Abstract
Introduction
While retrospective analyses support an association between early tumor recurrence and tumor suppressor gene (TSG) promoter methylation in early-stage non-small cell lung cancers (NSCLCs), few studies have investigated this question prospectively.
Methods
Primary tumor tissue from patients with resected pathologic stage I-IIIA NSCLCs was collected at the time of surgery and analyzed for promoter methylation via methylation-specific reverse-transcriptase polymerase chain reaction (MethyLight). The primary objective was to determine an association between promoter methylation of 10 individual TSGs (CDKN2A, CDH13, RASSF1, APC, MGMT, GSTP1, DAPK1, WIF1, SOCS3, and ADAMTS8) and recurrence-free survival (RFS), with the secondary objectives of determining association with overall survival (OS), and relation to clinical or pathologic features.
Results
107 patients had sufficient tumor tissue for successful promoter methylation analysis. Majority of patients were former/current smokers (88%) with lung adenocarcinoma (78%) and pathologic stage I disease (66%). Median follow-up was 4 years. When controlled for pathologic stage, promoter methylation of the individual genes CDKN2A, CDH13, RASSF1, APC, MGMT, GSTP1, DAPK1, WIF1, and ADAMTS8 was not associated with RFS. Promoter methylation of the same genes was not associated with OS except for DAPK1 which was associated with improved OS (p=0.03). The total number of genes with methylated promoters did not correlate with RFS (p=0.89) or OS (p=0.55).
Conclusions
Contrary to data established by previous retrospective series, TSG promoter methylation (CDKN2A, CDH13, RASSF1,APC, MGMT, GSTP1, DAPK1, WIF1, and ADAMTS8) was not prognostic for early tumor recurrence in this prospective study of resected NSCLCs.
INTRODUCTION
Lung cancer development is characterized by the acquisition of multiple methylation changes that drive the carcinogenic sequence.1,2 Many of these changes target tumor suppressor genes (TSGs) that control specific processes such as cell cycle regulation (CDKN2A), the development of an invasive phenotype (CDH13), and RAS and WNT signaling (RASSF1 and APC, respectively).3 While multiple retrospective series have demonstrated a negative prognostic association between TSG promoter methylation and outcomes in early-stage lung cancers,4,5 few studies have asked this question prospectively.
In 2008, a nested case-control study of stage I non-small cell lung cancers was published by Brock et al. in the New England Journal of Medicine.6 The primary objective of this retrospective study was to determine the association between tumor suppressor gene methylation and disease recurrence. Patients who had early recurrence of their cancer (≤ 40 months) after curative surgery were matched against a cohort of patients who did not have recurrent disease within 40 months. Tumor tissue was tested for promoter methylation of CDKN2A (P16), CDH13, RASSF1, and APC. The study showed that an increasing number of genes with methylated promoters (0, 1-2, 3-4 genes methylated) in primary tumor tissue was significantly associated with poorer recurrence-free survival (p=0.001). On multivariate analysis of the original cohort along with a separate validation cohort, when CDKN2A or CDH13 were methylated in the primary tumor, the odds ratios for recurrence were 3.55 (1.77-7.13, p<0.001) and 2.33 (1.16-4.69, p=0.02), respectively
Concurrent with this publication, we were conducting a prospective biomarker protocol with a similar objective of establishing an association between recurrence-free survival and promoter methylation. The same four genes and six other tumor suppressor genes (MGMT, GSTP1, DAPK1, WIF1, SOCS3, and ADAMTS8) were tested for promoter methylation in both resected tumor and serial plasma samples. We herein report the results of this study in an attempt to validate the findings published by Brock and colleagues in a prospective fashion in stage I-IIIA non-small cell lung cancers while providing additional data on the utility of promoter methylation of other tumor suppressor genes as potential biomarkers.
MATERIALS AND METHODS
Patients with clinical stage I-IIIA non-small cell lung cancers who were treated at Memorial Sloan Kettering Cancer Center and deemed to have resectable disease were eligible for enrollment onto this prospective, institutional review board-approved protocol. Subjects who received neoadjuvant therapy of any kind (chemotherapy, radiotherapy, or investigational agents) were excluded. All patients underwent surgical resection of their cancer with curative intent. Tumors from those with pathologic stage IIIIA disease and in whom an R0 resection was achieved were sent for promoter methylation analysis. Pathologic staging followed the 2009 TNM International System for Staging Lung Cancer.7 Stage-appropriate adjuvant therapy including chemotherapy and/or radiation therapy were administered as per the treating physician.
