Abstract
Chemotherapy-based treatment of patients with primary central nervous system lymphoma can lead to durable remissions and potentially cure in a fraction of patients. Accurate assessment of residual disease is necessary to determine the duration and success of treatment that cannot be achieved by contrast-enhanced imaging due to limited sensitivity and specificity. A tumor-derived blood-based biomarker, if detectable and quantifiable, could serve as a more specific and reliable marker for these patients. The goal of this study was to assess whether lymphoma-specific IgH rearrangements can be detected in plasma of patients with PCNSL. PCNSL tissue was analyzed by capturing and sequencing the IgH genomic regions (IgCap) using next generation sequencing with the Illumina platform. Plasma of patients with detected IgH rearrangement was then analyzed for presence of the respective rearrangement using polymerase chain reaction. Tumor tissue and matched plasma of five treatment-naïve patients with biopsy-proven PCNSL (mean age of 65.6 years; range 62–68 years) were analyzed. All patients had measurable contrast-enhancing disease on MRI at time of plasma collection. IgH rearrangements were identified in 4 of 5 analyzed PCNSL tissue samples. The respective rearrangement could be detected in the plasma of 1 patient (25 %) but not in the others. IgH rearrangements can be detected in tumor tissue of patients with PCNSL using IgCap, however, they are absent or only present in minimal quantities in plasma, even in treatment-natïve patients with bulky disease. Alternative strategies to develop circulating biomarkers for PCNSL patients need to be explored.
Keywords: Primary central nervous system lymphoma, Biomarkers, Gene rearrangements, Circulating tumor DNA
Introduction
Primary central nervous system lymphomas (PCNSL) account for about 4 % of tumors of the central nervous system. The incidence of PCNSL, however has been rising in the immunocompetent population over the last few decades, particularly in the elderly population [1, 2].
High-dose methotrexate-based therapies are the standard of care for newly diagnosed PCNSL. Several different regimens achieve response rates of greater than 50 % and durable remissions in many patients [3]. PCNSL is now considered a potentially curable disease; however, most patients still recur after initial therapy and many eventually die of their disease.
In order to choose the appropriate duration of treatment, it is important to determine when patients have achieved a complete response (CR), which is the prerequisite for a potential cure.
Response assessment in PCNSL has commonly been based on the same response criteria used for other primary brain tumors [4, 5], and patients are considered to have achieved a CR when all contrast-enhancing lesions have disappeared on MRI. However, PCNSL is known to diffusely involve non-contrast enhancing areas, CSF and the eyes, so contrast enhancement alone is not a reliable marker of a CR in these cancers. In addition, complete resolution of all contrast enhancing areas is not always achieved and residual contrast enhancement does not necessarily mean that there is residual disease [6]. As a result, investigators have started to focus on circulating tumor markers to assess disease status in patients with PCNSL [7]. To be clinically useful, such a marker needs to be exquisitely specific. A particularly attractive circulating biomarker is circulating tumor DNA (ctDNA), as such somatic alterations are unique to tumor cells.
The concept of mutated ctDNA as a specific and potentially sensitive circulating tumor marker has been demonstrated in several solid tumors [8–10]. Fusion genes are other genetic alterations that can function as circulating tumor markers in malignancies. Chronic myelogenous leukemia is an example of a disease in which fusion genes are routinely quantified as measure of disease burden and used for assessment of residual disease. Additionally, a recent study showed that IgH gene rearrangements could be detected in plasma of patients with non-Hodgkin’s lymphoma, suggesting this as a potentially useful and highly specific blood-based tumor marker for lymphomas [11]. Based on these data, we set out to determine whether a similar IgH-based approach would be feasible in patients with PCNSL, as described below.
Methods
Sample collection
Adult patients with newly diagnosed, previously untreated, biopsy-proven PCNSL with measurable areas of contrast enhancement were asked to participate in this study. Pretreatment MRI scans were reviewed. Histologic and immunohistochemical slides of the cases included in the study were reviewed by a neuropathologist (FJR). Informed consent was obtained using a protocol that was approved by the Institutional Review Board at Johns Hopkins University. Formalin-fixed paraffin-embedded (FFPE) tissue samples were collected and used for analysis. Plasma was collected in EDTA-containing tubes, centrifuged twice immediately after collection and stored at −80 °C. Plasma collection and processing was done the same way as in other circulating tumor DNA studies performed by our laboratory [8]. Plasma samples were only thawed once for DNA extraction and immediately processed to optimize sample quality.
