Abstract
Increased expression of cathepsins has diagnostic as well as prognostic value in several types of cancer. Here, we demonstrate a novel magnetic resonance imaging (MRI) method, which uses poly-L-glutamate (PLG) as an MRI probe to map cathepsin expression in vivo, in a rat brain tumor model. This noninvasive, high-resolution and non-radioactive method exploits the differences in the CEST signals of PLG in the native form and cathepsin mediated cleaved form. The method was validated in phantoms with known physiological concentrations, in tumor cells and in an animal model of brain tumor along with immunohistochemical analysis. Potential applications in tumor diagnosis and evaluation of therapeutic response are outlined.
Many tumors have been shown to exhibit elevated levels of proteases in incipient stages, which are thought to play a crucial role in tumor angiogenesis, invasion and metastasis1. Knowledge of the roles of proteases in tumor progression will potentially guide the development of novel diagnostic tools for early detection of tumors as well as disease modifying therapeutic strategies to combat tumors. Cathepsins, cysteine family proteases, are lysosomal proteases involved in cellular protein degradation and are over-expressed in many tumor cells as well as host cells associated with the tumor2,3. Out of eleven types of cathepsins, cathepsin B (CtB) and cathespin L (CtL) are predominantly involved in protein degradation within the lysosome4.These cathepsins have been shown to be prognostic indicators of many types of tumors5,6,7,8. The wide availability of cathepsins on tumor cells makes them attractive targets for tumor detection as well as for studying tumor evolution under therapy.
However, despite the tremendous significance, it is challenging to accomplish noninvasive detection of protease activity in vivo. Except for near infrared fluorescence (NIRF) optical imaging, there has been no known high resolution, noninvasive and nonradioactive methods to detect CtB activity in tumors in vivo9,10,11,12,13. In spite of its high sensitivity, NIRF suffers from poor depth penetration (few mm) and cannot be translated into a three dimensional (3D) imaging tool either in large animal models or in humans. Furthermore, it requires synthesis and evaluation of toxicity of the injected compounds. Therefore, there is an unmet need for a noninvasive, high-resolution imaging technique for measuring the expression of cathepsins in tumors.
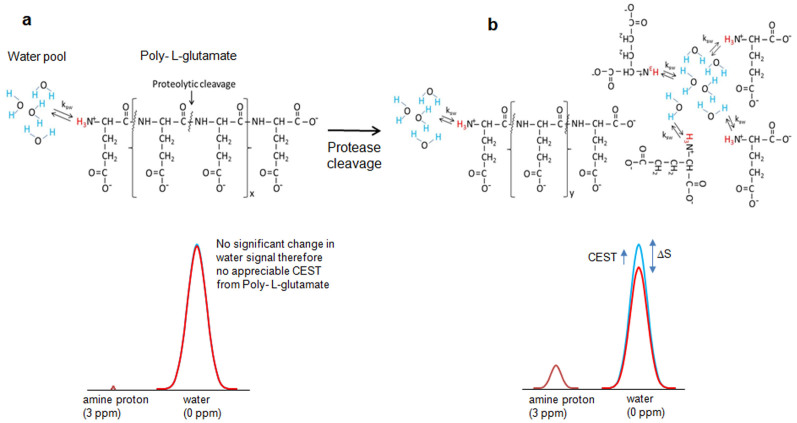
Here, for the first time, we demonstrate poly-L-glutamate (PLG) as a tumor protease activated magnetic resonance imaging (MRI) probe, which enables high resolution mapping of expression of cathepsins in tumors, in vivo. PLG is a nontoxic polymer that can be readily injected into animal models up to several millimoles (mM) without causing any adverse reactions13,14,15,16,17. This noninvasive, nonradioactive method is based on the chemical exchange saturation transfer (CEST) between protons attached to nitrogen in amine moieties with bulk water protons. Owing to very slow exchange rates of backbone amide protons (Fig. 1a), PLG in its native form does not exhibit appreciable CEST under physiological conditions. However, cleavage by the lysosomal enzymes such as cathepsins, results in individual glutamate (Glu) moieties as well as smaller Glu peptides, which readily exhibit CEST effect from their amine protons (Fig. 1b)18. Determination of the relationship between the protease expression and PLG degradation via CEST can establish a potential noninvasive MRI method for quantifying tumor protease expression.
