1.00 |
| Moderate differentiated | 17 | 3 (17.4) | 14 (82.4) | |
Follow-up data was available for 29 patients, with a median follow-up of 16.26 months (range, 0.69–63.36 months). In our data set, positive lymph node status (p = 0.014) and moderately-differentiated tumours (p = 0.028) were significantly associated with poor overall survival. Tumour stage III and IV showed a considerable trend (p = 0.062) toward unfavourable overall survival. Survival was not associated to primary tumour size and tumour site. None of the genes in our study were significantly associated with overall survival. KCNK9 amplifications showed a trend towards association with poor survival (p = 0.07).
Discussion
Around 50% of OSCC patients succumb to this disease within 5 years of diagnosis despite the advancements in treatments18. The interest therefore is to decrease the mortality rate of patients which can be done through early detection of OSCCs. Even before cells in a tumour lesion exhibit dysplasia histologically, genetic alterations of oncogenes and tumour suppressor genes may have already been found in the lesion. The alterations in these genes could be used as markers for early screening19,20.
Gains and losses in chromosome 8 have been reported in various types of cancers including breast cancers11, colorectal cancers21, prostate cancers22 and gastric cancers23. Many key oncogenes and tumour suppressor genes in the 8q and 8p arm have been found frequently altered in oral cancers. These genes may play a role in the carcinogenesis of oral cancer and can potentially be used in diagnosis and prognosis. In this study, we aim to detect the genes which are frequently altered in oral cancer and find the association between these genes and clinicopathological factors.
In our study, EIF3E (8q22–q23) was the most frequently amplified. EIF3E (also known as INT6 or EIF3S6) is one of the thirteen subunits of the eukaryotic translation initiation factor 3 complex (eIF3) and is involved in the initiation of protein synthesis. EIF3E was first discovered as a common integration site for mouse mammary tumour virus. Inserted viral genome causes the production of truncated chimeric mRNA24, where its overexpression has attributed to the malignant transformation of human mammary epithelial cells in both in vitro25 and in vivo26. However, the overexpression wild-type EIF3E did not have the same outcome. Both wild-type and truncated EIF3E can be expressed in the same cell which shows that truncated EIF3E acts as a dominant negative oncoprotein while the wild-type protein acts as a tumour suppressor27,28. Its involvement in tumourigenesis though is still unclear as some studies have proposed an oncogenic role29,30 for this gene while others have proposed a tumour suppressor role24,27. Our study showed that EIF3E was amplified in 27.3% of OSCCs which is consistent with its oncogenic role found in breast cancers29 and glioblastoma cells30. Moreover, the study on breast cancer cells found no substantial expression of truncated EIF3E which points towards a role for its wild-type form in promoting tumourigenesis29. EIF3E amplifications were also significantly associated with negative lymph node status. Essentially all 9 patients with amplification of this gene had a negative lymph node status. This implies that although EIF3E amplification might cause tumourigenesis, it seems to also prevent lymph node metastasis. In contrast, a study done on colorectal cancers showed that in stage D cancers, the amplification of EIF3E was significantly higher in liver metastasis than primary tumours. However, this study did not find a significant difference in the amplification of this gene between non-metasising and metasising tumours31. The amplification of EIF3E seems to affect tumourigenesis and metastasis of OSCC through a different mechanism when compared to colorectal cancers. To better understand the role of EIF3E in the carcinogenesis of OSCC, further studies have to be done on the expression of this gene and the involvement of the truncated EIF3E.
In addition, amplifications of EIF3E were significantly associated with OSCC patients who are non-betel quid chewers. Out of 15 non-betel quid chewers, 7 (46.7%) out had amplification of this gene compared to only 2 (11.1%) out of 18 chewers who had this amplification. This result implies that EIF3E amplification has a part to play in the carcinogenesis of OSCC in non-betel quid chewers.
MYC, located at 8q24.21, is a well-known oncogene that is involved in cell cycle progression, apoptosis and cellular transformation. Frequent amplifications of MYC in OSCCs were found in this study and are consistent with other results12,13. In a study on head and neck squamous cell carcinomas (HNSCC), amplifications of MYC has been associated with advanced tumours and late tumourigenesis32. MYC gene amplifications have also been associated with the progression of HNSCCs among Indians33. Our results showed that MYC amplifications could not predict survival which was similar to the results in other studies33. In our study, we found a trend associating MYC with negative lymph node status but we did not find any associations with other clinicopathological factors.
