Abstract
The regulation of cell cycle rate is essential for the correct timing of proliferation and differentiation during development. Changes to cell cycle rate can have profound effects on the size, shape and cell types of a developing organ. We previously identified a zebrafish mutant ceylon (cey) that has a severe reduction in T cells and hematopoietic stem/progenitor cells (HSPCs). Here we find that the cey phenotype is due to absence of the gene transducin (beta)-like 3 (tbl3). The tbl3 homologue in yeast regulates the cell cycle by maintaining rRNA levels and preventing p53-induced cell death. Zebrafish tbl3 is maternally expressed, but later in development its expression is restricted to specific tissues. Tissues expressing tbl3 are severely reduced in cey mutants, including HSPCs, the retina, exocrine pancreas, intestine, and jaw cartilage. Specification of these tissues is normal, suggesting the reduced size is due to a reduced number of differentiated cells. Tbl3 MO injection into either wild-type or p53-/- mutant embryos phenocopies cey, indicating that loss of tbl3 causes specific defects in cey. Progression of both hematopoietic and retinal development is delayed beginning at 3 days post fertilization due to a slowing of the cell cycle. In contrast to yeast, reduction of Tbl3 causes a slowing of the cell cycle without a corresponding increase in p53 induced cell death. These data suggest that tbl3 plays a tissue-specific role regulating cell cycle rate during development.
Keywords: cell cycle, p53, T cells, retina, tbl3, zebrafish
Introduction
Cell proliferation and differentiation are tightly linked processes. While parts of the proliferative process such as timing of cell cycle exit, cell cycle rate, and number of cells generated are specific to individual tissue types, all proliferating cell types need to exit the cell cycle in order to differentiate. This is particularly important during development. If proliferation is reduced, too few cells are generated, resulting in smaller tissues, lethality or diseases such as mircrophthalmia, anemia or immune deficiencies. (Agathocleous and Harris, 2009; Chasis and Mohandas, 2008; Fibach, 2011; Green et al., 2003; Kondo et al., 2003; Ng et al., 2009; Rhodes et al., 2008; Seita and Weissman, 2010; Uribe and Gross, 2010). Conversely, if overproliferation occurs and too many cells are generated, larger tissues or malignancies such as leukemia or retinoblastoma may ensue (Aifantis et al., 2008; Becker and Jordan, 2011; MacPherson et al., 2004; Mullighan and Downing, 2009; Seita and Weissman, 2010; Zhang et al., 2004). Moreover, a change in cell cycle speed during development can generate progenitors in an environment that may not be prepared to promote differentiation, as in the case of ectopic germ cells that degenerate into teratomas or testicular tumors (Bolande, 1979). These data reveal the importance of the cell cycle regulation for proper cellular differentiation and development.
Hematopoiesis requires tight control of the cell cycle to generate the required type and number of blood cells. During development, hematopoiesis occurs in two general waves that are subject to tight temporo-spatial regulation in mammals: primitive and definitive hematopoiesis (Orkin and Zon, 2008). Similarly, detailed analysis revealed the importance of timing and hematopoietic stem/progenitor cell (HSPC) environment in determining the blood cell types produced during zebrafish development (Ellett and Lieschke, 2010; Paik and Zon, 2010). Mutagenesis screens for hematopoietic defects in zebrafish frequently pinpointed mutants with dysregulated proliferation. For example, a mutation in the RNA helicase dead-box 18 (ddx18) results in anemia and neutropenia through p53-mediated G1 arrest and cell death (Payne et al., 2011). In addition, zebrafish models of Diamond-Blackfan anemia have been established demonstrating that loss of specific ribosomal proteins leads to loss of HSPCs and erythrocytes due to upregulation of p53 and cell cycle arrest (Danilova et al., 2008, 2011; Taylor and Zon, 2011; Uechi et al., 2008). These data underscore the importance of cell cycle control during hematopoiesis and suggest that further investigation into specific roles for genes that govern cell cycle speed and progression will be important for full understanding of hematopoietic development.
Another tissue where cell cycle control is essential for proper development is the retina (Agathocleous and Harris, 2009; Bilitou and Ohnuma, 2010; Dyer and Cepko, 2001; Livesey and Cepko, 2001). Hematopoiesis and retinal development are distinct processes that occur in very different environments. However, both processes begin with a stem cell population that generates varied types of tissue-specific differentiated cells. Both require tight regulation of the cell cycle to generate the differentiated cell type(s) needed at a particular stage of development. Retinal progenitor cells (RPCs) produce ganglion cells, amacrine cells, bipolar cells, horizontal cells, cones, rods and Müller glia. These retinal cell types are born in a particular order that is influenced by the environment, but is most highly regulated by intrinsic cues (Livesey and Cepko, 2001). In fact, it seems that environmental cues primarily regulate the number of cells generated by an RPC and have little influence over the types of retinal cells that an RPC can make at a particular time (Austin et al., 1995; Belliveau and Cepko, 1999; Belliveau et al., 2000; Cepko et al., 1996; Jusuf et al., 2011). Cell cycle timing is an important component regulating RPCs during development. For example, in both zebrafish and rat RPCs cycle is short during the early, proliferative phase of retinal development, but as development proceeds the cell cycle lengthens mostly through a slowing of S-phase (Alexiades and Cepko, 1996; Li et al., 2000). Mutations in genes that are regulators of the cell cycle during retinal development have underscored the importance of the cell cycle for eye formation. For example, mutations that disrupt de novo purine synthesis result in cell cycle exit defects and microphthalmia (Ng et al., 2009) and members of the nucleolar GTP-binding protein family are required for correct timing of cell cycle exit and differentiation (Paridaen et al., 2011). These data demonstrate the importance of cell cycle rate in regulating the number of retinal cells generated during development.