Figure 1. Poly L-glutamate (PLG) as a protease activated CEST contrast agent.
(a). Molecular structure of PLG shows three amine protons in exchange with the bulk water. Multiple amide protons are also present in the backbone of PLG. Due to the presence of significantly lower number of exchangeable amine protons, the saturation transfer of these protons does not reduce bulk water signal noticeably and therefore no appreciable CEST contrast is observed from PLG. No CEST contrast is observed from amide protons due to its slow exchange rate and saturation power and duration used in the current study. (b). Proteolytic cleavage of PLG into monomers (i.e. glutamate) and/or smaller peptide fragments exposes fair number of exchangeable amine protons. The exchange of saturated magnetization of these amine protons with bulk water significantly reduces the bulk water signal and results in appreciable CEST contrast.
Results and Discussion
The CEST effect from Glu and PLG with and without addition of CtB is evaluated in in vitro at frequency offset, Δω = 3 ppm downfield to bulk water (Fig. 2a). Glu showed a strong CEST contrast due to its exchangeable amine protons18 while no CEST contrast was observed from PLG due to very few amine protons. But the cleavage products of PLG showed an appreciable CEST contrast due to the exposure of amine protons interacting with bulk water after enzymatic CtB cleavage (Fig. 2a).
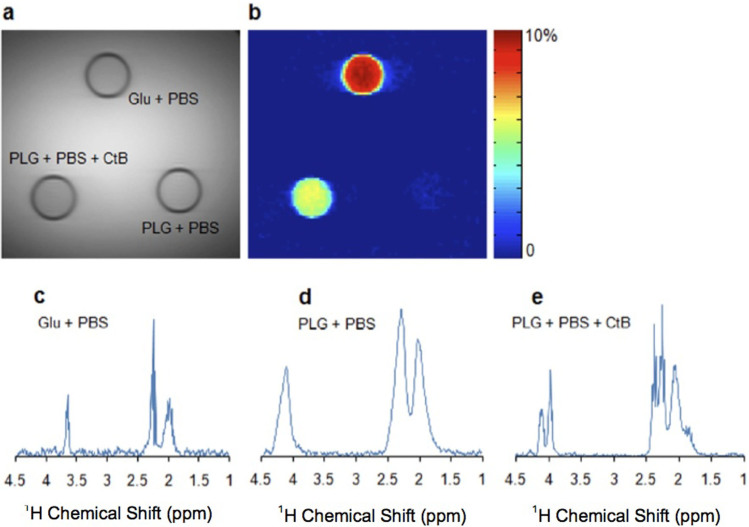
Figure 2. PLG as a marker of cathepsin B (CtB) enzyme activity.
(a). Anatomical proton density weighted MR image showing three different solutions (Glu, PLG, and PLG+Ctb) in NMR tubes immersed in a PBS phantom. (b). CEST contrast map at 3 ppm downfield to the bulk water from glutamate (Glu), PLG and PLG with CtB. Glu shows a strong CEST contrast. No CEST contrast was observed from PLG alone. After addition of CtB an appreciable CEST contrast was observed, this is due to the cleavage of PLG in to smaller fragments by CtB. This cleavage exposes significant number of labile –NH3+ protons, which can be easily detected using CEST MRI. (c-e). 1H MR spectra from phantoms containing Glu, PLG, and PLG + CtB. Spectrum from Glu shows multiple peaks related to aliphatic protons at ~2.02, ~2.25and ~3.66 ppm. Spectrum from PLG shows 3 peaks respectively at ~2.02, ~2.28 and ~4.1 ppm. The spectra are broad due to reduced mobility of PLG molecule and hence the fine structure is masked. Phantom with PLG and CtB is showing splitting of 2.28 and 4.1 ppm peaks, due to cleavage of PLG by CtB.
PLG cleavage by CtB can be monitored in vitro through proton magnetic resonance spectroscopy (1HMRS; Figs. 2 b–d). The 1HMRS spectrum from Glu showed three distinct peaks respectively at ~2.02 ppm, ~2.25 ppm and ~3.66 ppm (Fig. 2b). PLG also showed three peaks at ~2.01 ppm, ~2.28 ppm and ~4.1 ppm (Fig. 2c). Addition of CtB splits the 2.28 ppm and 4.1 ppm peaks (Fig. 2d). The splitting of PLG peaks denotes its cleavage by CtB.
