Abstract
The cardiac sarcomere is the functional unit for myocyte contraction. Ordered arrays of sarcomeric proteins held in stoichiometric balance with each other, respond to calcium to coordinate contraction and relaxation of the heart. Altered sarcomeric structure-function underlies the primary basis of disease in multiple acquired and inherited heart disease states. Hypertrophic and restrictive cardiomyopathies are caused by inherited mutations in sarcomeric genes and result in altered contractility. Ischemia mediated acidosis directly alters sarcomere function resulting in decreased contractility. In this review, we highlight the use of acute genetic engineering of adult cardiac myocytes through stoichiometric replacement of sarcomeric proteins in these disease states with particular focus on cardiac troponin I. Stoichiometric replacement of disease causing mutations has been instrumental in defining the molecular mechanisms of hypertrophic and restrictive cardiomyopathy in a cellular context. In addition, taking advantage of stoichiometric replacement through gene therapy is discussed, highlighting the ischemia-resistant histidine-button, A164H cTnI. Stoichiometric replacement of sarcomeric proteins offers a potential gene therapy avenue to replace mutant proteins, alter sarcomeric responses to pathophysiologic insults or neutralize altered sarcomeric function in disease.
Keywords: acute genetic engineering, myofilament, troponin, calcium sensitivity, sarcomere, molecular dynamics, adult cardiac myocytes
Introduction
The sarcomere is the subcellular functional unit of cardiac muscle. A near-liquid crystalline array of overlapping thin and thick filaments forms the sarcomere. Cardiac contractility is driven by the highly regulated, cyclical interaction of thin and thick filament proteins. By mass, the primary unit of the thin filament is actin. Actin monomers polymerize into an elongated filament of double helical strands. Tropomyosin (Tm), an elongated protein that spans seven actin monomers, polymerizes head to tail along the actin filament and resides in the groove between actin strands. Associated with every Tm subunit is the troponin complex (Tn) in a 1:1 molar ratio. Cardiac troponin is composed of troponin C (cTnC), the Ca sensitive subunit, troponin I (cTnI), the inhibitory subunit, and troponin T (cTnT), the tropomyosin binding subunit. The thin filament has a 1:1:7 stoichiometry of troponin:tropomyosin:actin that is strictly maintained (Figure 1). The molecular motor, myosin is the predominate protein of the thick filament. Myosin heavy chain dimers with the associated regulatory light chains and essential light chains align with the giant protein, titin, and the phospho-protein myosin binding protein C to make up the thick filament (Gordon et al., 2000). Cardiac contraction initiates with an action potential, activating the voltage gated L-type calcium channel, allowing a small influx of calcium across the sarcolemmal membrane (Bers, 2002). This small amount of calcium initiates calcium-induced calcium release from the sarcoplasmic reticulum through the ryanodine receptor resulting in a rapid and marked rise in intracellular calcium. TnC binds calcium which causes alterations in thin filament protein interactions allowing myosin to form strong cross-bridges with actin (Farah and Reinach, 1995; Filatov et al., 1999; Gordon et al., 2000; Vinogradova et al., 2005; Galińska-Rakoczy et al., 2008; Lehman et al., 2009). Hydrolysis of ATP by myosin produces force, and, if load permits, the relative sliding of myofilaments to cause contraction. The subsequent release of calcium from TnC and re-sequestration of calcium into the SR allows for relaxation to occur (Gordon et al., 2000; Bers, 2002). This highly regulated process of excitation-contraction coupling occurs more than a billion times over an average human lifespan.
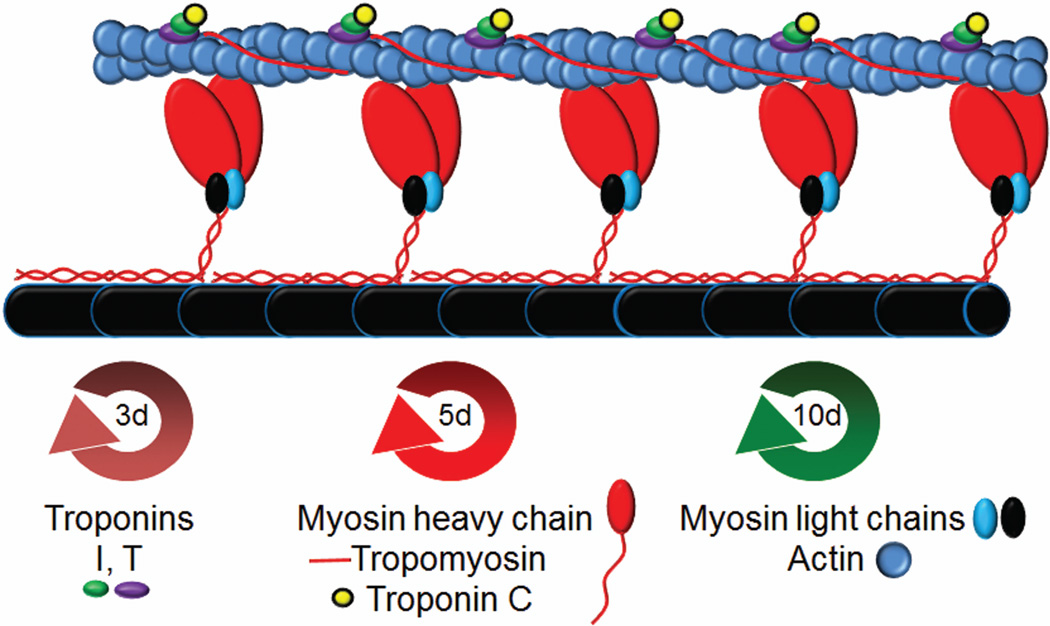
Figure 1.
Schematic representation of the structure of the cardiac sarcomere. The sarcomere is made up of the thin filament containing actin, troponin, and tropomyosin and the thick filament containing myosin, myosin light chains, and titin (black) along with myosin binding protein C (not depicted). Troponin interacts with tropomyosin and actin in 1:1:7 stochiometry. Myosin motor domains (red) interact with actin in a calcium dependent manner to produce force. Turnover half-lives of the depicted sarcomeric proteins are presented below (d=days).