Here we study the zebrafish mutant ceylon (cey) that was identified in a screen for defective T cell development (Trede et al., 2008). In addition to the T cell defect, cey mutants have defects in HSPCs, retina, cartilage, exocrine pancreas, and the intestine. These tissues are specified properly, but the number of differentiated cells is severely reduced. We find that the defects in cey are due to absence of the gene transducin (beta)-like 3 (tbl3) whose yeast homologue, utp13, has previously been implicated in rRNA biogenesis (Dosil and Bustelo, 2004; Dragon et al., 2002; Watkins et al., 2004). Loss of tbl3 in zebrafish causes a slowing of the cell cycle during tissue differentiation that is independent of p53 and cell death. These are the first data to implicate tbl3 in vertebrate development and suggest that tbl3 is required tissue specifically to regulate cell cycle rate during tissue growth.
Methods
Fish Husbandry
Zebrafish were bred and maintained using standard methods. The cey mutant was identified in a previously characterized early pressure screen (Trede et al., 2008). Fish were maintained on the WIK background for breeding. Mutants were also generated from a cey+/−CD41::eGFP+/+ (Lin et al., 2005) cross on the WIK/AB background and a cey+/− lck::eGFP+/+ (Langenau et al., 2004) cross on the WIK background. Mutants were identified between 3-5 dpf by their malformed jaw, small eye and/or genotyping. Homozygous p53I166T mutants (Parant et al., 2010) were incrossed for morpholino injections.
Mapping
cey linkage to chromosome 3 was described previously (Trede et al., 2008). For fine mapping cey heterozygotes were mated to wild-type Tü individuals. Resulting cey WIK/Tü hybrid individuals were crossed and resulting mutants and wild-type siblings were used for mapping as previously described (Trede et al., 2007).
In situ hybridization
Whole mount in situ hybridization was carried out as previously described (Trede et al., 2008). The tbl3 probe was generated from the MGC clone zgc:101778 (NM_001007402.1). The pME18S-FL3 plasmid was PCR amplified with pME_F (5′ – attgatttaggtgacactatagaacttctgctctaaaagctgcg – 3′) and pME_R (5′ – gtaatacgactcactatagggccgacctgcagctcgagcaca – 3′) primers containing Sp6 or T7 sequence respectively. T7 polymerase was used to generate antisense RNA. Riboprobes were also generated from the following plasmids: vsx2 (Batista et al., 2008), trypsin (Biemar et al., 2001), ifabp (Mudumana et al., 2004), c-myb (Thompson et al., 1998), insulin (Milewski et al., 1998), dlx2a (Akimenko et al., 1994), and nkx2.3 (Lee et al., 1996).
Immunohistochemistry
Alcian blue staining was performed as previously described (Laguerre et al., 2005; Trede et al., 2008). For antibody stains larvae at all stages were fixed overnight in 4% paraformaldehyde (PFA) with 1x PBS plus 0.1% tween-20 (PBST) at 4° C. Rabbit anti -p Histone H3 (Ser 10) from Santa Cruz Biotechnology (#sc-8656-R) was used at 1:500 diluted in block (1 x PBS, 0.5% triton x-100, and 1 x casein) and incubated overnight at 4°C. After washes in 1 x PBS + 0.1% tween-20, larvae were incubated overnight at 4°C in goat anti-rabbit Alexa-488 (#A11008) or Alexa 555 (# A21428) and TO-PRO-3 iodide (#T3605) from Invitrogen Molecular Probes diluted 1:1000 in block. Embryos were then washed and imaged using DIC light or fluorescence microscopy when ready. All antibodies were stored in 50% glycerol at −20°C.
The Roche In Situ Cell Death Detection Kit, TMR red (# 12 156 792 91) was used to label apoptotic cells with TUNEL. Larvae were fixed for 1 hour at room temperature (RT) in 4% PFA with 1 x PBS plus 0.1% triton x-100. Larvae were treated with Invitrogen proteinase K diluted 1:1000 in PBST for either 10 minutes (3 dpf) or 20 minutes (4 and 5 dpf) at RT then fixed for 10 minutes in 4% PFA +PBST. Larvae were incubated for 1 hour at 37°C in TUNEL solution then washed 4 times in PBST.
BrdU labeling was completed using10mM of 5-BrdU from Sigma (# B9285) as described previously (Laguerre et al., 2005) with the following modifications. Larvae were treated with BrdU for 30 minutes on ice at 72 hpf then recovered either 1, 2, 4 or 6 hours at 28°C in fish water. All larvae were fixed overnight at 4°C in 4% PFA and 1xPBST. Larvae were then stored in 100% MeOH until ready to use. Larvae were washed out of 100% MeOH and permeabilized with proteinase K as described above for TUNEL staining. To remove pigment larvae were bleached in 0.8% KOH, 0.9% H2O2, and 0.0125% tween-20 for 20-30 minutes at RT until the pigment was barely visible. Then larvae were washed four times in PBST and fixed for 10 minutes in 4% PFA. Larvae were washed two times in dH2O and then 1 hour at RT in 2N HCl to break open the nuclei. Larvae were washed out of HCl, and BrdU incorporation was revealed with mouse anti-BrdU IgG1,k at 1:400 from BD Biosciences (# 347580) as the primary antibody and goat anti-mouse Alexa 488 at 1:1000 from Invitrogen (#A-11001) as described above for anti-pH3 staining.