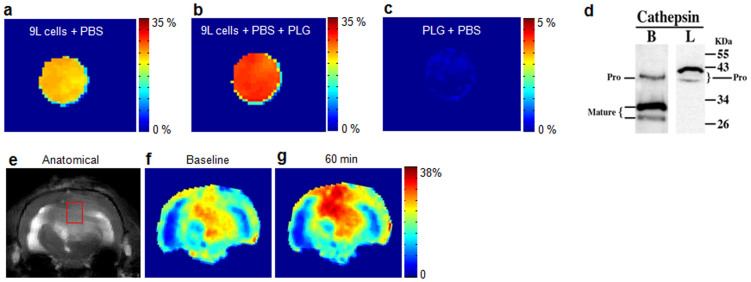
Further, we assessed the expression of CtB in 9L tumor cells. CEST imaging of 9L cells was performed with and without addition of PLG. With addition of PLG, the 9L cell line showed ~ 20% increase in CEST contrast compared to the cells without PLG, while there was no appreciable CEST contrast observed from PLG alone (Figs. 3a–c). The increased CEST contrast can be attributed to the cleavage of PLG by the cathepsins expressed in tumor cells. Since the two cathepsins i.e. CtB and CtL are primarily responsible for cleavage of PLG, Western blot analysis of 9L cell lysates was performed to determine the presence of CtB and CtL expression (Fig. 3d). Western blot analysis showed expression of CtB both in the mature and pro form. However, mature CtB was predominantly expressed than its pro form. The CtL was expressed only in the pro form. This suggests that cleavage of PLG in 9L cells is primarily due to the expression of mature CtB.
Figure 3. PLG as a marker of protease enzyme expression in cancer 9L cells.
(a). Cancer cells cultured without any PLG. This image shows ~22% CEST contrast, which is due to endogenous Glu in cultured cells. (b). Same cell line (as in a) cultured in the presence of PLG (20 mg/ml) for 1 hour shows an further elevation of ~17% of the CEST contrast over the baseline indicating that the CtB present in the tumor cells cleaved the PLG. (c). PLG alone in saline does not show any appreciable CEST effect owing to the slow exchange of amide protons present on the PLG. (d). Western blot analysis of tumor cell lysates shows both mature and pro form of CtB while cathepsin L (CtL) was expressed only in the pro form. The cleavage of PLG in this tumor cell line is predominantly due to CtB. (e), (f). Baseline anatomical CEST weighted image and CEST map from a rat brain with a 9L tumor. (g). At 60 minutes post intra-venous injection of PLG (160 mg/kg), a ~32% increase in GluCEST contrast was observed in the tumor region.
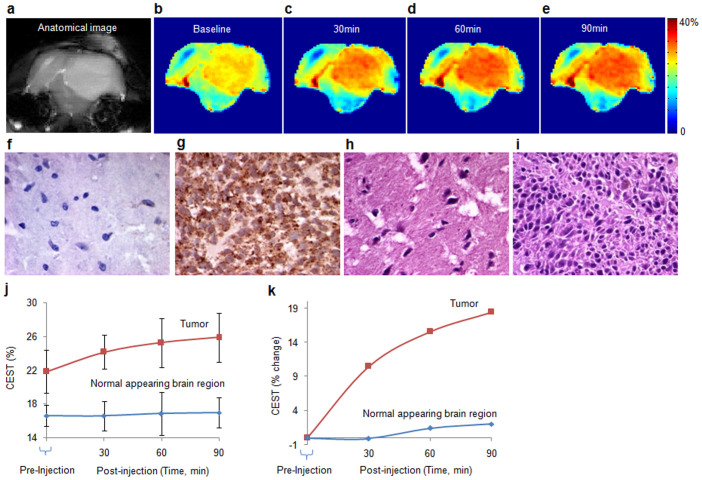
The feasibility of mapping CtB expression in in vivo was evaluated on a 9L rat brain gliosarcoma model (Figs. 3 e–g, Figs. 4 a–e). PLG (160 mg/kg) was injected in these rats through the tail vein and CEST imaging was performed up to 90 minutes with a 30 minutes time interval. A continuous increase in CEST contrast was observed in the tumor region due to the cleavage of PLG by protease enzyme (primarily CtB) expressed in tumor. We further performed immunohistochemistry of rat brain tumor tissues for CtB expression. A strong positive expression of CtB was observed in the tumor region while the normal appearing brain (NAB) showed negligible expression of CtB. The same tumor region showed higher nuclear density and mitosis reflecting increased cellular proliferation than NAB region on hematoxylin and eosin stain (Figs. 4f–i).The graph (Fig. 4j) shows the mean (±SD) CEST contrast from tumor (n = 6) as well as NAB region (n = 6). A progressive increase in CEST contrast from tumor region was observed over the period of 90 minutes, while the CEST contrast from NAB did not change appreciably over the time course of 90 minutes. An average of 19% increase in CEST contrast was observed from tumor region after 90 minutes injection of PLG (Fig. 4k).