Frequent amplifications of RECQL4 were found in our study. REQL4, located at 8q24.3, belongs to the RecQ family of DNA helicases and functions to maintain genome integrity. Deficiencies of RecQ helicases have been linked to cancer predisposition and premature aging disorders34. On the other hand, the amplification and overexpression of RECQL4 has been reported in colorectal31, breast35, laryngeal36 and cervical cancers37 which is consistent with what we found in this study. In a study on breast cancers, RECQL4 has been found to be a potential metastasis promoting gene35. RECQL4 overexpression has been associated with late stage laryngeal squamous cell carcinomas36. In our study, we did not find associations between the amplifications and late stage OSCCs which indicate that the gene expression of RECQL4 has not been affected in our samples even though it showed significant amplifications.
MYBL1, a strong transcriptional activator, was found to be amplified in 4 out of 33 patients. This gene which is also known as A-Myb, is located at 8q22 and is mainly expressed in spermatocytes, neuronal cells and B-lymphoid cell. MYBL1 plays a role in the proliferation and differentiation of these cells. The role of this gene in cancer progression is less clear as it is not commonly found amplified in cancers. We also found a study which showed that A-Myb overexpression promotes cell proliferation in the benign tumours of the smooth muscle wall of the uterus (uterine leiomyoma)38.
The most frequently deleted gene in our study was the GATA-binding protein 4 (GATA4) which belongs to the GATA family of zinc-finger transcription factors. This gene is located at the chromosomal region 8p23.1–p22 which has been shown to be among the frequent targets for deletion in colorectal cancers in concordance with our results39. The loss of GATA4 mRNA expression however, is attributed mostly to promoter hypermethylation as shown in various cancers40,41,42. This shows that GATA4 is a potential tumour suppressor43. In contrast, GATA4 was also found to be amplified and overexpressed in oesophageal adenocarcinomas, which suggests that this gene may have other roles in tumourigenesis44. In addition to the epigenetic factors, the deletion of this gene may have an effect its gene expression as shown by the frequent deletions found among OSCCs in our study45.
The fibroblast growth factor receptor 1 (FGFR1) gene, located on 8p12, acts as the cell surface receptor for fibroblast growth factors and is involved in the regulation of embryonic development, cell proliferation, differentiation and migration. FGFR1 belongs to a large complex family of signalling molecules that are known to be involved in cancer progression46. In different cancers, FGFR1 amplifications have been frequently described. Amplification of FGFR1 has been shown in 10% of breast cancers47, about 5% of ovarian cancers48 and 3% rhabdomyosarcomas49. A number of studies have also reported deletions of FGFR1 in addition to amplifications in bladder cancers50, lung cancers51 and breast cancers11. Our study showed a relatively high percentage of FGFR1 deletion (24.2%) in OSCCs which is in contrast with the study done by Freier et al.12 Although these FGFR1 copy number changes may not influence its gene expression, this difference in results indicates that FGFR1 could have both oncogenic and tumour protective functions as shown in a related receptor, FGFR252.
High level amplifications were rare among the 30 genes on chromosome 8 that we analysed in OSCCs. The most frequent was MYC (6.1%) with 2 cases out of 33 showing high-level amplifications which result is similar to the percentage of high level MYC found in breast cancers (8.0%)11. High level amplifications of another gene, FGFR1 was also found in OSCCs12. In our study, high level amplification of FGFR1 was not only absent, we found that this gene was commonly deleted among our samples.
A gene in our study, EXT1 (8q24.11) has been found to be gained in betel quid associated OSCC13. However, there were no associations for this gene among betel quid chewers from our study.
The amplifications of two separate genes in a tumour may interact to affect tumour development. However, some genes may just be simply passengers which are co-amplified with other genes which drive tumour development. We identified three most frequent co-amplifications between EIF3E-MYC (15%), EIF3E-MYBL1 (12%) and EIF3E-RECQL4 (9%). The only association found was between the co-amplification of EIF3E-RECQL4 and alcohol drinkers. The co-amplification of EIF3E-RECQL4 was found in 3 OSCC patients who were all alcohol drinkers. This suggests that the carcinogenesis in alcohol drinkers may involve the co-amplification of these two genes.