Morpholino injections
Morpholinos were designed to the translational start site of tbl3 (tbl3 ATG, 5′-TGAAGAGCAGCGTTTTCCCAGCCAT-3′) and to the donor site of exon 5 (tbl3 SB, 5′-GAATATAATCCAGCCTTCACCTGTA-3′). While injection of either tbl3 ATG or tbl3 SB MO phenocopied the cey mutant phenotype at 4 dpf, the ATG phenotype was more pronounced as early as 2 dpf. 1 nl of either ATG or SB MO was injected into the yolk of 1-cell stage *AB or p53−/− embryos at a 2.5 ng/nl concentration. The p53 morpholino was described previously (Robu et al., 2007). Embryos were raised at 28°C in E3 plus methyl blue until 4 dpf at which point they were fixed overnight at 4°C in 4% PFA. Embryos were then stored in 100% MeOH at −20°C until ready to use.
Northern Blot
Probes used for Northern blotting were generated according to Azuma et al., 2006. Total RNA was isolated from 4 dpf zebrafish using an RNeasy mini kit (QIAGEN). Northern analysis was performed as described previously (DeHart et al., 2005).
Imaging
A Nikon SMZ1000 stereo microscope with a reflected light illumination source was used to take live larvae images. A Nikon Eclipse E600 compound microscope with DIC was used to take all fixed brightfield images. A Spot Insight camera was used on either the dissecting microscope or compound microscope to take images. Photoshop was used to adjust brightness and contrast, as well as adjust the colors using variations on brightfield images. All photoshop maniupulations were done simultaneously for compared samples. An Olympus FV 1000 or Zeiss LSM 710 confocal was used to image fixed fluorescent tissues. Larvae were mounted on coverslips in 1% low-melt agarose to hold them in place during imaging. Confocal images were taken with 512 × 512 pixel resolution, 40 x magnification, with 2μm steps. Photoshop was used to adjust brightness and contrast of the fluorescent images.
Data quantification
Eye size was measured using dorsal (dorsal/ventral or medial/lateral measurements) or lateral (anterior/posterior measurements) view confocal z-sections. The center of the lens was identified by moving 10μm medially from the edge of the lens. The ImageJ line tool was then used to measure the number of pixels on the axis to be measured. The number of pixels was converted to μm using the conversion factor 100μm = 162 pixels. Standard deviation was calculated to determine variation and standard error of the mean. A student’s t-test determined the statistical significance of the data.
The average number of nuclei per section of a retina was calculated by counting the number of TO-PRO 3 stained nuclei in five confocal z – sections per eye. Dorsal view images were used. The five z - sections began at the dorsal edge of the lens and were analyzed every 10μm for 50μm of depth covered. The number of nuclei per section was then averaged and used as an N of 1. This method was also employed to calculate the number of nuclei per retinal layer. Standard deviation of the averages was used to determine the variation of the samples and the standard error of the mean. A student’s t-test determined the significance of the data. In addition, ANOVA analysis was completed on the entire data set of averages to further calculate the significance of the data.
The MO results were quantified by counting three separate clutches injected with tbl3SB MO for each type of analysis. Larvae were analyzed at 4 dpf either live by looking for a malformed jaw or fixed by looking for a reduction in either c-myb, trypsin, or ifabp staining. Then the percentage of larvae with wild-type characteristics was calculated.
Anti-pH3 positive cells in the caudal hematopoietic tissue (CHT) region and retina were counted by z-projecting z-stacks of lateral view tails or dorsal view retinas and then using the count function in ImageJ to count each anti-pH3 positive cell. We defined the CHT region as the vascular region ventral to the notochord and 5 somites wide starting at the first somite boundary posterior of the anus. The anti-pH3 cells were counted in the entire retina, excluding the lens. Standard deviation was calculated to determine variation and standard error of the mean. A student’s t-test determined the statistical significance of the sample.
To quantify S-phase cells in mutant and wild-type larvae the number of anti-BrdU positive cells in 5 sections, 20μm apart was counted, starting at the edge of the lens in lateral view 72 hpf retinas with 1 hour recovery post BrdU treatment. The average number of anti-BrdU positive cells per 5 sections per retina was calculated.
The percentage of anti-pH3 positive cells co-labeled with BrdU was determined by using ImageJ to count the number of anti-pH3 and co-labeled cells in 10 sections, 10μm apart, per retina. Counting began at the lateral edge of the lens in lateral view retinas. Excel was used to determine the percentage of anti-pH3 positive cells that were co-labeled with BrdU. Standard deviation was used to calculate the standard error of the mean and student’s t-test was used to determine the significance of the data.
The average number of TUNEL positive cells was calculated as described above for the average number of anti-pH3 positive cells, except lateral view z-stacks of retinas stained with TUNEL were used.
Results
Differentiated tissues in cey mutants are reduced
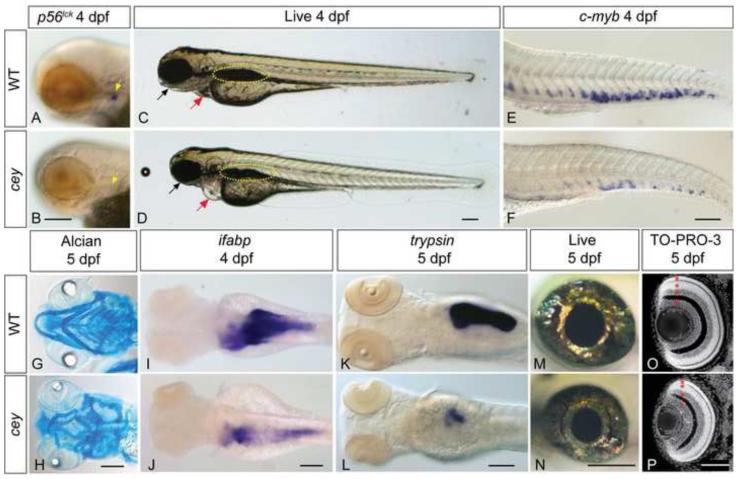
The cey mutant was previously identified in an early pressure screen for mutants with a severe reduction of T cells (Trede et al., 2008), evidenced by absent p56lck staining at 4 days post fertilization (dpf; Figure 1A, B, yellow arrows). cey mutants are morphologically indistinct from wild-type at 2 dpf, but have a malformed jaw, smaller eyes and slight cardiac edema by 3 dpf which becomes more pronounced by 4 dpf (black and red arrows Figure 1C, D, data not shown). The defects in cey are recessive, including the lack of swim bladder, visible by 4 dpf (yellow dotted circle Figure 1C, D), and homozygous mutants die by 7 dpf. Consistent with the severe reduction of T cells (Figure 1A, B) cey mutants have a severe reduction in HSPCs labeled with c-myb at 4 dpf (Figure 1E, F). Surprisingly, normal numbers of HSPCs are seen at 2 dpf, (Supplementary Figure 1A, B), suggesting the loss of T cells is secondary to a defect in definitive hematopoiesis.