Sample Acquisition and Promoter Methylation Analysis
Both primary tumor tissue and serial plasma samples were acquired for promoter methylation analysis. Fresh frozen tumor tissue was obtained at the time of surgical resection. Plasma samples were collected at four different time points during the study course: immediately prior to surgical intervention, 3 to 8 days post-surgery, 2 to 5 weeks post-surgery, and at 2 to 4 months post-surgery.
Tumor and plasma samples were analyzed via methylation-specific reverse transcriptase polymerase chain reaction (RT-PCR, MethyLight, Response Genetics, Los Angeles, CA). Assay sensitivity allowed the potential detection of a single methylated allele in the presence of a 10,000-fold excess of unmethylated alleles.8 The promoter regions of the following panel of 10 genes were analyzed: CDKN2A, CDH13, RASSF1, APC, MGMT, GSTP1, DAPK1, WIF1, SOCS3, and ADAMTS8. These genes were chosen as targets of interest based on their roles as regulators of cancer growth and their inclusion in previous retrospective series.3,4,9 DNA was isolated from fixed volumes of tumor and plasma and subjected to bisulfite treatment using a Qiagen Epitect Bisulfite kit. Fully methylated Qiagen EpiTect Control DNA was used as a positive control. Human genomic DNA from peripheral blood mononuclear cells (Ambion) was used as a negative control.
After bisulfite treatment, genomic DNA was amplified by fluorescence-based, real-time quantitative PCR using locus-specific PCR primers flanking an oligonucleotide probe with a 5′ fluorescent reporter dye (6FAM) and a 3′ quencher dye (TAMRA). 5′ to 3′ nuclease activity of Taq DNA polymerase resulted in cleavage of the 5′ probe, releasing the fluorescent reporter. Reporter fluorescence was detected by the laser ABI Prism 7900 Sequence Detection System (Perkin-Elmer, Foster City, CA). Primer and probe design for each of the 10 genes was based on previous reports and is detailed in Supplementary Table 1.10-13 Promoter methylation was reported as a methylation value percentage (MVP) with tumor suppressor gene levels normalized to ß-Actin in modified DNA.
Statistical Analysis
The primary objective of this study was to determine the association between promoter methylation of individual tumor suppressor genes (in tumor and plasma) and recurrence-free survival (RFS). Secondary objectives included determination of the association between promoter methylation and overall survival (OS) or clinicopathologic features. RFS and OS were calculated from the time of surgical resection using Kaplan-Meier estimates. Patients were followed for RFS until recurrence or death, whichever came first, and for OS until death of any cause. Patients who did not experience the event of interest during the study time were censored at the time of the last available follow-up.
For each individual tumor suppressor gene, patients whose tumors had methylated promoters were compared to those with unmethylated promoters with respect to RFS and OS using the log-rank test after adjusting for pathologic stage.14 In addition, the log-rank test was used to assess whether the total number of methylated tumor suppressor gene promoters per patient was associated with RFS and OS. Comparisons were performed within each stage and the results aggregated over all stages. In order to facilitate comparison with data presented by Brock et al.,6 we also undertook an analysis restricted to patients diagnosed with stage I disease.
For the purposes of this study, any non-zero MVP value for each individual tumor suppressor gene was deemed positive for promoter methylation. The incidence of promoter methylation of individual tumor suppressor genes was correlated with tumor morphology, histology, and pathologic stage. Group comparisons were performed with the log-rank test and Cox-proportional hazards.
RESULTS
A total of 346 patients with clinical stage I-IIIA non-small cell lung cancers who were deemed to have resectable disease at the time of diagnosis were identified between 2003 and 2008 at the Memorial Sloan Kettering Cancer Center. Subjects for whom neoadjuvant therapy was planned were excluded and 220 patients were enrolled onto this trial. Of these patients, 28 were excluded (23 found to have stage IIIB-IV during workup, and 5 patients with an R1 resection). 197 successfully underwent resection of all gross and microscopic disease (R0) and were found to have pathologic stage I-IIIA disease. As majority of patients had early-stage disease, only 156 had available tissue for further testing after pathologic review. After specimen processing, sufficient tumor tissue for successful promoter gene methylation analysis was available in 107 cases. Of these cases, plasma samples were drawn for all four time points (preoperatively and 3 to 8 days, 2 to 5 weeks, and 2 to 4 months post-surgery) in 74 patients.