Overall, the gains and losses we found at chromosome 8 in our OSCC samples were similar with those found in other cancers. For various cancers, gene losses were frequently found in the short arm of chromosome arm 8p while most gene amplifications were found in 8q53,54. The same pattern of gains and losses was also found in OSCCs7,8,10,55. Our results were consistent with these studies. There were some notable difference in the level of gains and losses of some genes when compared to other studies on OSCCs which could be due to the difference in ethnicity or aetiology2,56.
The heatmap (Figure 1) highlights a small part of the genetic heterogeneity of chromosome 8 in OSCCs. Out of the 30 genes we studied in chromosome 8, the number of alterations of each patient ranged from as high as 9 gene alterations to no alterations. It has long been known that cancer genomes are highly complex and unstable6. Heterogeneity within a single cancer type is known as inter-tumour heterogeneity. Recent studies have also shown genetic heterogeneity within a tumour itself57. It is known as intra-tumour heterogeneity and could occur due to both genetic and non-genetic factors58. As a result of this heterogeneity, small tissue sample obtained may not be representative of the whole tumour. Tumour heterogeneity may have a significant impact on the effectiveness of biomarkers used in diagnosis, prognosis and treatment of cancers.
Our data showed that both positive lymph node status and moderately-differentiated graded tumours were associated with poor overall survival. Lymph node status is shown to be an important prognostic factor for oral cancers59. Conversely, histological grading is known to have poor prognostic impact and poor reproducibility due to the heterogeneity of tumours59 which was in contrast to our results.
The KCNK9 gene amplifications showed a trend toward poor prognosis in our results. This gene has been shown to be amplified and overexpressed in some cancers including breast cancers60. However, out of the 33 patients in our study, only one patient had this amplification while another patient had a deletion. There were no prognostic significance on survival for the other genes in our study.
In conclusion, we were able to simultaneously detect gains and losses of genes in chromosome 8 in OSCCs using the MLPA technique. The genes found significantly amplified in were EIF3E, MYC, RECQL4 and MYBL1 while the genes significantly deleted were GATA4, FGFR1, MSRA and CSGALNACT1. The copy number gain of EIF3E was the most frequently found and was associated with non-betel quid chewers, indicating it has both an oncogenic and metastatic preventing role among non-betel quid chewers. Furthermore, EIF3E was also associated with the negative lymph node status, indicating its potential use as a biomarker for patients with lower risk of lymph node metastasis. However, this gene did not show prognostic value in predicting patient survival. The co-amplification of EIF3E-RECQL4 was found to be associated with alcohol drinking which suggest a role for this co-amplification in alcohol induced carcinogenesis. Our findings showed that genetic alterations could determine clinical characteristics. The alterations found in our study would assist in the development of markers for the early detection and prognosis of OSCC.
Methods
Tissue samples and DNA isolation
Fresh frozen tissues from 33 cases of oral squamous cell carcinoma (OSCC) were included in this study. Ten fresh frozen tissues of gingiva flaps taken from healthy normal non-cancer individuals during the minor impacted third permanent molar surgery were used as controls. All samples and related sociodemographic and clinical data were obtained from the Malaysian Oral Cancer Data and Tissue Bank System (MOCDTBS). The complete sociodemographic and clinicopathological details of all cases are shown in Table 4. Overall survival data was also collected. The tissues collected were snap frozen in liquid nitrogen (−100°C to −196°C) and were sectioned using a cryostat-microtome. A tissue section from each case was stained with haematoxylin and eosin (H&E) for histological assessment by oral pathologists. Only samples with more than 70% tumour cell content were further sectioned for DNA isolation. DNA was extracted from 750 μm of tissue sections using the DNEasy Blood & Tissue Kit (Qiagen GmBH, Hilden, Germany) according to the manufacturer's protocol. The concentration and quality of the DNA were measured using the Nanodrop ND-2000 spectrophotometer.