Figure 1. Many tissue types are severely reduced in cey mutants.
All figures are oriented anterior to the left. p56lck labels T cells in 4 dpf zebrafish larvae (A, yellow arrow). Cey mutants have severely reduced p56lck expression at 4 dpf (B, yellow arrow) suggesting a lack of T cells. In comparison to live wild-type larvae at 4 dpf (C), live cey mutants (D) have small eyes, heart edema (red arrow), and a deformed jaw (black arrow). Yellow dotted line designates the swim bladder (C, D). Although the tail is morphologically normal, there is a severe reduction of HSPCs in cey mutants at 4 dpf as shown by the severe reduction of c-myb positive cells in cey mutants (F) as compared to wild-type (E). Alcian blue staining indicates the mandibular and hyoid arches are smaller and malformed in cey mutants at 5 dpf (H) as compared to wild-type (G). Endodermal derivatives are severely reduced in cey mutants (I - L). Intestinal tissue labeled with ifabp is severely reduced in cey mutants at 4 dpf (I, J). In addition, exocrine pancreas labeled by trypsin is severely reduced in mutants at 5 dpf (K, L). The eyes of cey mutants are smaller than wild-type eyes at 5 dpf (M, N). Despite the smaller eye size, all cell layers are present in cey mutant retinas (O, P). Red asterisks label each retinal layer (O, P). Scale bars are 130 μm (A-L) and 95 μm (M-P).
Similar to the hematopoietic defect, other tissues affected in cey have normal specification at 2 dpf but reduced tissue mass at 3 dpf and thereafter. For example, there is severe reduction and malformation of jaw cartilage including a loss of all branchial arches in cey mutants at 5 dpf (Figure 1G, H). However, tissues that give rise to the jaw cartilage such as neural crest and endoderm are specified normally (Supplementary Figure 1C - F). Normal gata6 staining in cey mutants at 3 dpf indicates that specification of endodermal tissues is also unaffected (Supplementary Figure 1G, H). Differentiated cells in the intestine labeled with ifabp and exocrine pancreas labeled with trypsin are reduced at 4 and 5 dpf respectively (Figure1I - L). By contrast, beta cells in the endocrine pancreas labeled by insulin are normal in cey mutants (Supplementary Figure1I, J). Mutant eyes have normal pigmentation and shape but are smaller than wild-type eyes (Figure 1M, N). Despite the small eye size all retinal layers are present in cey mutants (Figure 1O, P). This suggests that, similar to the other tissue types, the different retinal cell types are properly specified and reduction of eye size is not due to loss of a retinal layer. Taken together, tissue specification is intact in cey mutants, while differentiation into organs is defective.
cey mutants have a telomeric deletion that includes the gene tbl3
We fine-mapped the lesion in cey mutants and identified a deletion at the right arm telomere of chromosome 3 that includes the gene transducin (beta)-like 3 (tbl3) (Figure 2A - C). According to the annotation on the zebrafish genome assembly zV9 sox9b is located proximal to the deletion (Figure 2A – C) and only four genes other than tbl3 are also included in the deletion (Figure 2A). Radiation hybrid mapping of the tbl3 gene indicates that it is located on the very end of the telomere (Figure 2A, data not shown). None of the five genes known to be present in the deletion (Figure 2A) are well characterized in zebrafish.
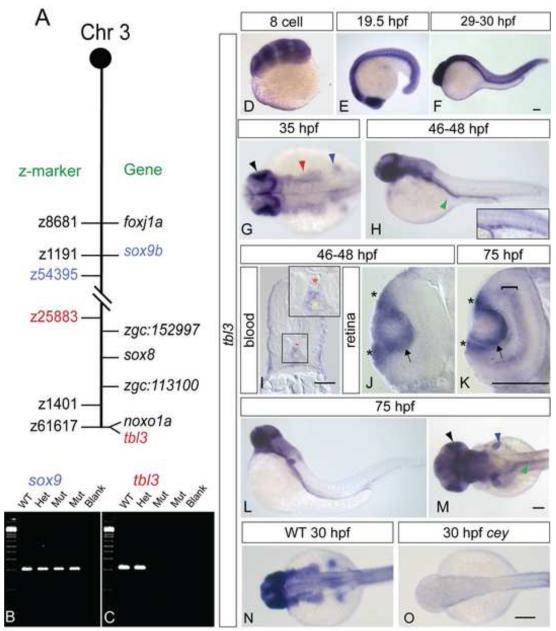
Figure 2. tbl3 is specifically expressed in wild-type and lost in cey mutants.