DNA purification
DNA was extracted from 10,000 µl of plasma with a QIAamp circulating nucleic acid kit following the manufacturer’s instructions (QIAGEN; Valencia, CA, USA). DNA was extracted from tumor tissues using a Qiagen AllPrep kit following the manufacturer’s protocol. The amount of amplifiable DNA was quantified by a real time PCR based assay that amplifies human repeated sequences [12].
Tumor illumina library preparation
Approximately 1.5–3 µg oftTumor genomic DNA from formalin-fixed paraffin-embedded (FFPE) samples was used to prepare libraries following Illumina’s (Illumina, San Diego, CA, USA) protocol.
IgCap capture
The targeted capture region included V-gene exons plus the first 36 bp of the downstream introns, six J-gene exons plus the first 36 bp of the upstream introns, and all the D-gene exons. Capture of the regions of interest was performed as described before [11, 13]. Captured DNA libraries were sequenced with the Illumina GAIIx Genome Analyzer, yielding 150 (2 × 75) base pairs from the final library fragments. Sequencing reads were analyzed and aligned to human genome hg18 with the Eland algorithm in CASAVA 1.6 software (Illumina). The sequence information was processed in silico to identify rearranged sequences as previously described [11].
Confirmation of IgH rearrangements in tumor and plasma samples
The full V-D-J or D-J joint region sequence and 40 bp from either side of the joint were used for primer design. The PCR mixture (50 µl) contained various amounts of 5 µl 10 × Platinum Taq buffer (Life Technologies), template DNA, 0.2 µM of forward-reverse primer mixture, 67 mM Tris (pH 8.8), 16.6 mM NH4SO4, 6.7 mM MgCl2, 10 mM b-mercaptoethanol, 0.1 mM dATP, 0.1 mM dCTP, 0.1 mM dGTP, 0.1 mM dTTP, 6 % (volyvol) DMSO, and 0.5 units of Invitrogen Taq polymerase. PCR was performed as follows: 96 °C for 4 min; 45 cycles of 96 °C for 10 s, 59 °C for 10 s, and 72 °C for 30 s. The primers used for PCR are listed in Supplement Table 1; some primers represented sequences present in the normal genome, while others represented sequences unique to particular patient’s rearranged IgH genes. PCR products were gel purified with a QIAquick Gel Extraction Kit (QIAGEN; Valencia, CA, USA) and cloned with a TA clone kit (Promega, Madison, WI, USA) according to the manufacture’s protocols. Plasmid DNA was evaluated by Sanger sequencing.
Results
Five patients with newly diagnosed, previously untreated PCNSL were studied. The patient demographics and representative MRI images are shown in Table 1 and Fig. 1. All patients had bulky, contrast enhancing, intraparenchymal brain lesions indicating disruption of the blood–brain barrier (BBB; Fig. 1).
Table 1.
Patient characteristics
| Patient ID | Age at diagnosis (years), gender (M/F) |
Location | Type of surgery | Diagnosis |
|---|---|---|---|---|
| Patient 1 | 67 F | Left frontal | Stereotactic biopsy | DLBCL |
| Patient 2 | 64 F | Left temporal | Partial resection | DLBCL |
| Patient 3 | 62 M | Cerebellar | Partial resection | DLBCL |
| Patient 4 | 67 F | Left frontotemporal | Stereotactic biopsy | DLBCL |
| Patient 5 | 68 M | Midbrain, pons | Stereotactic biopsy | DLBCL |
DLBCL diffuse large B-cell lymphoma
Fig. 1.
Contrast enhanced MRI images of patients prior to start of chemotherapy. Contrast enhancing areas have a disrupted BBB
DNA isolated from lymphoma tissue was used to construct libraries suited for massively parallel sequencing according to the IgCap protocol (see methods). Between 13,248,664 and 28,400,917 tags were generated per patient (Table 2). The sequence information obtained was then processed in silico to identify rearranged sequences at the IgH locus. IgH rearrangements were detected in the tumor tissue of four of the five patients and each rearrangement was validated by PCR with primers flanking the rearranged region (Table 3; Supplement Table 1).
Table 2.