Figure 4. In vivo imaging of cathepsin expression by monitoring PLG cleavage mediated CEST.
(a). Anatomical CEST weighted image showing the tumor as a hyper-intense region. (b). the base line CEST contrast map from the same slice. (c–e). CEST contrast maps of the same imaging slice respectively at 30, 60 and 90 minutes following PLG administration through tail vein. Following PLG injection a continuous increase in CEST contrast up to 90 minutes was observed mainly in tumor region due to cleavage of PLG into its monomer and/or smaller peptide fragments by cathepsins expressed in tumor. (f). (g). Immunohistochemistry of CtB from normal appearing brain (NAB) region and tumor region showing the expression of CtB (40x). NAB region shows very few cells positive for CtB expression while the tumor cells showed strong positivity for CtB. (h). (i). Hematoxylin and eosin staining from NAB and tumor regions show higher cell proliferation in tumor. (j). Graph showing PLG kinetics in the tumor (n = 6) and NAB (n = 6) for a period of 90 minutes following the injection of PLG. (k). The graph shows the mean percentage change in the CEST contrast following PLG injection. While no appreciable change in NAB is observed, tumor shows 19% increase in CEST contrast in about 90 minutes. One way analysis of variance showed a significant change (p = 0.048) in CEST contrast over the time period of 90 minutes.
Since PLG mediated CEST contrast directly depends on the expression of cathepsin proteases, monitoring changes in the CEST contrast in vivo will enable the quantitation of cathepsin expression in different tumor types.
Conjugation of polymers to anticancer drugs is an emerging approach for clinical development of combination therapy19. Such studies have the potential for advancing polymer therapeutics as a novel class of anticancer agents. These techniques primarily exploit the degradation of polymers by the lysosomal enzymes such as cysteine proteases, particularly CtB and CtL16,19,20,21,22. In particular, PLG has been used as a macromolecular carrier for drug delivery16 and PLG in combination with paclitaxel (PG-TXL) has advanced to clinical trials23,24. Since the development and application of targeted delivery of anticancer agents using PLG as the drug carrier is emerging23,24, we envisage that, in the long term, the proposed CEST imaging technology has the potential to monitor PLG conjugated drug delivery and therapeutic response of tumors.
This is the first study to show that PLG can be used as a protease activated MRI probe for tumor mapping in vivo and PLG degradation by cathepsins in tumors can be imaged with high resolution MRI. We expect that the current method will have numerous applications in cancer research including differentiation of radiation necrosis from tumor growth as well evaluating anti-autophagy therapies.
Methods
All animal experiments were performed according to an approved University of Pennsylvania Institutional Animal Care and Use Committee Protocol.
In vitro Study
Phantom Experiment
Solutions of 20 mg/ml PLG were prepared with and without addition of 0.5 mg/ml CtB enzyme (Sigma Aldrich, USA) in 10 mm NMR tubes. In addition, 10 mM sodium glutamate solution was also prepared. These phantoms were placed in a larger bottle containing phosphate buffered saline (PBS) and the CEST imaging of these phantoms was performed at room temperature on a 7T whole body MRI scanner (Siemens Medical Systems, Malvern, PA, USA).
For CEST imaging a frequency selective saturation pulse followed by a segmented radiofrequency spoiled gradient echo (GRE) readout sequence was used. The sequence parameters were: slice thickness = 10 mm, flip angle = 10°, TR = 5.6 ms, TE = 2.7 ms, field of view = 100 × 100 mm2, matrix size = 192 × 192, and one saturation pulse and 64 segments acquired every 10 s. CEST images were collected at peak B1rms of 3.64 µT and 1 s saturation duration for the frequencies (+2.4 to +3.6 ppm and −2.4 to −3.6 ppm in 0.2 ppm steps) from bulk water resonance. Radiofrequency field inhomogeneity (B1) and static magnetic field inhomogeneity (B0) maps were also obtained to correct for any inhomogeneities as described previously18.