Table 4. Sociodemographic and clinicopathological parameters of all 33 OSCC cases.
| Case | Sex | Ethnicity | Tobacco smoking | Alcohol drinking | Betel Quid chewing | Tumour site | pT | pN | T stage | Histological grade |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | Indian | Non-smoker | Non-drinker | Chewer | others/buccal | T4 | No | IV | Well |
| 2 | Female | Indian | Non-smoker | Non-drinker | Chewer | gingiva | T4 | No | IV | Well |
| 3 | Female | Malay | Non-smoker | Non-drinker | Non-chewer | others/buccal | T4 | No | IV | Moderately |
| 4 | Male | Chinese | Smoker | Non-drinker | Non-chewer | tongue | T1 | No | I | Well |
| 5 | Female | Malay | Non-smoker | Non-drinker | Non-chewer | others/buccal | - | - | IV | - |
| 6 | Female | Chinese | Non-smoker | Non-drinker | Non-chewer | tongue | T3 | No | III | Moderately |
| 7 | Female | Indian | Non-smoker | Non-drinker | Chewer | tongue | T4 | N1 | IV | Moderately |
| 8 | Male | Chinese | Smoker | Drinker | Non-chewer | tongue | T1 | No | I | Moderately |
| 9 | Male | Chinese | Non-smoker | Drinker | Non-chewer | others/buccal | Tx | N2B | IV | Moderately |
| 10 | Male | Chinese | Smoker | Drinker | Non-chewer | tongue | T2 | No | II | Well |
| 11 | Female | Indian | Non-smoker | Non-drinker | Chewer | others/buccal | T2 | N2C | IV | Well |
| 12 | Female | Indian | Non-smoker | Non-drinker | Chewer | others/buccal | T4 | No | IV | Moderately |
| 13 | Female | Chinese | Smoker | Drinker | Non-chewer | others/buccal | T4 | N2C | IV | Moderately |
| 14 | Female | Indian | Non-smoker | Drinker | Chewer | others/buccal | - | - | - | - |
| 15 | Male | Chinese | Non-smoker | Drinker | Non-chewer | tongue | T4 | No | IV | Moderately |
| 16 | Male | Indian | Smoker | Drinker | Chewer | tongue | T1 | No | I | Moderately |
| 17 | Male | Chinese | Smoker | Drinker | Non-chewer | tongue | T1 | No | I | Moderately |
| 18 | Female | Malay | Non-smoker | Non-drinker | Chewer | tongue | T1 | No | I | Well |
| 19 | Male | Malay | Non-smoker | Non-drinker | Non-chewer | others/buccal | T2 | No | II | Well |
| 20 | Female | Indian | Non-smoker | Non-drinker | Chewer | tongue | T3 | N2B | IV | Well |
| 21 | Female | Indian | Non-smoker | Non-drinker | Chewer | tongue | T1 | N2B | IV | Moderately |
| 22 | Male | Malay | Non-smoker | Non-drinker | Non-chewer | tongue | T1 | N2B | IV | Moderately |
| 23 | Female | Indian | Non-smoker | Non-drinker | Chewer | others/buccal | T2 | No | II | Moderately |
| 24 | Female | Malay | Non-smoker | Non-drinker | Non-chewer | tongue | T1 | N1 | III | Well |
| 25 | Female | Indian | Non-smoker | Non-drinker | Chewer | tongue | T2 | Nx | II | Well |
| 26 | Female | Indian | Non-smoker | Non-drinker | Chewer | tongue | T1 | No | I | Moderately |
| 27 | Female | Indian | Non-smoker | Drinker | Chewer | others/buccal | T2 | No | II | Moderately |
| 28 | Female | Indian | Non-smoker | Drinker | Chewer | tongue | T2 | N1 | III | Well |
| 29 | Male | Malay | Smoker | Non-drinker | Non-chewer | tongue | T2 | No | II | Well |
| 30 | Female | Indian | Non-smoker | Non-drinker | Chewer | tongue | T1 | N1 | III | Moderately |
| 31 | Female | Indian | Non-smoker | Non-drinker | Chewer | others/buccal | T4 | N1 | IV | Moderately |
| 32 | Male | Malay | Non-smoker | Non-drinker | Non-chewer | others/buccal | T4 | No | IV | Well |
| 33 | Male | Malay | Non-smoker | Non-drinker | Chewer | others/buccal | T3 | No | III | Well |
Multiplex ligation-dependant probe amplification
The P014-A1 MLPA probe-set (MRC-Holland, Amsterdam, The Netherlands) which specifically detects genes in chromosome 8 was used. The kit contains 32 probes which covers 30 genes on chromosome 8 and includes an additional 9 control probes from other chromosomes. The full list of genes can be found as Supplementary Table S1 online. All MLPA experiments were performed based on the supplier's protocol17 and in duplicates. In each reaction, 50 ng of DNA in a volume of 5 μl was denatured at 98°C for 5 minutes. Then, the DNA was cooled to 25°C and a mixture of 1.5 μl MLPA buffer and 1.5 μl probe-mix was added. The mixture was then incubated at 95°C for 1 minute and then for 16 hours at 60°C for the hybridization of the probes. Next, 32 μl ligation mix was added to each reaction and then incubated at 54°C followed by 5 minutes at 98°C to deactivate the Ligase-65 enzyme. PCR was performed on 10 μl of the ligation product with FAM-labelled primers. The amplified fragments were analysed using a DNA sequencer.