Sequencing summary
| Patient ID | Total tags | Tags matched uniquely to human genome |
Tags matched to the Ig region |
Tags matched to the Ig coding sequences |
Average coverage of the Ig region |
Target bases with more than 10 reads (number) |
Target bases with more than 10 reads (%)a |
|---|---|---|---|---|---|---|---|
| Patient 1 | 13,248,664 | 5,726,682 | 577,050 | 360,693 | 1,018 | 16,029 | 90 |
| 13,248,664 | 5,198,386 | 547,561 | 346,199 | 977 | 15,943 | 90 | |
| Patient 2 | 14,749,580.00 | 9,479,193 | 194,090 | 133,647 | 383 | 16,418 | 94 |
| 14,749,580 | 9,254,038 | 192,561 | 132,646 | 380 | 16,339 | 94 | |
| Patient 3 | 28,400,917 | 16,497,095 | 746,102 | 475,937 | 1,362 | 16,029 | 92 |
| 28,400,917 | 14,721,936 | 710,319 | 452,784 | 1,296 | 16,680 | 96 | |
| Patient 4 | 25,776,549 | 14,503,819 | 470,085 | 324,394 | 929 | 16,837 | 96 |
| 25,776,549 | 12,604,082 | 444,729 | 306,449 | 878 | 16,828 | 96 | |
| Patient 5 | 13,545,846 | 5,079,690 | 372,873 | 251,786 | 711 | 12,679 | 71 |
| 13,545,846 | 4,376,814 | 345,223 | 233,537 | 659 | 12,204 | 69 |
Percent of bases with more than 10 reads of all bases that were attempted to be captured
Table 3.
Rearrangements identified in this study
| Patient ID | Rearranged sequence | Rearranged genes |
Detected in plasma |
|---|---|---|---|
| Patient 1 | AAAAACCAGTTCTCCCTGATGTTAGTTTCTGTGACTGCCATGGA CACGGGTGTCTATTTTTGTGCGAAGGCCCCGCGATTTGACTAC TGGGGCCAGGGAAACCTGGTCACCGTCGCCTCAGGTGAGTC ATCACAAC | IGHV4–61: J1 | Yes |
| Patient 2 | TGGACCCTCCTTGCTCCCTGGGAAGCTCCTCCTGACAGCCCC GCCTCCAGTTCCAGGTGTGGTTATTGTCAGGGGGTGTCAGACT GTGGTGGATACAGCTATGTGTACTGATATCTGGGGCCAAGGG ACAATGGTCAC | IGHD5–18: J3 | No |
| Patient 3 | TGAAGGGCCGATTCACCATCTCCAGAGACAACTCCAAGAACA CTCTCTCTCTGCAAATGACCAGCCTGAAAACTGAAGACACGG CTGTCTATTACTGTGCGGGCGATGTTATAGCAGCTCGTCAGAT AGGATACTTCGTATACTGGGGCCGGGGCACCCTGGTCACCGT CTCTTCAGGTGA | IGHV3–7: J1 | No |
| Patient 4 | GTTTCCCCAGGCCTGGCGGTAGGTTTGAAGTGAGGTCTGTGTC ACTGTGGTATTACTATGATAGTAGTGGTTGCAACTAAACATAC GCTATCGTGGCTGTTGCCCGTTGGATCACGGCCTGGTACTTCG ATCTCTGGGGCCGTGGCACCCTGGTCACTGTCTCCT | IGHD3–22: J1 | No |
| Patient 5 | ND | ND | N/A |
ND not detected, N/A not applicable
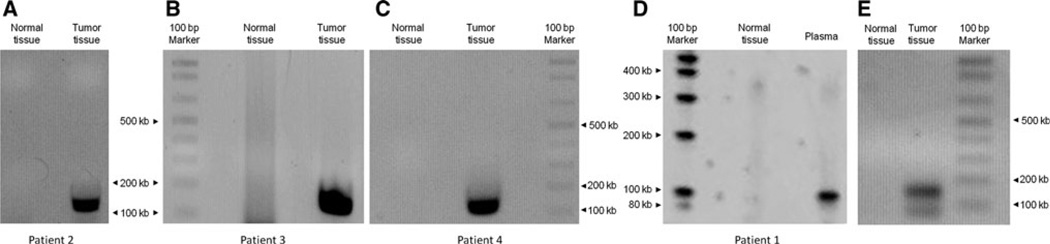
Next, we analyzed DNA from plasma of the four patients with an identifiable rearranged IgH gene in their lymphomas. Even after extensive cycles of PCR (50), easily adequate to detect single copies of rearranged sequences [14] rearrangements could only be detected in one of the four plasma specimens analyzed (Fig. 2). The one rearrangement that was found in plasma (Patient 1) was confirmed by direct sequencing of the PCR product; the DNA sequence was identical with the one that was detected in the corresponding lymphoma tissue.