Single voxel spectra (SVS) with point-resolved-spectroscopy (PRESS) acquisition mode were performed from each phantom using the following parameters: voxel size = 5 mm × 5 mm × 20 mm (Voxel volume 500 μL), spectral width = 4 kHz, number of points = 2048, averages = 64, TE = 20 ms, and TR = 3 s. Water suppression was achieved using the variable pulse power and optimized relaxation delays method (VAPOR).
Cell experiments
Rat 9L gliosarcoma cells were cultured in DMEM containing 10% Fetal Bovine Serum, containing Penicillin-streptomycin. 9L cells growing in log phase were trypsinized and used for in vitro and in vivo studies.
Ten million 9L cells were trypsinized and washed with sterile PBS and transferred to a 0.5 ml microfuge tube. Six tubes with equal volume of 9L cells were obtained for MR imaging. These tubes were categorized in two groups. In the first group (n = 3), 0.1 ml of normal saline was added, while in the 2nd group (n = 3) 0.1 ml of PLG solution was added making a total concentration of 20 mg/ml. CEST imaging was performed on a 9.4 T horizontal bore small animal MR scanner (Varian, Palo Alto, CA). CEST imaging was performed using a custom-programmed segmented radiofrequency GRE readout pulse sequence with a frequency selective continuous wave saturation preparation pulse. The sequence parameters were: field of view = 30 × 30 mm2, slice thickness = 2 mm, flip angle = 15 degree, TR = 6.2 ms, TE = 2.9 ms, matrix size = 128 × 128. For every 8 s one saturation pulse and 128 acquisition segments were applied. CEST images were collected using a 1 second saturation pulse at B1rms of 5.87 µT for multiple frequencies (+2.4 to +3.6 ppm and −2.4 to −3.6 ppm in 0.2 ppm steps) from bulk water. B1 and B0 field maps were also acquired and used to correct the CEST maps as described previously18.
Western blot analysis
9L cells were lysed in radioimmunoprecipitation assay buffer containing complete mini proteinase inhibitor cocktail (Roche Diagnostics, city, state, country) and protein lysate was quantitated using Pierce BCA protein assay kit (Thermo scientific). 9L cell lysate (40 μg) was boiled in sample buffer and loaded onto single lane of a 10% SDS-PAGE gel under reducing conditions. The separated proteins were blotted onto a PVDF membrane, which was blocked with 5% non-fat dry milk for 1 hour and then probed with rabbit anti-rat CtB polyclonal IgG (4 μg/ml; cross reacts with human and mouse; Abcam) or rabbit anti-human Ct L monoclonal IgG (1 μg/ml; Abcam) for 1 hour, followed by detection with HRP-conjugated goat anti-rabbit IgG (1:4000 dilution; Bio-Rad). The blot was stained with Amersham ECL western blotting substrate and imaged using Fujifilm LAS3000 imager.
In vivo Study
Rat brain tumor
To develop intracranial tumors, rat gliosarcoma cells (9L) were used. Syngeneic female Fisher rats (F344/NCR, four-six weeks old) weighing 130–150 g were used to generate tumor-bearing rats as described previously25. Briefly, general anesthesia was induced by isoflurane followed by intraperitonial injection of a ketamine (91 mg kg−1) and acepromazine (9.1 mg kg−1) mixture. A 10 μl suspension of 50,000 9L cells in phosphate buffered saline was injected into the cortex at a depth of 2 mm with a Hamilton syringe and a 30-gauge needle using a stereotactic apparatus (3 mm lateral and 3 mm posterior to the bregma). These rats were subjected to MR imaging four weeks after implantation of tumor cells.