Throughout the continuous, rhythmical beating of the heart the sarcomere maintains a lattice like architecture with strict stoichiometry. The sarcomere is often simplified to a static crystalline structure whereby its machine-like properties gives rise to force production and motion. This could not be further from the truth. The sarcomere is a highly dynamic multi-protein complex with constant alterations in protein-protein interactions, post-translational modifications and protein production/ turnover (Kleerekoper et al., 1995; Miki et al., 1998; Filatov et al., 1999; Dong et al., 2003; Takeda et al., 2003; Vinogradova et al., 2005; Lehman et al., 2009; Sanger et al., 2009). The ability of the myocyte to maintain contractility while replacing “old” or “damaged” sarcomeric proteins in a stoichiometric manner is amazing biology. While the mechanistic details by which this occurs are lacking new evidence suggests that the balance between chaperons and degradation factors, such as E3 ubiquitin ligases and calpains play a key role (Willis et al., 2009). It is likely that each of the sarcomeric proteins have unique signals and partners which determine its turnover and replacement.
Despite the absence of a clear mechanism, the regular turnover of sarcomeric proteins is well documented. The turnover rate for sarcomeric proteins in adult heart has been investigated using whole animal H3 leucine labeling experiments (Everett et al., 1981; Martin, 1981). These studies showed half-lives for troponin I and T ~3 days, troponin C, myosin and tropomyosin ~5.5 days, actin and myosin light chains ~7–10 days (Figure1). These experiments brought up many interesting questions which are still unresolved today. In the case of troponin, with troponin I and T having different half-lives compared to troponin C this suggests that TnC must recycle from “old” TnI-TnT to “new”. The same principle holds for myosin as the light chains which apparently must recycle from “old” to “new” in some regulated way. This also suggests that TnC and the myosin light chains have the ability to dissociate from their respective partners and could be in a more dynamic equilibrium with them than a static interaction. In vitro biochemistry experiments support the ability of TnC and the myosin light chains to dissociate from the sarcomere under certain conditions, albeit non-physiological (Ling et al., 1996; Preston et al., 2006).
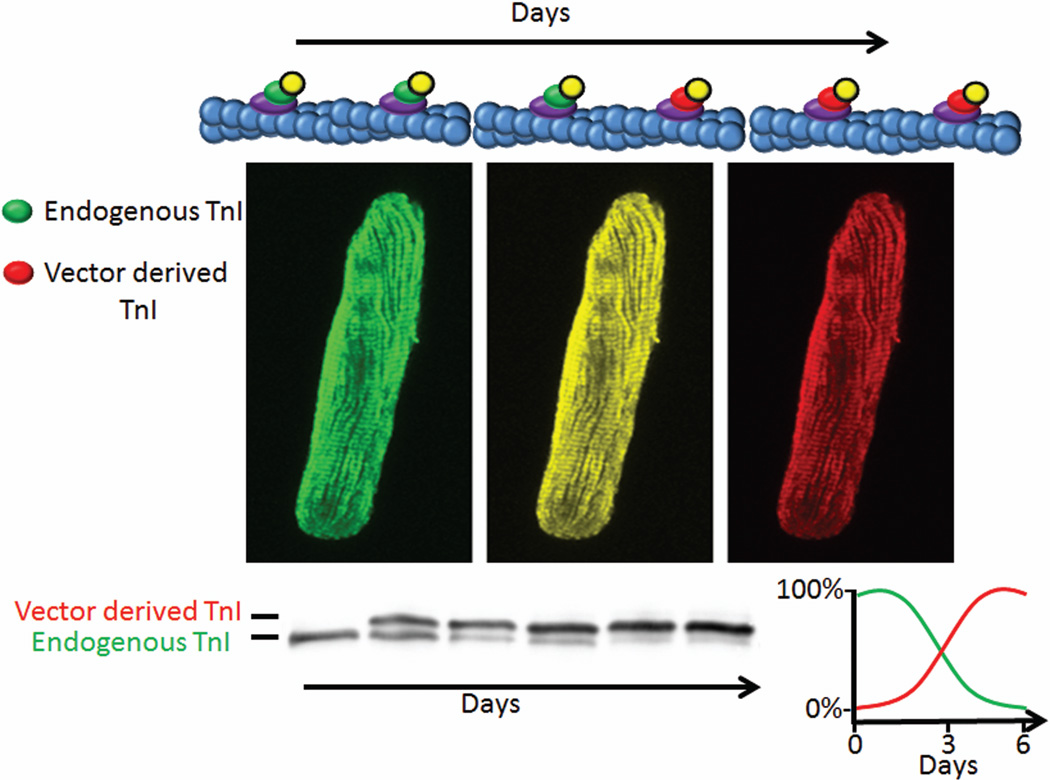
The use of adenovirus-mediated acute genetic engineering in adult cardiac myocytes has shed new light into the turnover and stoichiometry of the sarcomere. The principle of acute genetic engineering through adenoviral transduction of adult cardiac myocytes is depicted in figure 2. Treatment of adult cardiac myocytes with an adenovirus expressing an epitope tagged sarcomeric protein (i.e. TnI) results in a time dependent increase in expression and incorporation of the vector derived protein with a simultaneous stoichiometric decrease in the endogenous protein. This can be visualized through indirect immunofluorescence for proper incorporation and through Western blots which show shifted bands for the vector derived-epitope tagged protein for replacement. Many studies have confirmed stoichiometric replacement and proper incorporation into the sarcomere for myosin, TnI, TnT, TnC and Tm (Gulick et al., 1997; Michele et al., 1999; Rust et al., 1999; Tardiff et al., 1999; Robbins, 2000; Tardiff et al., 2000; Michele et al., 2002; Hernandez et al., 2005; Herron et al., 2007; Davis et al., 2008; Lim et al., 2008; Sadayappan et al., 2008).