Fig. 2.
a–c Agarose gels showing amplified tissue DNA in patients #2–4; rearranged DNA was not detected in plasma of these patients (not shown). d Agarose gel showing amplified rearranged DNA in plasma after 50 cycles of PCR (patient #1); the DNA sequence of the band was sequenced and was found to be identical with the DNA of the lymphoma tissue. e Amplified rearranged tissue DNA of patient #1. Of note, the designed PCR product is 153 bp; the size is different from the rearranged DNA fragment in plasma (d) as different primers were used for tissue and plasma (positions of tissue and plasma primers that were used are illustrated in Supplement Fig. 1). Normal tissue DNA was used as negative control
The detectability of IgH rearrangements in plasma did not correlate with presence or absence of contrast enhancement on MRI, as all 5 patients had visible contrast enhancement at time of plasma collection.
Discussion
This study shows that IgH rearrangements can be detected in tumor tissue of patients with PCNSL and that these rearrangements can occasionally be detected in the blood. However, it appears that the detectability of circulating rearrangements is limited, even in patients with bulky, contrast-enhancing disease prior to chemotherapy. The reason for the limited detectability of IgH rearrangements in plasma of these patients is unclear. The BBB in contrast enhancing regions of lymphoma is altered (as evidenced by presence of contrast enhancement on MRI), and one would assume that DNA can be shed into the bloodstream through gaps in the disrupted BBB, in particular in PCNSL that have not yet been treated with chemotherapy. Similar findings have been described previously by Jahnke et al. [15, 16] who analyzed 24 consecutive patients with PCNSL for presence of IgH rearrangements in blood and bone marrow using a three framework region (FR) assay. IgH rearrangements were detected in 4 of 24 patients with matched tumor and blood rearrangements were found in two patients, a similar frequency of detection to our observation [15].
Based on these combined findings, it appears that only limited quantities of tumor DNA find their way into the blood stream in patients with PCNSL and that circulating lymphoma DNA of rearranged IgH does not appear to be a useful marker of disease burden due to its low frequency of detection.
The question remains if this biomarker may be of higher yield when analyzing other compartments such as the patients’ CSF. ctDNA may be present at higher concentrations in CSF; however, this assay would be more difficult to use clinically as it would require repeat lumbar punctures.
Supplementary Material
Acknowledgments
This work was supported by the Robert H. Gross Memorial Fund, The Virginia and D.K. Ludwig Fund for Cancer Research and NIH grants CA96888, P01CA015396, and P30CA006973. M.H. was recipient of a Conquer Cancer Center Young Investigator Award by the American Society of Clinical Oncology. We thank Cherie Blair and Katharine Judge for sample acquisition and Dr. Stuart A. Grossman for critical review of this manuscript.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11060-013-1182-7) contains supplementary material, which is available to authorized users.
Conflict of interest K.W.K., N.P. and B.V. are co-founders of Inostics and Personal Genome Diagnostics (PGDx), own stock, and are members of their Scientific Advisory Boards. Inostics and PGDx have licensed several patent applications from Johns Hopkins, on which K.W.K., N.P. and B.V. are inventors.
Contributor Information
Jian He, Ludwig Center for Cancer Genetics and Therapeutics, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore, MD, USA.
Jian Wu, Ludwig Center for Cancer Genetics and Therapeutics, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore, MD, USA.
Yuchen Jiao, Ludwig Center for Cancer Genetics and Therapeutics, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore, MD, USA.
Fausto J. Rodriguez, Brain Cancer Program, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, 1550 Orleans Street, 1M16, Baltimore, MD 21287, USA
Jaishri O. Blakeley, Brain Cancer Program, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, 1550 Orleans Street, 1M16, Baltimore, MD 21287, USA
Kenneth W. Kinzler, Ludwig Center for Cancer Genetics and Therapeutics, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore, MD, USA
Nickolas Papadopoulos, Ludwig Center for Cancer Genetics and Therapeutics, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore, MD, USA.
Bert Vogelstein, Ludwig Center for Cancer Genetics and Therapeutics, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore, MD, USA; Howard Hughes Medical Institute, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore, MD, USA.