Mapping the cey lesion revealed a telomeric deletion on chromosome 3 (A, not to scale). Double dashed lines indicate the approximate location of the deletion. Z-markers are listed on the left of the chromosome schematic and genes are on the right. The gene sox9b is excluded from the deletion (B), while tbl3 is included in the deletion (C). Radiation hybrid mapping indicates that tbl3 is located on the very end of the telomere on chromosome 3 and is closely linked to the noxo1a (GenBank ID: NM_001077584.1) gene (A). tbl3 is maternally expressed at the 8-cell stage (D). By 19.5 hpf tbl3 is still widely expressed in most tissues with high expression in the eye (E). tbl3 is expressed in most tissues between 29-30 hpf (F). At 35 hpf tbl3 expression is high in the eye and brain (E) and by 46-48 hpf (F) expression is present in the intestinal region (green arrowhead) and the HSPCs (inset – larvae in inset is different than larvae in the panel). Cross sections confirm that tbl3 is expressed between the dorsal aorta (red asterisk) and the caudal vein (yellow asterisk) at 45-48 hpf where HSPCs are found (I). Cross sections of the retina at 45-48 hpf (J) and 75 hpf (K) show that tbl3 is expressed around the lens (arrow) and in the retinal stem cell region, also known as the ciliary marginal zone (CMZ; asterisks) at the lateral edge of the retina adjacent to the lens. At 75 hpf (K) tbl3 is also expressed in the inner nuclear layer (bracket). tbl3 expression remains high in the eye (black), brain, fin bud (blue), and gut (green) at 75 hpf (L, M). Consistent with the loss of the tbl3 gene in cey, tbl3 expression is absent in cey mutant embryos (N, O). Accession numbers for other genes on the chromosome in (A) are as follows: foxj1a (GenBank ID: NM_001076706.2), zgc:152997 (GenBank ID: NM_001077319.1), sox8 (GenBank ID: NM_001025465.1), and zgc:113100 (GenBank ID: NM_001031844.1). Scale bars are 60 μm (I), 130 μm (D - H, J – O), and 245μm (N, O).
Contrary to the other genes in the cey deletion, tbl3’s expression pattern is both dynamic and specific in all tissues affected in cey mutants suggesting that loss of tbl3 could cause the defects observed in cey (Figure 2D – M). Tbl3 is maternally expressed (Figure 2D) and continues to be maintained at high levels in most tissues during early embryonic development (Figure 2E, F). From 35 to 75 hours post fertilization (hpf) tbl3 is expressed strongly in the eye, hindbrain, the location of the neural crest (which gives rise to the jaw cartilage), and the gut primordium (Figure 2G - M). In particular, tbl3 is expressed in the same region as HSPCs at 48 and 75 hpf, correlating with the defect in definitive hematopoiesis in cey mutants (Figure 2H inset, H, I). The specific expression in the eye and brain continues until at least 4 dpf (data not shown).
Sections of the eye show that tbl3 is expressed in the presumptive ciliary marginal zone (CMZ) that contains retinal stem cells (asterisks Figure 2J, K), and in tissue surrounding the lens at 48 and 75 hpf (arrows Figure 2J, K). tbl3 is also expressed in the inner nuclear layer at 75 hpf, coinciding with expression of vsx2, an RPC marker (bracket Figure 2J, K; Figure 5C). These data indicate that temporo-spatial expression of tbl3 is initially widespread before it becomes restricted to defined cell types. The cell types expressing tbl3 give rise to the structures affected in cey mutants. As expected, tbl3 expression is undetectable in cey mutants (Figure 2N, O) further in keeping with the presence of tbl3 in the deletion.
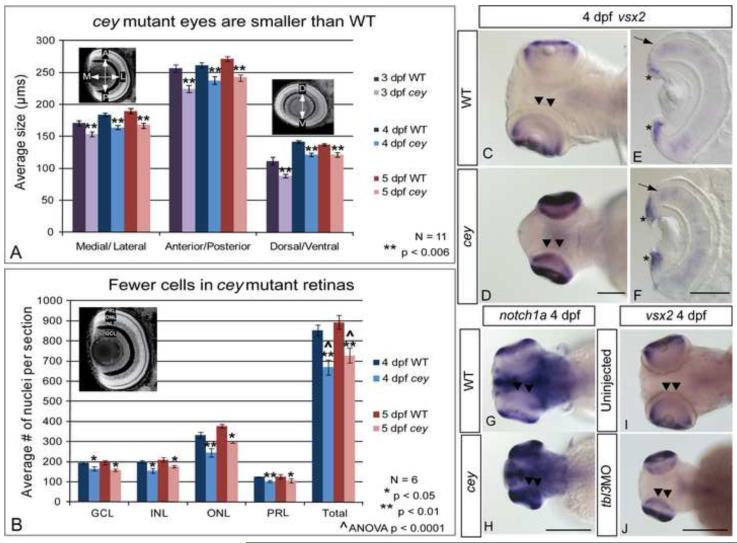
Figure 5. RPC markers are expanded in central retina despite smaller eyes and fewer cells in cey mutants.
The eyes of cey mutants are smaller than wild-type eyes by 3 dpf in the medial/lateral, anterior/posterior and dorsal/ventral axes (A). This difference is maintained at all time points (A). In addition, the number of cells in each layer is significantly reduced in cey mutants (B), resulting in a reduction of eye size. The number of cells in each layer was estimated by averaging number of TO-PRO-3 stained nuclei in five z-sections 10 μm apart in six different larvae (see Materials and Methods). Retinal stem cell markers are maintained longer in the central retina (black arrowheads) of cey mutants than in wild-types (C-H). vsx2 and notch1a are expressed in the peripheral retina of the wild-type 4 dpf retina (C, G), but are expressed throughout the central (black arrowheads) and peripheral retina in cey (D, H). Cross sections of 4 dpf retinas expressing vsx2 show that vsx2 is expressed in more cells in the CMZ region of mutant retinas (asterisks) and throughout the outer nuclear layer (black arrow E, F). The small eye and expansion of vsx2 expression into the central retina (black arrowheads) is phenocopied in tbl3 morphants at 4 dpf (I, J). The abbreviations in (B) are as follows: ganglion cell layer (GCL), inner nuclear cell layer (INL), outer nuclear cell layer (ONL), photoreceptor layer (PRL). The error bars in (A) and (B) represent the standard error of the mean. Scale bars are130 m.