Rat MR imaging
MRI imaging was performed on tumor bearing rats three weeks after 9L cell implantation. Rats were transferred to a 9.4T horizontal bore small animal MR scanner (Varian, Palo Alto, CA) and placed in a 20-mm diameter commercial quadrature proton coil (m2m Imaging Corp., Cleveland, OH). Animals were kept under anesthesia (1.5% isoflurane in 1 liters/min oxygen) and their body temperature maintained with the air generated and blowing through a heater (SA Instruments, Inc., Stony Brook, NY). Respiration and body temperature were continuously monitored using a MRI compatible small animal monitoring system (SA Instruments, Inc., Stony Brook, NY). The Institutional Animal Care and Use Committees of the University of Pennsylvania approved experimental protocols.
CEST imaging of rat brain tumor was performed using the pulse sequence and parameters as described in case of 9L cell line imaging except field of view 35 × 35 mm2. After baseline imaging the rats were injected with 1 ml PLG (160 mg/Kg) solution at a rate of 0.1 ml per minute through the catheter inserted in a tail vein. After PLG administration, CEST imaging was performed at each 30 minute time interval for a total period of 90 minutes.
At the end of the study the rats were euthanized by CO2 asphyxiation and brain tissue was collected for immunohistochemical studies.
Immunohistochemistry
All the excised rat brains were fixed in 10% buffered formalin and embedded in paraffin. The whole brain specimen was blocked and 6 µm sections were taken for immunohistochemistry analysis. The slides were deparaffinized in Xylene (Surgipath, 03672) followed by rehydration with 100%, 95%, 70% ethanol. Antigen retrieval was performed in the digital decloaking chamber (Biocare medical, DC2002) by using citric acid based antigen unmasking solution (Vector laboratories, H-3300). The sections were blocked with 4% BSA (Sigma, A-7906) and 2% goat or horse serum in TT buffer (10 mM of Tric-HCI, 500 mM NaCI, and 0.05% Tween-20) at room temperature for 30 minutes. Sections were then incubated with primary antibody diluted in blocking buffer at 4°C overnight. Following washing in Tris buffer, secondary antibody (Vector laboratories, BA-1000 or BA-9200) in blocking buffer was added and incubated at room temperature for 45 minutes. After three washes, the sections were incubated with Vectastain ABC reagent (Vector laboratories, PK-6100) for 40 minutes at room temperature. The sections were again washed thrice in Tris buffer and incubated with the chromogendiaminobenzidine-tetrahydrochloride (DAB) ((Sigma, D4293) substrate followed by counterstaining in hematoxylin. The sections were mounted in DPX mountant. The immunostained slides were digitized with a 40× objective using Olympus camera.
Image Processing
All image processing and data analysis were performed using software routines written in MATLAB (version 7.5, R2007b). Acquired CEST images were corrected for B0 and used to generate CEST contrast map using Equation [1].
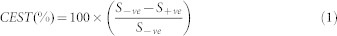 |
where S-ve and S+ve are the B0 corrected MR signals at −3 ppm and +3 ppm respectively. CEST contrast was further corrected for B1 and mapped as false-colors onto anatomical proton image. To compute the CEST contrast, region of interests were manually drawn on tumor and NAB regions.
Author Contributions
M.H. designed and performed experiments, analyzed data and wrote the manuscript; A.S. helped with experiments, image processing, data analysis and contributed to manuscript editing. M.I. helped with cell line Western blotting; R.I. helped with rat brain tumor model development; K.N., R.P.R.N., C.D. and F.K. helped with phantom imaging, animal handling and manuscript editing; K.C. provided technical support with cell lines and animal imaging; H.P. advised on tumor model development and contributed to manuscript editing; D.R. provided technical support with handling the cells line and tumor model development; H.H. provided technical guidance and contributed to the manuscript editing; R.R. provided the concept, experimental design and contributed to manuscript writing and editing. All authors reviewed the manuscript.
Acknowledgments
This project was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health through Grant Numbers: P41-EB015893, P41-EB015893S1. The authors acknowledge W. Liu and S. Pickup for their technical assistance in using the 9.4T NMR spectrometer.