Data analysis
The relative copy number ratio for each gene in every sample was calculated by dividing the normalized mean peak areas of each gene in a sample tissue with the normalized mean peak areas of the same gene in a tissue from non-cancer patients. Copy number ratios of below 0.7 were considered as loss while copy number ratios of 1.3 and above were considered as a gain. The data were interpreted using the Coffalyser software (MRC-Holland) and Microsoft Excel.
Statistical analysis
Statistical analysis was conducted using an Excel spreadsheet and IBM SPSS version 20. Significant amplifications and deletions were identified using Fisher's exact test. Copy number associations with sociodemographic and clinicopathological parameters were calculated using Chi-square or Fisher's exact test, whenever appropriate. Survival analysis was conducted using the Kaplan Meier method and log-rank test. P-values < 0.05 were considered to be significant. The data were grouped as follows: amplified vs non-amplified, male vs female, Indians vs others (Malays and Chinese), tobacco smokers vs non-smokers, alcohol drinkers vs non-drinkers, betel quid chewers vs non-chewers, tongue vs others (buccal), primary tumour size (T1/T2 vs T3/T4), lymph node (LN+ vs LN−), tumour stage (Stage I/II vs Stage III/IV), and histological grading (well vs moderate).
Author Contributions
Z.W.E.Y., Z.M.Z., L.P.K. and R.B.Z. designed this research. Z.W.E.Y. conducted the experiments and drafted the manuscript. Z.M.Z., T.G.K., Z.A.A.R., S.M.I., N.A.S., W.M.W.M., M.T.A., K.K.T. and R.B.Z. provided the clinical expertise and contributed in the acquisition of samples. R.B.Z. and T.G.K. provided the pathological expertise for sample selection. Z.W.E.Y., L.P.K. and R.B.Z. edited the final manuscript.
Supplementary Material
Supplementary Table S1
Acknowledgments
This research is supported by High Impact Research MoE Grant UM.C/625/1/HIR/MoE/DENT/06 from the Ministry of Education Malaysia. Postgraduate Research Fund (PV101-2011A) and University Malaya Research Grant (UMRG101/09HTM). We thank Prof. Dr. Yang Yi-Hsin for her help with the statistical analysis of associations between gene alterations and various parameters. We thank Oral Cancer Research and Coordinating Centre (OCRCC) for providing tissue and data.