Patient Characteristics and Promoter Methylation Frequency in Tumors
The characteristics of patients whose tumors were successfully analyzed for promoter gene methylation (n=107) are presented in Table 1. The majority of patients were current or former smokers (88%, n=84) with lung adenocarcinoma (78%, n=83) and pathologic stage I disease (66%, n=62). Patients with pathologic stage II and IIIA disease comprised 25% (n=27) and 13% (n=14) of the population, respectively. The median duration of follow-up on this study was 4 years. In relation to the primary endpoint of recurrence-free survival, the large majority of recurrences would have been expected to occur within this period.
Table 1. Demographics.
The clinical characteristics of 107 patients with resected non-small cell lung cancers whose tumors successfully underwent promoter methylation gene analysis are summarized.
| Patient Characteristics | n | % | |
|---|---|---|---|
| Age | Median 68 years (43-85) |
||
|
| |||
| Sex | Male | 55 | 51% |
| Female | 52 | 49% | |
|
| |||
| Smoking | Current | 20 | 19% |
| Former | 74 | 69% | |
| Never | 13 | 12% | |
|
| |||
| Stage | IA | 37 | 35% |
| IB | 29 | 27% | |
| IIA | 17 | 16% | |
| IIB | 10 | 9% | |
| IIIA | 14 | 13% | |
|
| |||
| Pathology | Adenocarcinoma | 83 | 78% |
| Squamous cell carcinoma | 20 | 19% | |
| Large cell carcinoma | 4 | 12% | |
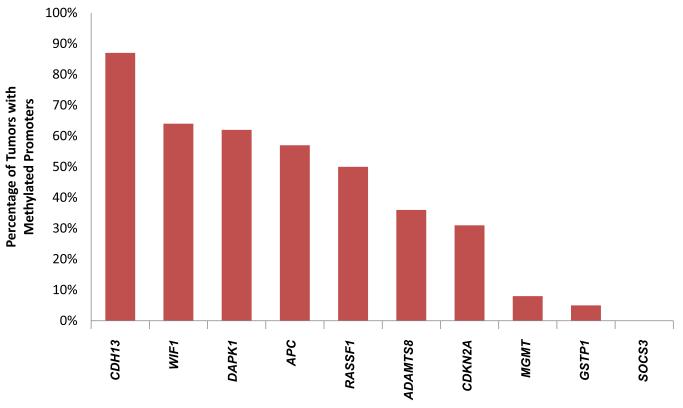
Promoter methylation in primary tumor tissue was a frequently observed event for the following tumor suppressor genes: CDH13, WIF1, DAPK1, APC, and RASSF1 (occurring in 87%, 64%, 62%, 57%, and 50% of tumors, respectively). Four of the remaining genes on the panel including ADAMTS8, CDKN2A, MGMT, and GSTP1 were less commonly methylated (36%, 31%, 8%, and 5%, respectively). SOCS3 was not found to be methylated in any samples (Figure 1). Consequently, the association between SOCS3 and either recurrence-free or overall survival could not be analyzed. A significant variability in absolute MVP values was noted between individual tumor suppressor genes.
Figure 1. Frequency of promoter gene methylation in resected stage I-IIIA NSCLCs.
The percentage of tumors with methylated promoter regions of ten tumor suppressor genes is depicted. Genes whose promoter regions were found to be methylated in at least 50% of samples tested include CDH13 (87%), WIF1 (64%), DAPK (62%), APC (57%), and RASSF1A (50%). ADAMTS8 (36%), CDKN2A (31%), MGMT (8%), and GSTP1 (5%) were less commonly methylated. SOCS3 was not found to be methylated in any samples.