Loss of tbl3 phenocopies cey mutants
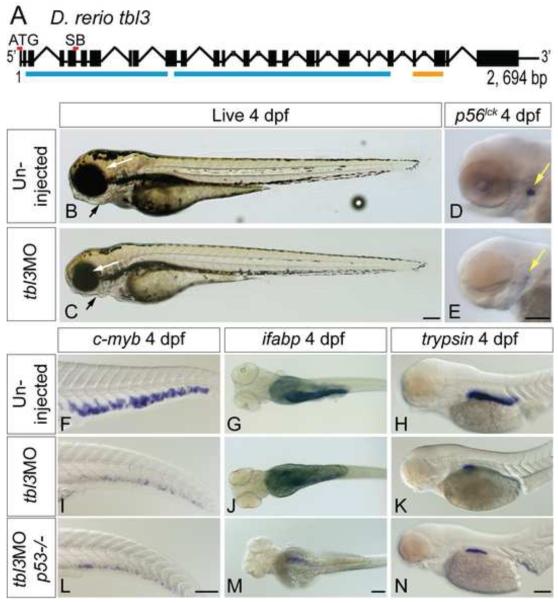
Tbl3 is a highly conserved WD-repeat protein whose C terminus contains a domain homologous to yeast Utp13 (Supplementary Figure 2). Yeast Utp13 has been implicated in regulation of 18S rRNA biogenesis (Dosil and Bustelo, 2004; Dragon et al., 2002; Tschochner and Hurt, 2003), suggesting a potential role for Tbl3 in nucleolar 18S rRNA processing. Maternal expression suggests a potential housekeeping function for zebrafish Tbl3 (Figure 2D). However, restricted expression later in development indicates zebrafish Tbl3 may also have more specific functions (Figure 2G-M). To test if absence of tbl3 in mutant embryos explains the cey phenotype, we injected either a translation blocking anti-sense morpholino (ATG MO) or a splice blocking morpholino (SB MO Figure 3A) into wild-type embryos to disrupt Tbl3 protein expression. Reverse transcriptase PCR confirmed that tbl3 SB MO disrupts splicing (data not shown). Reduction of Tbl3 protein using either the ATG or the SB MO results in a phenocopy of cey mutants at 4 dpf (Figure 3B, C). Tbl3 morphant larvae have small eyes (white arrows), slight heart edema and malformed jaws (black arrows) similar to cey mutants (Figure 3B, C; Supplementary Figure 3A). T cells, HSPCs, the exocrine pancreas and intestinal cells are also severely reduced in tbl3 morphants at 4 dpf, matching the cey mutant phenotype (Figure 3D – K; Supplementary Figure 3B - D). Injection of tbl3MO into p53−/− mutants also results in a phenocopy of both cey mutants and tbl3 morphants (Figure 3L - N; Supplementary Figure 3A - D). These data indicate that the tbl3MO phenotype is specific and p53 independent. The tbl3 morphant phenocopy of cey mutants, together with the loss of genomic tbl3 DNA, the specific tbl3 expression pattern, and the loss of tbl3 expression in cey mutants, suggests that the defects seen in cey are due to the loss of tbl3. Therefore, according to the current zebrafish nomenclature guidelines, we assign cey the name Df(Chr03)tbl3cz26, but for simplicity we will continue to refer to the mutant as cey.
Figure 3. tbl3MO phenocopies the cey mutant.
The tbl3 gene is 2,694 base pairs and 25 exons long (A). Black boxes represent exons and lines joining boxes are introns. Lines with hash marks indicate introns of unknown length. Red lines above exons represent the location of the MO sequence used in this study. Blue lines represent WD repeats. Orange line represents UTP13 domain (A). SB MO is directed against the splice donor site at the 3′ end of exon 5 (A). Injection of MOs designed against the tbl3 gene results in a phenocopy of the cey mutant phenotype at 4 dpf. tbl3 morphant 4 dpf larvae have malformed jaws (black arrows), small eyes (white arrows; B, C) and a reduction of T cells labeled by p56lck (D, E) as compared to uninjected controls. HSPCs labeled by c-myb (F, I), intestinal cells labeled by ifabp (G, J), and exocrine pancreas labeled by trypsin (H, K) are all severely reduced as compared to controls and similar to cey mutants (Figure 1). Images of larvae labeled with trypsin are pictures of the right side of the embryo reflected so that anterior is to the right (H, K, N). Injection of the tbl3MO into p53−/− mutants resulted in similar phenotypes (L – N) as compared to injection of tbl3MO into wild-type (I – K). Scale bars are 130μm.