Figure 2.
Guiding principles of acute genetic engineering of adult cardiac myocytes. Adenoviral expression of vector-derived cTnI-Flag (Red) results in a time dependent increase in replacement of the endogenous cTnI (Green). Myocyte images are artistic renderings for discussion purposes only. The pseudo colored change from green (endogenous cTnI) to red (vector derived cTnI-Flag) of the sarcomere are used to visually demonstrate incorporation as cTnI-Flag replaces the endogenous cTnI over time. Generic Western blot of cTnI shows replacement percent with the endogenous band decreasing over time with the vector derived-epitope tagged band increasing over time as it replaces endogenous TnI while total TnI is unchanged. For TnI, replacement reaches ~100% 6 days after adenoviral transduction.
Cell biology studies on incorporation
Adenoviral transduction of adult cardiac myocytes has been used to study the fundamental aspects of incorporation of thin filament proteins into the sarcomere. Early studies first showed stoichiometric replacement with ssTnI, the fetal TnI isoform in the heart (Westfall et al., 1997a). These studies showed replacement was time dependent and increased to nearly 100% replacement of cTnI with ssTnI, seven days after viral transduction. Permeabilization of the cells followed by Western blots showed the viral expressed protein is incorporated into the sarcomere and that there is no accumulation of expressed protein in the cytosol. This result can be interpreted as the endogenous cTnI turning over while the new vector derived protein competes for incorporation. The timing of replacement fits well with the documented half-life of cTnI of ~3 days. Transgenic (Tg) expression of sarcomeric proteins results in a similar replacement as is seen in the acute genetic engineering of adult cardiac myocytes (Robbins, 2000). Studies show that replacement can be from 10–90% (Ling et al., 1996; Gulick et al., 1997; Miki et al., 1998; Fentzke et al., 1999; Day et al., 2006; Sadayappan et al., 2008; Davis et al., 2012). Feng et al. probed the regulation of TnI incorporation through transgenesis (Feng et al., 2009). Homozygous knockout of cTnI is post-natal lethal at day ~19 (Liu et al., 2007). Transgenic rescue with an N-terminal truncated cTnI results is complete restoration of TnI. Through the analysis of multiple Tg lines with different mRNA expression of the transgene the authors were able to conclude that total TnI protein was unchanged from wild type mice. They also showed that heterozygous cTnI-KO mice have normal levels of cTnI protein in the face of 50% the mRNA level. These first studies help inform a stoichiometric system with defined kinetics that correlate well with the endogenous turnover of sarcomeric proteins. They also reveal that in the face of increased mRNA expression of TnI that the overall amount of TnI protein in the myocyte is constant. This suggests a limited amount of “parking spots” for TnI and competition for those spots is what dictates replacement. It also suggests that there are cellular mechanisms which limit the amount of unincorporated TnI in the cytosol.
Further studies helped to elucidate the spatial aspect to incorporation. Michele et al showed cTnI-Flag incorporates in a time dependent manner and through indirect immunofluorescences that incorporation is stochastic across the sarcomere (Michele et al., 1999). Through dual labeling experiments, using α-actinin as a Z-line marker, cTnI-Flag incorporated with even distribution between Z-lines. In time this staining became brighter and filled the area between Z-lines. In contrast, Flag-tropomyosin staining showed a restricted initiate pattern of incorporation, with staining at early time points being at the M-line and progressing towards the Z-lines with time. Thus tropomyosin has an ordered incorporation at the pointed-end of actin progressing towards the barbed end and Z-line. This study also documented that cTnI-flag replacement occurs faster that Flag-Tm consistent with the increased half-life of Tm protein of ~5days. The difference in incorporation suggests that thin filament proteins turnover with different temporal and spatial patterns. This points to specific partners and mechanisms of turnover for cTnI and Tm. Further studies are necessary to determine the mechanism of this difference and to reveal if all troponins incorporate in stochastic manner or if there is some ordered incorporation. In addition to revealing the spatial and temporal dependence of sarcomere turnover this study confirmed that function was unaltered by replacement of "self" proteins. The ability to replace sarcomeric proteins while maintaining contractile function suggests that the endogenous mechanisms of sarcomere maintenance are unaltered by adenoviral transduction of sarcomeric proteins.