Association Between Tumor Promoter Gene Methylation and Survival
When controlled for pathologic stage, promoter methylation of the individual genes APC, CDH13, MGMT, RASSF1, WIF1, ADAMTS8, GSTP1, and CDKN2A in primary tumor tissue was not significantly associated with RFS. Similarly, promoter methylation of the same genes in primary tumor tissue was not associated with OS. For each of these genes, median RFS and OS for patients with patients with either methylated or unmethylated promoters are detailed in Table 2. These lack of an association between promoter methylation of these genes and RFS and OS was confirmed in a subset analysis of patients with stage I disease.
Table 2. Association between tumor suppressor gene promoter methylation and survival.
Median recurrence-free survival (RFS) and overall survival (OS) for patients with both methylated and unmethylated promoters of 10 tumor suppressor genes are shown. SOCS3 was not found to be methylated in any samples and could not be analyzed in relation to RFS or OS. NR - not reached.
| Gene | Methylation Results | Median RFS |
p-value | Median OS |
p- value |
|---|---|---|---|---|---|
| CDH13 | Methylated (87%, n=93) Unmethylated (13%, n=14) |
4.5 mo 2.7 mo |
p=0.43 | 6.2 mo 4.4 mo |
p=0.59 |
| WIF1 | Methylated (64%, n=69) Unmethylated (36%, n=38) |
4.5 mo 3.9 mo |
p=0.49 | 6.2 mo NR |
p=0.57 |
| DAPK1 | Methylated (62%, n=66) Unmethylated (38%, n=41) |
4.9 mo 2.8 mo |
p=0.09 | NR 4.2 mo |
p=0.03 |
| APC | Methylated (57%, n=62) Unmethylated (43%, n=45) |
3.5 mo 5.0 mo |
p=0.56 | 4.3 mo NR |
p=0.07 |
| RASSF1 | Methylated (50%, n=54) Unmethylated (50%, n=53) |
4.0 mo 4.5 mo |
p=0.86 | 5.3 mo NR |
p=0.67 |
| ADAMTS8 | Methylated (36%, n=38) Unmethylated (64%, n=69) |
5.0 mo 3.8 mo |
p=0.49 | 6.2 mo 5.3 mo |
p=0.71 |
| CDKN2A | Methylated (31%, n=33) Unmethylated (68%, n=74) |
4.9 mo 3.9 mo |
p=0.60 | 6.1 mo 6.2 mo |
p=0.72 |
| MGMT | Methylated (8%, n=9) Unmethylated (92%, n=98) |
3.4 mo 4.3 mo |
p=0.92 | 4.2 mo 6.2 mo |
p=0.65 |
| GSTP1 | Methylated (5%, n=6) Unmethylated (95%, n=101) |
3.6 mo 4.5 mo |
p=0.95 | 4.2 mo 6.2 mo |
P=0.81 |
| SOCS3 | Methylated (0%, n=0) Unmethylated (100%, n=107) |
- - |
- | - - |
- |
Across the entire panel of 10 genes, the total number of methylated tumor suppressors per tumor (0-2, 3, 4, 5, or 6-8 methylated genes per sample) did not correlate with either RFS (p=0.89) or OS (p=0.55).
A non-significant trend towards improved RFS was noted in patients whose tumors harbored a methylated vs unmethylated DAPK1 promoter (4.9 vs 2.8 mo, p=0.09). DAPK1 promoter methylation, however, was significantly associated with improved OS. Patients who had tumors with an unmethylated DAPK promoter had a median OS of 4.2 months while median OS was not reached for those whose tumors harbored a methylated DAPK1 promoter (p=0.03). This was confirmed in a subset analysis of patients with stage I disease (median OS of patients with unmethylated vs methylated DAPK promoters: 5.2 months vs. not reached, p=0.03).
Tumor Promoter Gene Methylation and Pathologic Features
RASSF1 promoter methylation was more likely to be present in tumors of poorly-differentiated or undifferentiated morphology versus tumors of moderately-differentiated or well-differentiated morphology (64% vs. 4%, p=0.03).
Promoter methylation of CDKN2A was more prevalent in SQCLCs (55%) in comparison to ADCLs (26%) and large cell carcinomas (17%, p=0.03). ADCLs had a higher frequency of APC promoter methylation (65%) compared to large cell carcinomas (50%) and SQCLCs (30%, p=0.02).