Reduction of HSPCs in cey mutants leads to a loss of T cell and red blood cells
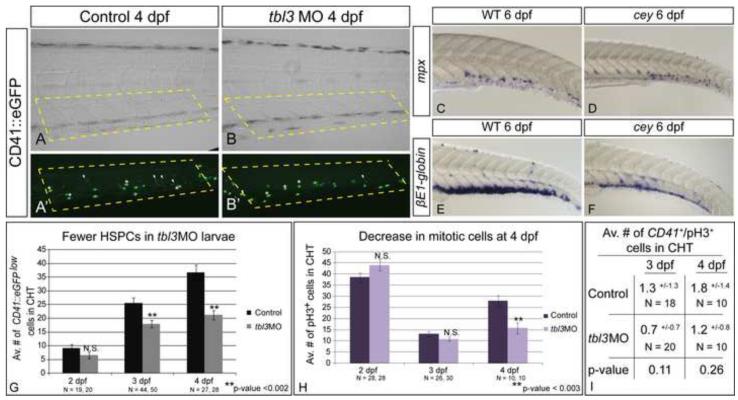
The thymus is a specialized hematopoietic tissue and is indispensable for T cell development (Carpenter and Bosselut, 2010; Yang et al., 2010). Surprisingly, despite the presence of foxn1 labeling the thymus, T cells are severely reduced at 4 dpf in cey mutants (Trede et al., 2008). This suggests that the loss of HSPCs (Figure 1E, F) that give rise to T cell progenitors may be responsible for the reduction of T cells in cey mutants (Kissa et al., 2008). Previous data indicated that CD41::eGFPlow cells in the caudal hematopoietic tissue (CHT) are HSPCs (Murayama et al., 2006). Since the first T cells reach the thymus between 2 and 3 dpf (Kissa et al., 2008; Trede et al., 2001; Willett et al., 1999) we quantified CD41::eGFPlow cells in the CHT in control and tbl3 morphants at 2, 3 and 4 dpf to determine if decreased numbers of T cells correlate with loss of HSPCs (Figure 4A, A’, B, B’,G; white arrowheads indicate HSPCs). Consistent with normal c-myb expression at 2 dpf (Supplementary Figure 1A, B), there is no significant difference between the number of CD41::eGFPlow HSPCs at 2dpf in tbl3 morphants and controls (Figure 4G). However, by 3 dpf there is a marked decrease in the number of HSPCs in tbl3 morphants as compared to controls (Figure 4G). The difference between control and morphant larvae is even more severe at 4 dpf (Figure 4A, A’, B, B’, G). These data show that the loss of HSPCs begins between 2 and 3 dpf, correlating with the generation of the first T cells in wild-type larvae (Kissa et al., 2008; Murayama et al., 2006).
Figure 4. Loss of HSPCs differentially affects hematopoietic lineages.
CD41::eGFPlow labels HSPCs (A, A’ white arrowheads) while CD41::eGFPhigh labels thrombocytes (A, A’ white asterisks) in the CHT of 4 dpf larvae. HSPCs (white arrowheads) are severely reduced in tbl3 morphants at 4 dpf (B, B’) as compared to uninjected wild-type controls (A, A’). In contrast, thrombocytes in tbl3 morphants are unaffected (A, A’, B, B’). There are 24.9+/− 10.2 thrombocytes in the CHT in wild-type (N = 28 larvae), and 29+/−11.8 in the CHT of cey (N = 28) which is an insignificant difference (p-value = 0.16). Brightfield images in (A) and (B) are the same larvae as (A’) and (B’) respectively. Dotted yellow boxes indicate area in CHT counted in G. Neutrophils labeled by mpx are unaffected in cey larvae (D) at 6 dpf as compared to wild-type (C), while red blood cells labeled by βE1-globin at 6 dpf are severely reduced in cey (E, F). The average number of HSPCs in the CHT of tbl3 morphants is normal at 2 dpf, but significantly reduced at 3 and 4 dpf (G). The average number of mitotic cells in the CHT of tbl3 morphant larvae is not significantly affected at 2 or 3 dpf, but is severely reduced at 4 dpf (H). The number of CD41::eGFPlow+/pH3+ cells is not significantly changed in the CHT of tbl3 morphant larvae at 3 or 4 dpf (I). The area of the CHT counted in (G - I) is labeled by the yellow dotted lines in (A), (A’), (B) and (B’). The error bars in (G) and (H) represent standard error of the mean. The error in (I) represents standard deviation. Scale bars are 130μm.
The loss of HSPCs led us to hypothesize that other blood cell types should also be reduced in cey mutants. To our surprise, thrombocytes, identified by high levels of CD41::eGFP, are normal at 4 dpf in tbl3 morphants (Figure 4A, A’, B, B’, white asterisks label representative thrombocytes – see figure legend for quantification). Neutrophils are also unaffected in cey mutants showing little change in numbers even by 6 dpf (Figure 4C, D). By contrast, at 6 dpf erythrocytes labeled by βE1-globin expression are severely reduced in cey mutants (Figure 4E, F). Therefore, despite global reduction in HSPCs some blood cell types, such as thrombocytes and neutrophils, are less affected than erythrocytes and T cells when Tbl3 is reduced.
While the number of HSPCs in the CHT of tbl3 morphants is markedly lower than in wild-type larvae, it continues to increase from 2 to 4 dpf showing that new HSPCs are being generated during that time, albeit at a lower rate than wild-type (Figure 4G). In addition while T cells and red blood cells are severely reduced, they are not completely lost, even by 6 dpf (Figure 4F, data not shown). This suggests that the remaining HSPCs are still actively cycling. To test this possibility, we quantified the number of mitotic cells in the CHT labeled by the phosphohistone H3 antibody (anti-pH3), a marker for late G2 and M phase of the cell cycle (Hendzel et al., 1997; Sansam et al., 2006). In contrast to the number of HSPCs, the number of mitotic cells in tbl3 morphants remains normal until 4 dpf when they markedly decrease (Figure 4H). To address more directly the possibility that the remaining HSPCs are actively cycling we counted the number of CD41::eGFPlow+/pH3+ co-labeled cells. While only a few co-labeled cells were present in either control or tbl3 morphants, there was no significant change in the number of co-labeled cells in the CHT at 3 or 4 dpf (Figure 4I), suggesting that the remaining HSPCs are indeed still proliferating.