References
- Jemal A. et al. Global cancer statistics. CA Cancer J. Clin. 61, 69–90, 10.3322/caac.20107 (2011). [DOI] [PubMed] [Google Scholar]
- Scully C. & Bedi R. Ethnicity and oral cancer. Lancet Oncol. 1, 37–42, 10.1016/S1470-2045(00)00008-5 (2000). [DOI] [PubMed] [Google Scholar]
- Petti S. Lifestyle risk factors for oral cancer. Oral Oncol. 45, 340–350, 10.1016/j.oraloncology.2008.05.018 (2009). [DOI] [PubMed] [Google Scholar]
- Califano J. et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 56, 2488–2492 (1996). [PubMed] [Google Scholar]
- Braakhuis B. J., Tabor M. P., Kummer J. A., Leemans C. R. & Brakenhoff R. H. A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res. 63, 1727–1730 (2003). [PubMed] [Google Scholar]
- Nowell P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). [DOI] [PubMed] [Google Scholar]
- Pathare S., Schaffer A. A., Beerenwinkel N. & Mahimkar M. Construction of oncogenetic tree models reveals multiple pathways of oral cancer progression. Int. J. Cancer 124, 2864–2871, 10.1002/ijc.24267 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. et al. Allelic imbalance analysis of oral tongue squamous cell carcinoma by high-density single nucleotide polymorphism arrays using whole-genome amplified DNA. Hum. Genet. 118, 504–507, 10.1007/s00439-005-0069-x (2005). [DOI] [PubMed] [Google Scholar]
- Yoshioka S. et al. Genomic profiling of oral squamous cell carcinoma by array-based comparative genomic hybridization. PLoS One 8, e56165, 10.1371/journal.pone.0056165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent-Chong V. K. et al. Genome-wide analysis of oral squamous cell carcinomas revealed over expression of ISG15, Nestin and WNT11. Oral Dis. 18, 469–476, 10.1111/j.1601-0825.2011.01894.x (2012). [DOI] [PubMed] [Google Scholar]
- Moelans C. B., de Weger R. A., Monsuur H. N., Vijzelaar R. & van Diest P. J. Molecular profiling of invasive breast cancer by multiplex ligation-dependent probe amplification-based copy number analysis of tumor suppressor and oncogenes. Mod. Pathol. 23, 1029–1039, 10.1038/modpathol.2010.84 (2010). [DOI] [PubMed] [Google Scholar]
- Freier K. et al. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC). Oral Oncol. 43, 60–66, 10.1016/j.oraloncology.2006.01.005 (2007). [DOI] [PubMed] [Google Scholar]
- Chen Y. J. et al. Genome-wide profiling of oral squamous cell carcinoma. J. Pathol. 204, 326–332, 10.1002/path.1640 (2004). [DOI] [PubMed] [Google Scholar]
- Wu C. L. et al. Deletion mapping defines three discrete areas of allelic imbalance on chromosome arm 8p in oral and oropharyngeal squamous cell carcinomas. Genes Chromosom. Cancer 20, 347–353 (1997). [DOI] [PubMed] [Google Scholar]
- El-Naggar A. K. et al. Localization of chromosome 8p regions involved in early tumorigenesis of oral and laryngeal squamous carcinoma. Oncogene 16, 2983–2987, 10.1038/sj.onc.1201808 (1998). [DOI] [PubMed] [Google Scholar]
- Ono K. et al. Down-regulation of FEZ1/LZTS1 gene with frequent loss of heterozygosity in oral squamous cell carcinomas. Int. J. Oncol. 23, 297–302 (2003). [PubMed] [Google Scholar]
- Schouten J. P. et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 30, e57 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans C. R., Braakhuis B. J. & Brakenhoff R. H. The molecular biology of head and neck cancer. Nat. Rev. Cancer 11, 9–22, 10.1038/nrc2982 (2011). [DOI] [PubMed] [Google Scholar]
- Bremmer J. F. et al. Screening for oral precancer with noninvasive genetic cytology. Cancer Prev. Res. (Phila.) 2, 128–133, 10.1158/1940-6207.CAPR-08-0128 (2009). [DOI] [PubMed] [Google Scholar]
- Sethi S., Benninger M. S., Lu M., Havard S. & Worsham M. J. Noninvasive molecular detection of head and neck squamous cell carcinoma: an exploratory analysis. Diagn. Mol. Pathol. 18, 81–87, 10.1097/PDM.0b013e3181804b82 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S., Williams R. D., Webb E. & Houlston R. S. Meta-analysis and pooled re-analysis of copy number changes in colorectal cancer detected by comparative genomic hybridization. Anticancer Res. 26, 3439–3444 (2006). [PubMed] [Google Scholar]