References
- Koblinski J. E., Ahram M. & Sloane B. F. Unraveling the role of proteases in cancer. Clin Chim Acta 291, 113–135 (2000). [DOI] [PubMed] [Google Scholar]
- Jedeszko C. & Sloane B. F. Cysteine cathepsins in human cancer. Biol Chem 385, 1017–1027 (2004). [DOI] [PubMed] [Google Scholar]
- Mohamed M. M. & Sloane B. F. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer 6, 764–775 (2006). [DOI] [PubMed] [Google Scholar]
- Keppler D. & Sloane B. F. Cathepsin B: multiple enzyme forms from a single gene and their relation to cancer. Enzyme Protein 49, 94–105 (1996). [DOI] [PubMed] [Google Scholar]
- Foekens J. A. et al. Prognostic significance of cathepsins B and L in primary human breast cancer. J Clin Oncol 16, 1013–1021 (1998). [DOI] [PubMed] [Google Scholar]
- Nouh M. A. et al. Cathepsin B: a potential prognostic marker for inflammatory breast cancer. J Transl Med 9, 1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strojan P. et al. Prognostic significance of cysteine proteinases cathepsins B and L and their endogenous inhibitors stefins A and B in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res 6, 1052–1062 (2000). [PubMed] [Google Scholar]
- Gong F. et al. Cathepsin B as a potential prognostic and therapeutic marker for human lung squamous cell carcinoma. Mol Cancer 12, 125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G. et al. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nat Chem Biol 1, 203–209 (2005). [DOI] [PubMed] [Google Scholar]
- Blum G., von Degenfeld G., Merchant M. J., Blau H. M. & Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat Chem Biol 3, 668–677 (2007). [DOI] [PubMed] [Google Scholar]
- Bremer C. et al. Optical imaging of spontaneous breast tumors using protease sensing ‘smart' optical probes. Invest Radiol 40, 321–327 (2005). [DOI] [PubMed] [Google Scholar]
- Weissleder R., Tung C. H., Mahmood U. & Bogdanov A. Jr. In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol 17, 375–378 (1999). [DOI] [PubMed] [Google Scholar]
- Melancon M. P. et al. A novel method for imaging in vivo degradation of poly(L-glutamic acid), a biodegradable drug carrier. Pharm Res 24, 1217–1224 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny A. D. Evaluation of sodium poly-alpha, L-glutamate as a plasma expander. Proc Soc Exp Biol Med 100, 778–780 (1959). [DOI] [PubMed] [Google Scholar]
- Kishore B. K. et al. Mechanism of protection afforded by polyaspartic acid against gentamicin-induced phospholipidosis. II. Comparative in vitro and in vivo studies with poly-L-aspartic, poly-L-glutamic and poly-D-glutamic acids. J Pharmacol Exp Ther 255, 875–885 (1990). [PubMed] [Google Scholar]
- Li C. Poly(L-glutamic acid)--anticancer drug conjugates. Adv Drug Deliv Rev 54, 695–713 (2002). [DOI] [PubMed] [Google Scholar]
- Sumi H., Kawabe K. & Nakajima N. Effect of various polyamino acids and D- and L-amino acids on the blood fibrinolytic system. Comp Biochem Physiol B 102, 159–162 (1992). [DOI] [PubMed] [Google Scholar]
- Cai K. et al. Magnetic resonance imaging of glutamate. Nat Med 18, 302–306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R. Polymer conjugates as anticancer nanomedicines. Nat Rev Cancer 6, 688–701 (2006). [DOI] [PubMed] [Google Scholar]
- Li C. et al. Antitumor activity of poly(L-glutamic acid)-paclitaxel on syngeneic and xenografted tumors. Clin Cancer Res 5, 891–897 (1999). [PubMed] [Google Scholar]
- Li C. et al. Complete regression of well-established tumors using a novel water-soluble poly(L-glutamic acid)-paclitaxel conjugate. Cancer Res 58, 2404–2409 (1998). [PubMed] [Google Scholar]
- Yang Y., Hong H., Zhang Y. & Cai W. Molecular Imaging of Proteases in Cancer. Cancer Growth Metastasis 2, 13–27 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S. et al. Comparison of the efficacy and safety of paclitaxel-eluting coroflex please stents and paclitaxel-eluting stents in patients with coronary artery disease: a randomized PIPA trial. Catheter Cardiovasc Interv 80, 799–806 (2012). [DOI] [PubMed] [Google Scholar]
- Li C. & Wallace S. Polymer-drug conjugates: recent development in clinical oncology. Adv Drug Deliv Rev 60, 886–898 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Pickup S., Hsu O. & Poptani H. Diffusion tensor MRI in rat models of invasive and well-demarcated brain tumors. NMR Biomed 21, 208–216 (2008). [DOI] [PubMed] [Google Scholar]