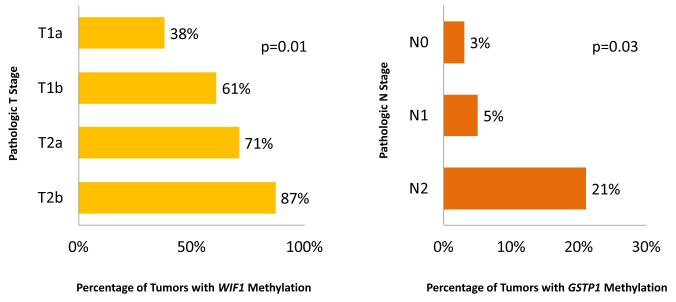
Promoter methylation of WIF1 was significantly associated with increasing pathologic T stage (38%, 61%, 71%, and 87% for pT1a, pT1b, pT2a, and pT2b, respectively, p=0.01). In addition, promoter methylation of GSTP1 was significantly associated with increasing pathologic N stage (3%, 5%, and 21% for pN0, pN1, and pN2, respectively, p=0.03). These are depicted in Figure 2.
Figure 2. Correlation between WIF1 and GSTP1 promoter methylation and pathologic stage.
The percentage of tumors with methylated WIF1 and GSTP1 promoter regions is shown in relation to pathologic T and N stage, respectively. Increasing WIF1 promoter methylation was associated with increasing pathologic T stage (p=0.01). Increasing GSTP1 methylation correlated with increasing pathologic N stage (p=0.03).
Promoter methylation of the remaining individual genes APC, MGMT, GSTP1, DAPK, CDH1, and ADAMTS8 was not associated with tumor morphology, histology, or pathologic stage.
Promoter Gene Methylation in Blood
For plasma tumor-suppressor gene methylation analysis, the number of patients with non-zero MVP values at each pre-specified time point was exceedingly low. Because of the lack of variability in blood promoter gene methylation values, we could not formally evaluate the association between methylation status and survival outcomes or clinical features.
DISCUSSION
Epigenetic changes have long been touted as ideal biomarker candidates due to their early acquisition15 and roles in tumorigenesis.16 Our findings confirm that tumor suppressor gene promoter methylation is a common event in NSCLCs,5,17,18 the prevalence of which exceeds half of all tumors tested for CDH13, WIF1, DAPK1, and APC. While these events are common, data regarding their roles as strong predictors of outcome in early-stage lung cancers has relied largely on retrospective series, limiting the utility of these observations.
In this prospective series of resected, early-stage lung cancers, we did not detect an association between recurrence-free survival and tumor promoter methylation of CDKN2A, CDH13, RASSF1, and APC in addition to five other genes on our panel (MGMT, GSTP1, DAPK, WIF-1, and METH-2). These results were confirmed in a subset analysis of patients with stage I disease, comparable but not completely similar to the patient population analyzed in the Brock series.6 In addition, we did not confirm that an increasing number of genes with methylated promoters was associated with poorer recurrence-free survival. Promoter methylation was not associated with overall survival, except for DAPK where methylation was found to be associated with improved survival. The significance of this finding remains unclear as previous studies have reported a negative impact of DAPK methylation on survival outcomes.5,19 Similarly, the number of methylated tumor suppressor gene promoters did not correlate with overall survival. Future confirmatory studies would benefit from the inclusion of a validation set to add strength to these conclusions.