Disease modeling through stoichiometric replacement
In addition to using acute genetic engineering to understand the turnover and stoichiometric replacement of sarcomeric proteins, this technique has been instrumental in understanding structure-function of sarcomeric proteins in disease. Hypertrophic and restrictive cardiomyopathies (HCM and RCM) are common inherited diseases of the sarcomere. Single amino acid mutations in thin and thick filament proteins are associated with HCM and RCM in an autosomal dominant manner (Seidman and Seidman, 2001; Teekakirikul et al., 2012). HCM is characterized by left ventricle hypertrophy, arrhythmias, and sudden cardiac death (Maron and Maron, 2013). More than 1000 mutations in 11 sarcomeric genes have been linked to HCM, with the highest prevalence being in β-myosin heavy chain, myosin binding protein C and cardiac troponin T (Lopes et al., 2013). Troponin was studied extensively early on due to its ability to be reconstituted and exchanged in permeabilized myofibers and its high turnover rate in intact cells which allows for high replacement in the time frame of viable adult cardiac myocytes in culture. Early myofibrillar ATPase assays along with calcium-force measurements in reconstituted myofibers indicated that cTnI HCM and RCM mutations result, in general, to increased calcium sensitivity of the sarcomere (Parvatiyar et al., 2010; Willott et al., 2010; Liu et al., 2012). RCM mutations tended to be hypersensitive compared to HCM mutations and were predicted to give rise to a more deleterious disease state. To fully elucidate the cellular implications of these mutations acute genetic engineering of adult cardiac myocytes with membrane intact functional measures of sarcomere shortening and calcium transient analysis were necessary. Davis et al. compared cTnI HCM and RCM mutations for replacement and cellular function (Davis et al., 2008). R145G, an HCM mutation, resulted in a limited contractile phenotype with slowed relaxation. This effect was replacement dependent, with increased phenotype with increased replacement. Two RCM mutations in cTnI, A172T and R193H showed decreased diastolic sarcomere length and very slow relaxation. Although the phenotypes of the HCM and RCM mutations mirror each other qualitatively, the RCM mutations quantitatively have hyper-phenotypes. This study also suggested that different mutations have altered ability to compete for replacement with wild type cTnI. Using unlabeled cTnI adenovirus with increasing dosage to compete with the Flag labeled mutant adenovirus showed that R145G competes less well with wild type cTnI while R193H has increased competition (Davis et al., 2007, 2008). These findings suggest that the disease phenotype is dependent on the replacement percentage of the mutant protein. In addition they suggest that the intrinsic alteration of calcium sensitivity does not fully determine the disease phenotype as replacement percentage can influence the cellular function. Further experiments with R193H cTnI transgenic mice showed similar dose-dependent functional alterations. R193H cTnI mice have ~10–15% R193H in two independent lines suggesting that the heart can’t tolerate higher levels of replacement (Du et al., 2008; Davis et al., 2012; Li et al., 2013). Indeed recent studies confirmed this through crossing R193H transgenics to the cTnI knockout. These mice have ~80% R193H cTnI incorporated in the sarcomere and die at post-natal day 30 (Li et al., 2013). This marked phenotype again suggests that the extent of replacement in addition to the intrinsic calcium sensitivity determines the disease phenotype. The ability to study cTnI mutations in the context of living cells has advanced our understanding of the genotype-phenotype relationship. In addition to the contractile phenotypes these studies also advanced the idea of calcium alterations in HCM. Calcium transient analysis for the HCM and RCM mutations showed increased diastolic calcium, decreased calcium transient amplitude and slow reuptake of calcium into the SR (Davis et al., 2007, 2008). All together this suggests that the heightened calcium sensitivity allows troponin to “buffer” calcium in the sarcomere and stay in a partially active state. This has been shown with cTnT HCM mutations as well and may be a common feature of HCM and RCM mutations (Knollmann et al., 2003; Haim et al., 2007; Guinto et al., 2009).
The ability to stochiometrically replace sarcomeric proteins has the potential as a therapy for HCM and RCM. As discussed above, the extent of replacement of the mutant polypeptide determines the phenotype. It follows that decreasing mutant peptide content in the sarcomere would be therapeutic. This could be through specifically targeting the transcript of the mutant allele for degradation through shRNA as was shown recently for a MHC mutation which causes HCM (Jiang et al., 2013). Another avenue is to out-compete the mutant protein through viral-mediated transduction and expression of the wild type protein (Davis and Metzger, 2010). Both avenues hold promise because the myocyte has the intrinsic ability to dictate the level of incorporated protein in the face of decreased or increased levels of transcripts by preservation of overall sarcomeric protein stoichiometry.
Therapeutic use of stoichiometric replacement
Altered sarcomeric function is a hallmark of many disease states, including ischemic cardiomyopathy and myocardial infarction. Ischemia causes an alteration of the intracellular milieu of the myocyte resulting in acidosis. This acidosis results in decreased contractility of the cardiac myocyte which a significant portion is due to calcium desensitization of the sarcomere (Orchard and Kentish, 1990). Early biochemical studies showed that the isoform of TnI is a critical determinate of acidosis-induced desensitization of the sarcomere (Metzger and Westfall, 2004). The fetal cardiac TnI, slow skeletal TnI (ssTnI), shows resiliency to acidosis in comparison to the adult isoform cTnI. Acute gene transfer experiments in adult cardiac myocytes with ssTnI results in stoichiometric replacement of cTnI with ssTnI (Westfall et al., 1997b). Cellular tension-calcium measurements showed that ssTnI confers increased calcium sensitivity at pH 7 and maintains calcium sensitivity at pH 6.2 in comparison to cTnI. Independently, an ssTnI transgenic mouse showed a similar gain in calcium sensitivity and pH resiliency (Fentzke et al., 1999; Wolska et al., 2001).
Chimeras of cTnI and ssTnI were used to decipher the critical domains which confer these attributes. cTnI has a unique N-terminal extension of 32 amino acids that has a critical PKA phosphorylation site necessary for β-adrenergic stimulated calcium desensitization and relaxation that is not present in ssTnI (Guo et al., 1994; Metzger and Westfall, 2004; Yasuda et al., 2007; Feng et al., 2009). Other than the N-terminal extension ssTnI and cTnI differ only in amino acid composition while having preserved domains (Palpant et al., 2010). Through the analysis of an N-cardiac/slow-C and an N-slow/cardiac-C in tension-pCa measurements it was revealed that the C-terminal portion of ssTnI confers protection from pH-mediated calcium desensitization (Westfall et al., 2000). Single amino acid changes between cTnI and ssTnI were analyzed through substitution of the ssTnI for the cTnI residue. These studies showed that a single residue, H132 in ssTnI confers protection from acidosis (Westfall and Metzger, 2007). cTnI has an alanine at this same position. In an effort to confer the pH resiliency to cTnI, Day et al made the opposite substitution, creating A164H cTnI (Day et al., 2006). A164H cTnI transgenic mice conferred protection from ischemia/reperfusion injury, hypoxia, and age-related cardiac dysfunction (Day et al., 2006; Palpant et al., 2008; Palpant et al., 2009). Importantly they also showed no deleterious effects under physiologic conditions. Cellular studies showed a resiliency to pH dependent decrease in tension-pCa measures as well as maintenance of contractility at acidic pH in membrane intact myocytes (Day et al., 2006; Palpant et al., 2012). The cellular studies also showed that unlike ssTnI, A164H cTnI did not increase tension-pCa measurements at physiologic pH. This was confirmed in contractility measures in membrane intact cells (Palpant et al., 2012). This finding suggests that A164H is a titratable molecular inotrope which behaves like cTnI under normal physiologic conditions but reacts to pathophysiologic conditions such as acidosis to maintain contractile function. This is an important distinction between ssTnI and A164H cTnI. As discussed above in HCM most of the mutations result in increased calcium sensitivity of the sarcomere much like ssTnI. In accordance, ssTnI transgenic hearts show significant diastolic dysfunction at baseline similar to HCM mutant mice (Fentzke et al., 1999). Additionally, A164H cTnI maintains the N-terminal extension and responds like wild type cTnI to β-adrenergic stimulation. Thus A164H cTnI, by its titratable function has the attributes to be a viable therapy for ischemia induced disease states such as ischemic cardiomyopathy.