Matthias Holdhoff, Email: mholdho1@jhmi.edu, Ludwig Center for Cancer Genetics and Therapeutics, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore, MD, USA; Brain Cancer Program, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, 1550 Orleans Street, 1M16, Baltimore, MD 21287, USA.
References
- 1.Villano JL, Koshy M, Shaikh H, Dolecek TA, McCarthy BJ. Age, gender, and racial differences in incidence and survival in primary CNS lymphoma. Br J Cancer. 2011;105(9):1414–1418. doi: 10.1038/bjc.2011.357. doi: 10.1038/bjc.2011.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olson JE, Janney CA, Rao RD, Cerhan JR, Kurtin PJ, Schiff D, Kaplan RS, O’Neill BP. The continuing increase in the incidence of primary central nervous system non-Hodgkin lymphoma: a surveillance, epidemiology, and end results analysis. Cancer. 2002;95:1504–1510. doi: 10.1002/cncr.10851. doi: 10.1002/cncr.10851. [DOI] [PubMed] [Google Scholar]
- 3.Swinnen LJ. Primary central nervous system lymphoma: recent progress, many remaining questions. Curr Opin Oncol. 2009;21:393–396. doi: 10.1097/CCO.0b013e32832f3cb7. doi: 10.1097/CCO.0b013e32832f3cb7. [DOI] [PubMed] [Google Scholar]
- 4.Abrey LE, Batchelor TT, Ferreri AJ, Gospodarowicz M, Pulczynski EJ, Zucca E, Smith JR, Korfel A, Soussain C, DeAngelis LM, et al. Report of an international workshop to standardize baseline evaluation and response criteria for primary CNS lymphoma. J Clin Oncol. 2005;23(22):5034–5043. doi: 10.1200/JCO.2005.13.524. [DOI] [PubMed] [Google Scholar]
- 5.Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, Degroot J, Wick W, Gilbert MR, Lassman AB, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 6.KukerW,Nagele T, Thiel E, Weller M, Herrlinger U. Primary central nervous system lymphomas (PCNSL):MRI response criteria revised. Neurology. 2005;65:1129–1131. doi: 10.1212/01.wnl.0000178894.51436.54. doi: 10.1212/01.wnl.0000178894.51436.54. [DOI] [PubMed] [Google Scholar]
- 7.Hottinger AF, Iwamoto FM, Karimi S, Riedel E, Dantis J, Park J, Panageas KS, Lassman AB, Abrey LE, Fleisher M, et al. YKL-40 and MMP-9 as serum markers for patients with primary central nervous system lymphoma. Ann Neurol. 2011;70:163–169. doi: 10.1002/ana.22360. doi: 10.1002/ana.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holdhoff M, Schmidt K, Donehower R, Diaz LA., Jr Analysis of circulating tumor DNA to confirm somatic KRAS mutations. J Natl Cancer Inst. 2009;101:1284–1285. doi: 10.1093/jnci/djp240. doi: 10.1093/jnci/djp240. [DOI] [PubMed] [Google Scholar]
- 10.Angenendt P, Juhl DH, Diehl F. Detection of phosphoinositide-3-kinase, catalytic, and alpha polypeptide (PIK3CA) mutations in matched tissue and plasma samples from patients with metastatic breast cancer. J Clin Oncol. 2010;28:10502. [Google Scholar]
- 11.He J, Wu J, Jiao Y, Wagner-Johnston N, Ambinder RF, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. IgH gene rearrangements as plasma biomarkers in non- Hodgkin’s lymphoma patients. Oncotarget. 2011;2:178–185. doi: 10.18632/oncotarget.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rago C, Huso DL, Diehl F, Karim B, Liu G, Papadopoulos N, Samuels Y, Velculescu VE, Vogelstein B, Kinzler KW, et al. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 2007;67(19):9364–9370. doi: 10.1158/0008-5472.CAN-07-0605. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Matthaei H, Maitra A, Dal Molin M, Wood LD, Eshleman JR, Goggins M, Canto MI, Schulick RD, Edil BH. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3(92):92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jahnke K, Hummel M, Korfel A, Burmeister T, Kiewe P, Klasen HA, Muller HH, Stein H, Thiel E. Detection of subclinical systemic disease in primary CNS lymphoma by polymerase chain reaction of the rearranged immunoglobulin heavy-chain genes. J Clin Oncol. 2006;24:4754–4757. doi: 10.1200/JCO.2006.06.7165. doi: 10.1200/JCO.2006.06.7165. [DOI] [PubMed] [Google Scholar]
- 16.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.