- El Gammal A. T. et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin. Cancer Res. 16, 56–64, 10.1158/1078-0432.CCR-09-1423 (2010). [DOI] [PubMed] [Google Scholar]
- Buffart T. E. et al. High resolution analysis of DNA copy-number aberrations of chromosomes 8, 13, and 20 in gastric cancers. Virchows Arch. 455, 213–223, 10.1007/s00428-009-0814-y (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A., Buttitta F., Pellegrini S., Bertacca G. & Callahan R. Reduced expression of INT-6/eIF3-p48 in human tumors. Int. J. Oncol. 18, 175–179 (2001). [DOI] [PubMed] [Google Scholar]
- Rasmussen S. B., Kordon E., Callahan R. & Smith G. H. Evidence for the transforming activity of a truncated Int6 gene, in vitro. Oncogene 20, 5291–5301, 10.1038/sj.onc.1204624 (2001). [DOI] [PubMed] [Google Scholar]
- Mayeur G. L. & Hershey J. W. Malignant transformation by the eukaryotic translation initiation factor 3 subunit p48 (eIF3e). FEBS Lett. 514, 49–54 (2002). [DOI] [PubMed] [Google Scholar]
- Buttitta F. et al. Int6 expression can predict survival in early-stage non-small cell lung cancer patients. Clin. Cancer Res. 11, 3198–3204, 10.1158/1078-0432.CCR-04-2308 (2005). [DOI] [PubMed] [Google Scholar]
- Dong Z. & Zhang J. T. Initiation factor eIF3 and regulation of mRNA translation, cell growth, and cancer. Crit. Rev. Oncol. Hematol. 59, 169–180, 10.1016/j.critrevonc.2006.03.005 (2006). [DOI] [PubMed] [Google Scholar]
- Grzmil M. et al. An oncogenic role of eIF3e/INT6 in human breast cancer. Oncogene 29, 4080–4089, 10.1038/onc.2010.152 (2010). [DOI] [PubMed] [Google Scholar]
- Sesen J. et al. Int6/eIF3e is essential for proliferation and survival of human glioblastoma cells. Int J Mol Sci 15, 2172–2190, 10.3390/ijms15022172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffart T. E. et al. DNA copy number changes at 8q11-24 in metastasized colorectal cancer. Cell. Oncol. 27, 57–65 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo J. P., Lazo P. S., Ramos S., Alvarez I. & Suarez C. MYC amplification in squamous cell carcinomas of the head and neck. Arch. Otolaryngol. Head Neck Surg. 122, 504–507 (1996). [DOI] [PubMed] [Google Scholar]
- Bhattacharya N., Roy A., Roy B., Roychoudhury S. & Panda C. K. MYC gene amplification reveals clinical association with head and neck squamous cell carcinoma in Indian patients. J. Oral Pathol. Med. 38, 759–763, 10.1111/j.1600-0714.2009.00781.x (2009). [DOI] [PubMed] [Google Scholar]
- Mohaghegh P. & Hickson I. D. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 10, 741–746 (2001). [DOI] [PubMed] [Google Scholar]
- Thomassen M., Tan Q. & Kruse T. A. Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res. Treat. 113, 239–249, 10.1007/s10549-008-9927-2 (2009). [DOI] [PubMed] [Google Scholar]
- Saglam O., Shah V. & Worsham M. J. Molecular differentiation of early and late stage laryngeal squamous cell carcinoma: an exploratory analysis. Diagn. Mol. Pathol. 16, 218–221, 10.1097/PDM.0b013e3180d0aab5 (2007). [DOI] [PubMed] [Google Scholar]
- Narayan G. et al. Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: identification of candidate amplified and overexpressed genes. Genes Chromosom. Cancer 46, 373–384, 10.1002/gcc.20418 (2007). [DOI] [PubMed] [Google Scholar]
- Swartz C. D., Afshari C. A., Yu L., Hall K. E. & Dixon D. Estrogen-induced changes in IGF-I, Myb family and MAP kinase pathway genes in human uterine leiomyoma and normal uterine smooth muscle cell lines. Mol. Hum. Reprod. 11, 441–450, 10.1093/molehr/gah174 (2005). [DOI] [PubMed] [Google Scholar]
- Fujiwara Y. et al. Evidence for the presence of two tumor suppressor genes on chromosome 8p for colorectal carcinoma. Cancer Res. 53, 1172–1174 (1993). [PubMed] [Google Scholar]
- Hellebrekers D. M. et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin. Cancer Res. 15, 3990–3997, 10.1158/1078-0432.CCR-09-0055 (2009). [DOI] [PubMed] [Google Scholar]