Mechanistic detail as to how an engineered histidine button in cTnI can confer protection under acidic conditions comes from a combination of cellular functional studies with molecular dynamic (MD) simulations. Histidine is a unique amino acid in that its pKa for protonation is in the physiologic range. At normal physiologic pH most histidines are deprotonated while a slight shift to acidic conditions will result in increased protonation of a histidine. Codon A164 is located next to the switch peptide of cTnI. The switch region of TnI interacts with cTnC N-terminal domain in a calcium dependent manner (Li et al., 1999). cTnC binds calcium which partially opens a hydrophobic patch, the switch region of TnI interacts with the hydrophobic patch inducing the full opening of the hydrophobic patch. This interaction is modeled to pull the inhibitory region of TnI away from tropomyosin allowing myosin strong binding sites on actin to be revealed (Gordon et al., 2000). Thus cTnI switch region binding to cTnC allows contraction to occur. Protonation of the histidine in close proximity to cTnC lead to the hypothesis that the histidine could interact with an acidic residue on cTnC.
MD simulations of sTnI (a.a.115–141) from the chicken fast skeletal troponin crystal structure (PDB 1YTZ)(Vinogradova et al., 2005) or A164H cTnI (a.a.148–174) from the cardiac troponin crystal structure (PDB 1J1E)(Takeda et al., 2003) bound to cTnC (a.a.1–90) provided evidence for a histidine (cognate codon 164 in cTnI) to glutamic acid (codon 19 in cTnC) interaction in acidic conditions. The sTnI simulations showed H132 had an electrostatic interaction with cTnC E19 (Palpant et al., 2010). Substitution of sTnI with the cTnI residue H132A showed this electrostatic interaction is eliminated and sTnI moves away from cTnC. In agreement, cTnI shows no interaction around A164 with cTnC and adopts a significantly different conformation than sTnI. Simulations with A164H cTnI showed the potential for a similar electrostatic interaction between H164 and E19 of cTnC (Palpant et al., 2012). NMR studies using similar portions of sTnI and A164H bound to cTnC in acidic conditions support the potential for a histidine to E19 interaction (Pineda-Sanabria et al., 2012; Robertson et al., 2012). The NMR studies also indicate that there is increased affinity of sTnI and A164H for cTnC under acidic conditions. These results in combination with the cellular functional studies suggest that under acidic conditions H164 is protonated, increasing TnI to cTnC affinity through the electrostatic interaction of H164 with E19 which in turn increases contractile function. Further evidence supports this electrostatic interaction as a determinate of contractile function. An engineered cTnI A164R was used to mimic the charge on H164 during acidosis (Palpant et al., 2012). A164R MD simulations showed R164 to E19 cTnC interaction is present and overall its confirmation is similar to sTnI and A164H. In adult cardiac myocytes A164R results in increased tension-pCa calcium sensitivity and increased contractility in live cells under physiologic pH conditions. Under acidic conditions A164R has similar function as A164H. These studies support the idea that A164H titratable gain in function is due to an electrostatic interaction between H164 and E19 of cTnC, increasing TnI-TnC affinity and thereby altering contractility in acidotic conditions.
The titratable gain-in-function of A164H cTnI under pathophysiologic stimuli while maintaining normal function under physiologic conditions makes it an attractive potential therapeutic strategy. In addition, due to the cell intrinsic capacity of stoichiometric replacement, “molecular torture” due to extraneous protein production or accumulation is not a concern. This makes A164H cTnI a viable gene therapy for conditions which result in ischemia, whether transient or chronic.
Sarcomeric replacement offers a suitable system to exploit for therapeutic intervention in diseases of the sarcomere. HCM, RCM and ischemia result in alterations of sarcomere function as the prime mechanism of the disease phenotype. As such, therapeutic intervention at the level of the sarcomere is necessary to alleviate the disease phenotype. Whether this is through gene therapy, taking advantage of the stoichiometric replacement of sarcomeric proteins or through small molecules targeting the sarcomere, this therapeutic avenue needs to be explored. Acute genetic engineering of adult cardiac mocytes offers a viable system to test both therapeutic avenues as a disease model and test bed for beneficial engineered sarcomeric proteins.
Acknowledgements
We thank Joshua Martindale, Anthony Vetter and members of the Metzger lab for helpful discussions and editing of the manuscript. We thank the Minnesota Supercomputing Institute for computational resources. This work was supported in part by NIH (BRT (T32) and JMM).