Despite their lack of prognostic significance, the profile of tumor suppressor gene promoter methylation in early-stage lung cancers continues to contribute important information regarding tumor biology. We herein confirm a high frequency of promoter methylation of CDKN2A in squamous cell lung carcinomas in comparison to other histologies. These results echo the findings of genomic characterization of squamous lung cancers by The Cancer Genome Atlas where inactivation of the CDKN2A locus via a variety of mechanisms (epigenetic silencing, inactivating mutation, exon skipping, and homozygous deletion) was found in 72% of cases analyzed.20 CDKN2A encodes the protein p16INK4A, a CDK inhibitor that blocks the actions of CDK4 and CDK6 that are important for cell cycle G1 phase progression. Silencing of CDKN2A results in increased CDK4/6 activity and dysregulation of cell cycling that may contribute significantly to the pathogenesis of these tumors. A large prospective clinical trial for squamous lung cancers is planned to contain an arm with a CDK4/6 inhibitor for tumors with genomic aberrations thought to contribute to cell cycle dysregulation.21
We similarly demonstrate that promoter methylation of the tumor suppressor genes RASSF1, and WIF-1 and GSTP1 is associated with a poorly-differentiated tumors and more advanced disease in early-stage lung cancers, respectively. RASSF1 encodes the RAS association domain family protein 1A that mediates the apoptotic effects of the RAS protein.22 Consistent with the results we present here, several studies have established a correlation between gene hypermethylation and poorly-differentiated histology.23-25 WIF-1 (Wnt-inhibitory factor-1) hypermethylation has previously been described in lung cancers and results in increased activation of the Wnt pathway that plays a critical role in stem cell regulation and carcinogenesis.26,27 The latter may play a role in the increase in nodal disease seen in patients with methylated WIF-1 promoters in our series. In contrast, we found GSTP1 promoter methylation to be associated with increasing pathologic T stage. GSTP1 encodes glutathione S-transferase P, an enzyme involved in the metabolism of xenobiotic agents.28,29 The mechanism by which silencing of this gene and a putative decrease in the activity of the enzyme relate to increasing tumor size remains to be determined.
Lastly, while all non-zero MVP values were taken to represent evidence of promoter gene methylation in this report, the range of absolute MVP values varied significantly between genes. This heterogeneity and the need for both laboratory and clinical validation of existing assays are important issues that need to be recognized as we move forward. Whereas the presence of a mutation or fusion involving a driver oncogene is, for practical purposes, an ‘all-or-none’ phenomenon, the degree of promoter methylation varies significantly between tumors with a lack of test-specific cutoff values for ‘positive’ methylation in quantitative assays. While the assay that we used in this trial had the ability to provide semi-quantitative data regarding the degree of promoter methylation, with ten genes and two outcomes investigated, an exploratory analysis of the relationship between the degree of methylation and RFS or OS would have had a high risk of generating false positive results. Investigations into epigenetic markers of tumor biology and patient outcome will benefit from an increased focus on standardization of available assays of gene methylation.
In summary, contrary to data published in previous retrospective reports, we failed to demonstrate an association between promoter methylation of APC, CDKN2A, MGMT, GSTP1, DAPK, CDH1, RASSF1, WIF1, and ADAMTS8 and recurrence-free survival in this prospective study of resected early-stage non-small cell lung cancers.
Supplementary Material
Acknowledgments
Support: The project described was supported by Grant Number RO1CA092315 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
REFERENCES
- 1.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 2.Shenker NS, Polidoro S, van Veldhoven K, et al. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22:843–51. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 3.Vendetti FP, Rudin CM. Epigenetic therapy in non-small-cell lung cancer: targeting DNA methyltransferases and histone deacetylases. Expert Opin Biol Ther. 2013;13:1273–85. doi: 10.1517/14712598.2013.819337. [DOI] [PubMed] [Google Scholar]
- 4.Wen J, Fu J, Zhang W, Guo M. Genetic and epigenetic changes in lung carcinoma and their clinical implications. Mod Pathol. 2011;24:932–43. doi: 10.1038/modpathol.2011.46. [DOI] [PubMed] [Google Scholar]