- Lassus H. et al. Comparison of serous and mucinous ovarian carcinomas: distinct pattern of allelic loss at distal 8p and expression of transcription factor GATA-4. Lab. Invest. 81, 517–526 (2001). [DOI] [PubMed] [Google Scholar]
- Bai Y. et al. Distinct expression of CDX2 and GATA4/5, development-related genes, in human gastric cancer cell lines. Mol. Carcinog. 28, 184–188 (2000). [DOI] [PubMed] [Google Scholar]
- Akiyama Y. et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol. Cell. Biol. 23, 8429–8439 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. et al. A minimal critical region of the 8p22–23 amplicon in esophageal adenocarcinomas defined using sequence tagged site-amplification mapping and quantitative polymerase chain reaction includes the GATA-4 gene. Cancer Res. 60, 1341–1347 (2000). [PubMed] [Google Scholar]
- Yang S. et al. Identification of genes with correlated patterns of variations in DNA copy number and gene expression level in gastric cancer. Genomics 89, 451–459, 10.1016/j.ygeno.2006.12.001 (2007). [DOI] [PubMed] [Google Scholar]
- Dailey L., Ambrosetti D., Mansukhani A. & Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 16, 233–247, 10.1016/j.cytogfr.2005.01.007 (2005). [DOI] [PubMed] [Google Scholar]
- Courjal F. et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 57, 4360–4367 (1997). [PubMed] [Google Scholar]
- Gorringe K. L. et al. High-resolution single nucleotide polymorphism array analysis of epithelial ovarian cancer reveals numerous microdeletions and amplifications. Clin. Cancer Res. 13, 4731–4739, 10.1158/1078-0432.CCR-07-0502 (2007). [DOI] [PubMed] [Google Scholar]
- Missiaglia E. et al. Genomic imbalances in rhabdomyosarcoma cell lines affect expression of genes frequently altered in primary tumors: an approach to identify candidate genes involved in tumor development. Genes Chromosom. Cancer 48, 455–467, 10.1002/gcc.20655 (2009). [DOI] [PubMed] [Google Scholar]
- Simon R. et al. High-throughput tissue microarray analysis of 3p25 (RAF1) and 8p12 (FGFR1) copy number alterations in urinary bladder cancer. Cancer Res. 61, 4514–4519 (2001). [PubMed] [Google Scholar]
- Kohler L. H. et al. FGFR1 expression and gene copy numbers in human lung cancer. Virchows Arch. 461, 49–57, 10.1007/s00428-012-1250-y (2012). [DOI] [PubMed] [Google Scholar]
- Turner N. & Grose R. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer 10, 116–129, 10.1038/nrc2780 (2010). [DOI] [PubMed] [Google Scholar]
- Meijer G. A. et al. Progression from colorectal adenoma to carcinoma is associated with non-random chromosomal gains as detected by comparative genomic hybridisation. J. Clin. Pathol. 51, 901–909 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbieva Z. H. et al. High-resolution physical map and transcript identification of a prostate cancer deletion interval on 8p22. Genome Res. 10, 244–257 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparano A. et al. Genome-wide profiling of oral squamous cell carcinoma by array-based comparative genomic hybridization. Laryngoscope 116, 735–741, 10.1097/01.mlg.0000205141.54471.7f (2006). [DOI] [PubMed] [Google Scholar]
- Paterson I. C., Eveson J. W. & Prime S. S. Molecular changes in oral cancer may reflect aetiology and ethnic origin. Eur. J. Cancer. B Oral Oncol. 32B, 150–153 (1996). [DOI] [PubMed] [Google Scholar]
- Navin N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94, 10.1038/nature09807 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A., Almendro V. & Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323–334, 10.1038/nrc3261 (2012). [DOI] [PubMed] [Google Scholar]
- Walker D. M., Boey G. & McDonald L. A. The pathology of oral cancer. Pathology 35, 376–383, 9NLGYYNG3DJ0DN0R [pii] (2003). [DOI] [PubMed] [Google Scholar]
- Mu D. et al. Genomic amplification and oncogenic properties of the KCNK9 potassium channel gene. Cancer Cell 3, 297–302 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1