Literature Cited
- Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- Davis J, Metzger JM. Combinatorial Effects of Double Cardiomyopathy Mutant Alleles in Rodent Myocytes: A Predictive Cellular Model of Myofilament Dysregulation in Disease. PloS one. 2010;5:e9140. doi: 10.1371/journal.pone.0009140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J, Wen H, Edwards T, Metzger JM. Thin filament disinhibition by restrictive cardiomyopathy mutant R193H troponin I induces Ca2+-independent mechanical tone and acute myocyte remodeling. Circ Res. 2007;100:1494–1502. doi: 10.1161/01.RES.0000268412.34364.50. [DOI] [PubMed] [Google Scholar]
- Davis J, Wen H, Edwards T, Metzger JM. Allele and species dependent contractile defects by restrictive and hypertrophic cardiomyopathy-linked troponin I mutants. Journal of molecular and cellular cardiology. 2008;44:891–904. doi: 10.1016/j.yjmcc.2008.02.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J, Yasuda S, Palpant NJ, Martindale J, Stevenson T, Converso K, Metzger JM. Diastolic dysfunction and thin filament dysregulation resulting from excitation-contraction uncoupling in a mouse model of restrictive cardiomyopathy. Journal of molecular and cellular cardiology. 2012;53:446–457. doi: 10.1016/j.yjmcc.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day SM, Westfall MV, Fomicheva EV, Hoyer K, Yasuda S, La Cross NC, D'Alecy LG, Ingwall JS, Metzger JM. Histidine button engineered into cardiac troponin I protects the ischemic and failing heart. Nat Med. 2006;12:181–189. doi: 10.1038/nm1346. [DOI] [PubMed] [Google Scholar]
- Dong WJ, Robinson JM, Stagg S, Xing J, Cheung HC. Ca2+-induced conformational transition in the inhibitory and regulatory regions of cardiac troponin I. J Biol Chem. 2003;278:8686–8692. doi: 10.1074/jbc.M212886200. [DOI] [PubMed] [Google Scholar]
- Du J, Liu J, Feng H-Z, Hossain MM, Gobara N, Zhang C, Li Y, Jean-Charles P-Y, Jin J-P, Huang X-P. Impaired relaxation is the main manifestation in transgenic mice expressing a restrictive cardiomyopathy mutation, R193H, in cardiac TnI. American Journal of Physiology - Heart and Circulatory Physiology. 2008;294:H2604–H2613. doi: 10.1152/ajpheart.91506.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett AW, Prior G, Zak R. Equilibration of leucine between the plasma compartment and leucyl-tRNA in the heart, and turnover of cardiac myosin heavy chain. The Biochemical journal. 1981;194:365–368. doi: 10.1042/bj1940365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah CS, Reinach FC. The troponin complex and regulation of muscle contraction. The FASEB Journal. 1995;9:755–767. doi: 10.1096/fasebj.9.9.7601340. [DOI] [PubMed] [Google Scholar]
- Feng H-Z, Hossain MM, Huang X-P, Jin JP. Myofilament incorporation determines the stoichiometry of troponin I in transgenic expression and the rescue of a null mutation. Archives of Biochemistry and Biophysics. 2009;487:36–41. doi: 10.1016/j.abb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fentzke RC, Buck SH, Patel JR, Lin H, Wolska BM, Stojanovic MO, Martin AF, Solaro RJ, Moss RL, Leiden JM. Impaired cardiomyocyte relaxation and diastolic function in transgenic mice expressing slow skeletal troponin I in the heart. The Journal of physiology. 1999;517(Pt 1):143–157. doi: 10.1111/j.1469-7793.1999.0143z.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov VL, Katrukha AG, Bulargina TV, Gusev NB. Troponin: structure, properties, and mechanism of functioning. Biochemistry (Mosc) 1999;64:969–985. [PubMed] [Google Scholar]
- Galińska-Rakoczy A, Engel P, Xu C, Jung H, Craig R, Tobacman LS, Lehman W. Structural Basis for the Regulation of Muscle Contraction by Troponin and Tropomyosin. Journal of Molecular Biology. 2008;379:929–935. doi: 10.1016/j.jmb.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AM, Homsher E, Regnier M. Regulation of Contraction in Striated Muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- Guinto PJ, Haim TE, Dowell-Martino CC, Sibinga N, Tardiff JC. Temporal and mutation-specific alterations in Ca2+ homeostasis differentially determine the progression of cTnT-related cardiomyopathies in murine models. American Journal of Physiology - Heart and Circulatory Physiology. 2009;297:H614–H626. doi: 10.1152/ajpheart.01143.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick J, Hewett TE, Klevitsky R, Buck SH, Moss RL, Robbins J. Transgenic Remodeling of the Regulatory Myosin Light Chains in the Mammalian Heart. Circulation Research. 1997;80:655–664. doi: 10.1161/01.res.80.5.655. [DOI] [PubMed] [Google Scholar]
- Guo X, Wattanapermpool J, Palmiter KA, Murphy AM, Solaro RJ. Mutagenesis of cardiac troponin I. Role of the unique NH2-terminal peptide in myofilament activation. J Biol Chem. 1994;269:15210–15216. [PubMed] [Google Scholar]
- Haim TE, Dowell C, Diamanti T, Scheuer J, Tardiff JC. Independent FHC-related cardiac troponin T mutations exhibit specific alterations in myocellular contractility and calcium kinetics. Journal of molecular and cellular cardiology. 2007;42:1098–1110. doi: 10.1016/j.yjmcc.2007.03.906. [DOI] [PubMed] [Google Scholar]
- Hernandez OM, Szczesna-Cordary D, Knollmann BC, Miller T, Bell M, Zhao J, Sirenko SG, Diaz Z, Guzman G, Xu Y, Wang Y, Kerrick WGL, Potter JD. F110I and R278C Troponin T Mutations That Cause Familial Hypertrophic Cardiomyopathy Affect Muscle Contraction in Transgenic Mice and Reconstituted Human Cardiac Fibers. Journal of Biological Chemistry. 2005;280:37183–37194. doi: 10.1074/jbc.M508114200. [DOI] [PubMed] [Google Scholar]
- Herron TJ, Vandenboom R, Fomicheva E, Mundada L, Edwards T, Metzger JM. Calcium-independent negative inotropy by beta-myosin heavy chain gene transfer in cardiac myocytes. Circ Res. 2007;100:1182–1190. doi: 10.1161/01.RES.0000264102.00706.4e. [DOI] [PubMed] [Google Scholar]