- 5.Buckingham L, Penfield Faber L, Kim A, et al. PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer. 2010;126:1630–9. doi: 10.1002/ijc.24896. [DOI] [PubMed] [Google Scholar]
- 6.Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–28. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 7.Detterbeck FC, Boffa DJ, Tanoue LT. The new lung cancer staging system. Chest. 2009;136:260–71. doi: 10.1378/chest.08-0978. [DOI] [PubMed] [Google Scholar]
- 8.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shinjo K, Kondo Y. Clinical implications of epigenetic alterations in human thoracic malignancies: epigenetic alterations in lung cancer. Methods Mol Biol. 2012;863:221–39. doi: 10.1007/978-1-61779-612-8_13. [DOI] [PubMed] [Google Scholar]
- 10.Eads CA, Lord RV, Wickramasinghe K, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–8. [PubMed] [Google Scholar]
- 11.Brabender J, Usadel H, Metzger R, et al. Quantitative O(6)-methylguanine DNA methyltransferase methylation analysis in curatively resected non-small cell lung cancer: associations with clinical outcome. Clin Cancer Res. 2003;9:223–7. [PubMed] [Google Scholar]
- 12.Lehmann U, Langer F, Feist H, Glockner S, Hasemeier B, Kreipe H. Quantitative assessment of promoter hypermethylation during breast cancer development. Am J Pathol. 2002;160:605–12. doi: 10.1016/S0002-9440(10)64880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann AC, Kaifi JT, Vallbohmer D, et al. Lack of prognostic significance of serum DNA methylation of DAPK, MGMT, and GSTPI in patients with non-small cell lung cancer. J Surg Oncol. 2009;100:414–7. doi: 10.1002/jso.21348. [DOI] [PubMed] [Google Scholar]
- 14.Heller G, Venkatraman ES. A nonparametric test to compare survival distributions with covariate adjustment. J R Stat Soc. 2004;66:719–33. [Google Scholar]
- 15.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 16.Helman E, Naxerova K, Kohane IS. DNA hypermethylation in lung cancer is targeted at differentiation-associated genes. Oncogene. 2012;31:1181–8. doi: 10.1038/onc.2011.307. [DOI] [PubMed] [Google Scholar]
- 17.Castro M, Grau L, Puerta P, et al. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med. 2010;8:86. doi: 10.1186/1479-5876-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moran A, Fernandez-Marcelo T, Carro J, et al. Methylation profiling in non-small cell lung cancer: clinical implications. Int J Oncol. 2012;40:739–46. doi: 10.3892/ijo.2011.1253. [DOI] [PubMed] [Google Scholar]
- 19.Lu C, Soria JC, Tang X, et al. Prognostic factors in resected stage I non-small-cell lung cancer: a multivariate analysis of six molecular markers. J Clin Oncol. 2004;22:4575–83. doi: 10.1200/JCO.2004.01.091. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research N Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox J. Master Protocol for squamous cell lung cancer readies for launch. Nat Biotechnol. 2014;32:116–8. doi: 10.1038/nbt0214-116b. [DOI] [PubMed] [Google Scholar]
- 22.Underhill-Day N, Hill V, Latif F. N-terminal RASSF family: RASSF7-RASSF10. Epigenetics. 2011;6:284–92. doi: 10.4161/epi.6.3.14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Lee JJ, Wang L, et al. Value of p16INK4a and RASSF1A promoter hypermethylation in prognosis of patients with resectable non-small cell lung cancer. Clin Cancer Res. 2004;10:6119–25. doi: 10.1158/1078-0432.CCR-04-0652. [DOI] [PubMed] [Google Scholar]
- 24.Choi N, Son DS, Song I, et al. RASSF1A is not appropriate as an early detection marker or a prognostic marker for non-small cell lung cancer. Int J Cancer. 2005;115:575–81. doi: 10.1002/ijc.20916. [DOI] [PubMed] [Google Scholar]
- 25.Tomizawa Y, Kohno T, Kondo H, et al. Clinicopathological significance of epigenetic inactivation of RASSF1A at 3p21.3 in stage I lung adenocarcinoma. Clin Cancer Res. 2002;8:2362–8. [PubMed] [Google Scholar]
- 26.Mazieres J, He B, You L, et al. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:4717–20. doi: 10.1158/0008-5472.CAN-04-1389. [DOI] [PubMed] [Google Scholar]
- 27.Yang TM, Leu SW, Li JM, et al. WIF-1 promoter region hypermethylation as an adjuvant diagnostic marker for non-small cell lung cancer-related malignant pleural effusions. J Cancer Res Clin Oncol. 2009;135:919–24. doi: 10.1007/s00432-008-0527-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arai T, Yasuda Y, Takaya T, et al. Immunohistochemical expression of glutathione transferase-pi in untreated primary non-small-cell lung cancer. Cancer Detect Prev. 2000;24:252–7. [PubMed] [Google Scholar]
- 29.Grimminger PP, Maus MK, Schneider PM, et al. Glutathione S-transferase PI (GST-PI) mRNA expression and DNA methylation is involved in the pathogenesis and prognosis of NSCLC. Lung Cancer. 2012;78:87–91. doi: 10.1016/j.lungcan.2012.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.