- Jiang J, Wakimoto H, Seidman JG, Seidman CE. Allele-Specific Silencing of Mutant Myh6 Transcripts in Mice Suppresses Hypertrophic Cardiomyopathy. Science. 2013;342:111–114. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerekoper Q, Howarth JW, Guo X, Solaro RJ, Rosevear PR. Cardiac troponin I induced conformational changes in cardiac troponin C as monitored by NMR using site-directed spin and isotope labeling. Biochemistry. 1995;34:13343–13352. doi: 10.1021/bi00041a010. [DOI] [PubMed] [Google Scholar]
- Knollmann BC, Kirchhof P, Sirenko SG, Degen H, Greene AE, Schober T, Mackow JC, Fabritz L, Potter JD, Morad M. Familial hypertrophic cardiomyopathy-linked mutant troponin T causes stress-induced ventricular tachycardia and Ca2+-dependent action potential remodeling. Circ Res. 2003;92:428–436. doi: 10.1161/01.RES.0000059562.91384.1A. [DOI] [PubMed] [Google Scholar]
- Lehman W, Galińska-Rakoczy A, Hatch V, Tobacman LS, Craig R. Structural Basis for the Activation of Muscle Contraction by Troponin and Tropomyosin. Journal of Molecular Biology. 2009;388:673–681. doi: 10.1016/j.jmb.2009.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MX, Spyracopoulos L, Sykes BD. Binding of cardiac troponin-I147–163 induces a structural opening in human cardiac troponin-C. Biochemistry. 1999;38:8289–8298. doi: 10.1021/bi9901679. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang L, Jean-Charles P-Y, Nan C, Chen G, Tian J, Jin JP, Gelb IJ, Huang X. Dose-dependent diastolic dysfunction and early death in a mouse model with cardiac troponin mutations. Journal of molecular and cellular cardiology. 2013;62:227–236. doi: 10.1016/j.yjmcc.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CC, Yang H, Yang M, Wang C-K, Shi J, Berg EA, Pimentel DR, Gwathmey JK, Hajjar RJ, Helmes M, Costello CE, Huo S, Liao R. A Novel Mutant Cardiac Troponin C Disrupts Molecular Motions Critical for Calcium Binding Affinity and Cardiomyocyte Contractility. Biophysical journal. 2008;94:3577–3589. doi: 10.1529/biophysj.107.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling N, Shrimpton C, Sleep J, Kendrick-Jones J, Irving M. Fluorescent probes of the orientation of myosin regulatory light chains in relaxed, rigor, and contracting muscle. Biophysical journal. 1996;70:1836–1846. doi: 10.1016/S0006-3495(96)79749-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Tikunova SB, Kline KP, Siddiqui JK, Davis JP. Disease-Related Cardiac Troponins Alter Thin Filament Ca<sup>2+</sup>Association and Dissociation Rates. PloS one. 2012;7:e38259. doi: 10.1371/journal.pone.0038259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Du J, Zhang C, Walker JW, Huang X. Progressive troponin I loss impairs cardiac relaxation and causes heart failure in mice. American Journal of Physiology - Heart and Circulatory Physiology. 2007;293:H1273–H1281. doi: 10.1152/ajpheart.01379.2006. [DOI] [PubMed] [Google Scholar]
- Lopes LR, Rahman MS, Elliott PM. A systematic review and meta-analysis of genotype–phenotype associations in patients with hypertrophic cardiomyopathy caused by sarcomeric protein mutations. Heart. 2013 doi: 10.1136/heartjnl-2013-303939. [DOI] [PubMed] [Google Scholar]
- Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet. 2013;381:242–255. doi: 10.1016/S0140-6736(12)60397-3. [DOI] [PubMed] [Google Scholar]
- Martin AF. Turnover of cardiac troponin subunits. Kinetic evidence for a precursor pool of troponin-I. Journal of Biological Chemistry. 1981;256:964–968. [PubMed] [Google Scholar]
- Metzger JM, Westfall MV. Covalent and noncovalent modification of thin filament action: the essential role of troponin in cardiac muscle regulation. Circ Res. 2004;94:146–158. doi: 10.1161/01.RES.0000110083.17024.60. [DOI] [PubMed] [Google Scholar]
- Michele DE, Albayya FP, Metzger JM. Thin filament protein dynamics in fully differentiated adult cardiac myocytes: toward a model of sarcomere maintenance. J Cell Biol. 1999;145:1483–1495. doi: 10.1083/jcb.145.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michele DE, Gomez CA, Hong KE, Westfall MV, Metzger JM. Cardiac dysfunction in hypertrophic cardiomyopathy mutant tropomyosin mice is transgene-dependent, hypertrophy-independent, and improved by beta-blockade. Circ Res. 2002;91:255–262. doi: 10.1161/01.res.0000027530.58419.82. [DOI] [PubMed] [Google Scholar]
- Miki M, Kobayashi T, Kimura H, Hagiwara A, Hai H, Maeda Y. Ca2+-induced distance change between points on actin and troponin in skeletal muscle thin filaments estimated by fluorescence energy transfer spectroscopy. J Biochem. 1998;123:324–331. doi: 10.1093/oxfordjournals.jbchem.a021940. [DOI] [PubMed] [Google Scholar]
- Orchard CH, Kentish JC. Effects of changes of pH on the contractile function of cardiac muscle. Am J Physiol. 1990;258:C967–C981. doi: 10.1152/ajpcell.1990.258.6.C967. [DOI] [PubMed] [Google Scholar]
- Palpant NJ, D'Alecy LG, Metzger JM. Single histidine button in cardiac troponin I sustains heart performance in response to severe hypercapnic respiratory acidosis in vivo. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:1529–1540. doi: 10.1096/fj.08-121996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant NJ, Day SM, Herron TJ, Converso KL, Metzger JM. Single histidine-substituted cardiac troponin I confers protection from age-related systolic and diastolic dysfunction. Cardiovascular research. 2008;80:209–218. doi: 10.1093/cvr/cvn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant NJ, Houang EM, Delport W, Hastings KE, Onufriev AV, Sham YY, Metzger JM. Pathogenic peptide deviations support a model of adaptive evolution of chordate cardiac performance by troponin mutations. Physiological genomics. 2010;42:287–299. doi: 10.1152/physiolgenomics.00033.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant NJ, Houang EM, Sham YY, Metzger JM. pH-responsive titratable inotropic performance of histidine-modified cardiac troponin I. Biophysical journal. 2012;102:1570–1579. doi: 10.1016/j.bpj.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvatiyar MS, Pinto JR, Dweck D, Potter JD. Cardiac Troponin Mutations and Restrictive Cardiomyopathy. Journal of Biomedicine and Biotechnology. 2010 doi: 10.1155/2010/350706. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda-Sanabria SE, Robertson IM, Li MX, Sykes BD. Interaction between the regulatory domain of cardiac troponin C and the acidosis-resistant cardiac troponin I A162H. Cardiovascular research. 2012 doi: 10.1093/cvr/cvs348. [DOI] [PubMed] [Google Scholar]
- Preston L, Watkins H, Redwood C. A revised method of troponin exchange in permeabilised cardiac trabeculae using vanadate: functional consequences of a HCM-causing mutation in troponin I. J Muscle Res Cell Motil. 2006;27:585–590. doi: 10.1007/s10974-006-9079-0. [DOI] [PubMed] [Google Scholar]
- Robbins J. Remodeling the Cardiac Sarcomere Using Transgenesis. Annual Review of Physiology. 2000;62:261–287. doi: 10.1146/annurev.physiol.62.1.261. [DOI] [PubMed] [Google Scholar]
- Robertson IM, Holmes PC, Li MX, Pineda-Sanabria SE, Baryshnikova OK, Sykes BD. Elucidation of isoform-dependent pH sensitivity of troponin i by NMR spectroscopy. J Biol Chem. 2012;287:4996–5007. doi: 10.1074/jbc.M111.301499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust EM, Albayya FP, Metzger JM. Identification of a contractile deficit in adult cardiac myocytes expressing hypertrophic cardiomyopathy-associated mutant troponin T proteins. J Clin Invest. 1999;103:1459–1467. doi: 10.1172/JCI6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadayappan S, Finley N, Howarth JW, Osinska H, Klevitsky R, Lorenz JN, Rosevear PR, Robbins J. Role of the acidic N′ region of cardiac troponin I in regulating myocardial function. The FASEB Journal. 2008;22:1246–1257. doi: 10.1096/fj.07-9458com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JW, Wang J, Holloway B, Du A, Sanger JM. Myofibrillogenesis in skeletal muscle cells in zebrafish. Cell Motility and the Cytoskeleton. 2009;66:556–566. doi: 10.1002/cm.20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman JG, Seidman C. The Genetic Basis for Cardiomyopathy: from Mutation Identification to Mechanistic Paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- Tardiff JC, Hewett TE, Factor SM, Vikstrom KL, Robbins J, Leinwand LA. Expression of the β (slow)-isoform of MHC in the adult mouse heart causes dominant-negative functional effects. American Journal of Physiology - Heart and Circulatory Physiology. 2000;278:H412–H419. doi: 10.1152/ajpheart.2000.278.2.H412. [DOI] [PubMed] [Google Scholar]
- Tardiff JC, Hewett TE, Palmer BM, Olsson C, Factor SM, Moore RL, Robbins J, Leinwand LA. Cardiac troponin T mutations result in allele-specific phenotypes in a mouse model for hypertrophic cardiomyopathy. The Journal of Clinical Investigation. 1999;104:469–481. doi: 10.1172/JCI6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teekakirikul P, Padera RF, Seidman JG, Seidman CE. Hypertrophic cardiomyopathy: Translating cellular cross talk into therapeutics. The Journal of Cell Biology. 2012;199:417–421. doi: 10.1083/jcb.201207033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ. Ca(2+)-regulated structural changes in troponin. Proc Natl Acad Sci U S A. 2005;102:5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall MV, Albayya FP, Turner II, Metzger JM. Chimera analysis of troponin I domains that influence Ca(2+)-activated myofilament tension in adult cardiac myocytes. Circ Res. 2000;86:470–477. doi: 10.1161/01.res.86.4.470. [DOI] [PubMed] [Google Scholar]
- Westfall MV, Metzger JM. Single amino acid substitutions define isoform-specific effects of troponin I on myofilament Ca2+ and pH sensitivity. Journal of molecular and cellular cardiology. 2007;43:107–118. doi: 10.1016/j.yjmcc.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall MV, Rust EM, Albayya F, Metzger JM. Adenovirus-mediated myofilament gene transfer into adult cardiac myocytes. Methods Cell Biol. 1997a;52:307–322. [PubMed] [Google Scholar]
- Westfall MV, Rust EM, Metzger JM. Slow skeletal troponin I gene transfer, expression, and myofilament incorporation enhances adult cardiac myocyte contractile function. Proc Natl Acad Sci U S A. 1997b;94:5444–5449. doi: 10.1073/pnas.94.10.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Schisler JC, Portbury AL, Patterson C. Build it up-Tear it down: protein quality control in the cardiac sarcomere. Cardiovascular research. 2009;81:439–448. doi: 10.1093/cvr/cvn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willott RH, Gomes AV, Chang AN, Parvatiyar MS, Pinto JR, Potter JD. Mutations in Troponin that cause HCM, DCM AND RCM: What can we learn about thin filament function? Journal of molecular and cellular cardiology. 2010;48:882–892. doi: 10.1016/j.yjmcc.2009.10.031. [DOI] [PubMed] [Google Scholar]
- Wolska BM, Vijayan K, Arteaga GM, Konhilas JP, Phillips RM, Kim R, Naya T, Leiden JM, Martin AF, de Tombe PP, Solaro RJ. Expression of slow skeletal troponin I in adult transgenic mouse heart muscle reduces the force decline observed during acidic conditions. The Journal of physiology. 2001;536:863–870. doi: 10.1111/j.1469-7793.2001.00863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S-i, Coutu P, Sadayappan S, Robbins J, Metzger JM. Cardiac transgenic and gene transfer strategies converge to support an important role for troponin I in regulating relaxation in cardiac myocytes. Circ Res. 2007;101:377–386. doi: 10.1161/CIRCRESAHA.106.145557. [DOI] [PubMed] [Google Scholar]