Prolonged maintenance of RPC markers in cey mutants
Similar to the CHT, the retina is another tissue where tbl3 is expressed in a stem cell region and has developmental defects in cey mutants. Corresponding to the reduction of blood cell numbers, the eye size of cey mutants is smaller than in wild-type siblings (Figure 5A). Both the lens and the retina are visibly smaller in cey mutants as early as 3 dpf and remain smaller until they die at 7 dpf (Figure 1M - P; Figure 5A, data not shown). While the mutant eyes are consistently smaller than wild-type they continue to grow in size in all axes until at least 5 dpf (Figure 5A). The continued growth of the mutant eyes is similar to the continuous production of HSPCs and blood cells, suggesting slower, but not arrested growth.
Despite the continuous growth of the eye, 5 dpf mutant eyes are still smaller than wild-type eyes at 3 dpf in both the medial/lateral and anterior/posterior axes (Figure 5A). To determine whether the reduction in eye size was due to smaller or fewer cells we counted the number of nuclei in sections of wild-type and mutant eyes to obtain an estimate of the number of cells in each eye and each retinal layer. We found that cey mutant eyes have fewer cells than wild-type in all retinal layers (Figure 5B). In addition, despite having a reduced total cell number, cells continue to be generated between day 4 and 5 dpf in cey mutants, correlating with the sustained growth of mutant eyes (Figure 5B). These data indicate that the small eye phenotype is due to a reduced production of retinal cells in cey mutants and mirror the reduction of HSPCs and blood cells seen in the CHT of mutant larvae.
Proliferation is high in zebrafish retinas at 3 dpf and then drops precipitously at 4 dpf (Marcus et al., 1999). In addition, while mitosis occurs in both the peripheral and the central retina at 2 and 3 dpf (Baye and Link, 2007; Hu and Easter, 1999), by 4 dpf it is primarily restricted to the peripheral CMZ region. In wild-type zebrafish there is a significant increase in proliferation and expansion of the retinal progenitor cell (RPC) population in the central retina at 3 dpf that results in a large increase in eye size. This is resolved by 4 dpf when RPC markers and proliferation becomes restricted to the peripheral retina (Marcus et al., 1999). Expression of RPC markers vsx2, notch1a, and pax6a in cey mutants is maintained in the central retina until 4 dpf, longer than in wild-type (black arrowheads; Figure 5C - H, data not shown). The RPC phenotype is due to loss of tbl3 as injection of tbl3MO into wild-type and p53−/− larvae phenocopies the vsx2 expansion in cey mutants at 4 dpf (Figure 5I, J; Supplementary Figure 3E). By 5 dpf the expression of vsx2 and notch1a in mutant retinas is restricted to the peripheral eye, similar to wild-type (data not shown). The prolonged maintenance of RPC markers in the central retina in cey mutants (Figure 5C – H; data not shown) suggests that this proliferative phase of retinal development is protracted in mutants compared to wild-type.
Increased proliferation in retinas with reduced Tbl3
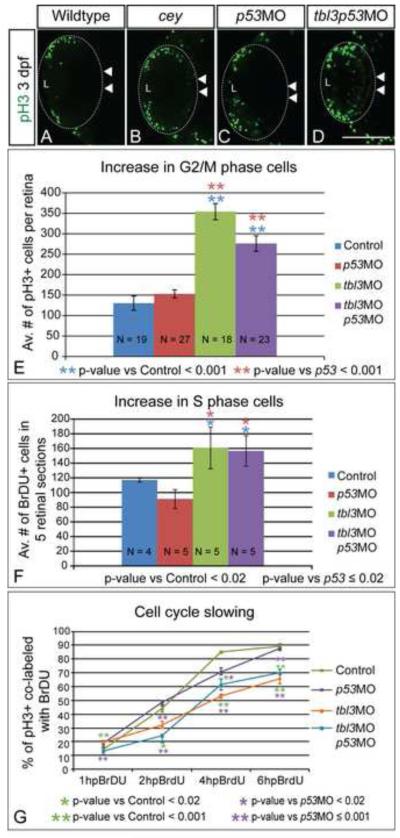
Since the RPC markers are maintained in the central retina of cey mutants at 4 dpf we predicted a corresponding increase in proliferative markers in the absence of Tbl3. To test this prediction we labeled G2/M cells with anti-pH3 and counted the number of pH3+ cells in each retina (Figure 6A - E). The number of G2/M cells was significantly increased in cey and tbl3 morphant larvae at 3 dpf (Figure 6A - E; Supplementary Figure 4E). Larvae co-injected with tbl3 and p53MO had a significant increase in G2/M cells, indicating that the defect is due to loss of Tbl3, independent of p53 (Figure 6A - E). In addition to their increased number, pH3+ cells were found throughout both the central and peripheral retina of tbl3 morphants and cey mutants at 3 and 4 dpf (white arrowheads; Figure 6A – D; Supplementary Figure 4A, B). Consistent with the expression of RPC markers, pH3 labeling became restricted to a more peripheral region by 5 dpf in mutant larvae (Supplementary Figure 4C, D). The localization of G2/M cells in the central retina correlates with the prolonged maintenance of stem cell markers in the central retina beyond age-appropriate stages. Taken together, the increased number and the abnormal localization of G2/M cells in both cey and tbl3 morphant larvae indicate that Tbl3 regulates proliferation during retinal development.
Figure 6. Prolonged proliferative phase in retina results from slowing of the cell cycle.
Proliferation is increased in cey mutant retinas. At 3 dpf there are more pH3+ (G2/M) cells in cey mutants in the central retina (white arrowheads; B) as compared to wild-type (A). Injection of both tbl3 MO and p53 MO also results in an increase in pH3+ cells in the central retina (white arrowheads) at 3 dpf (D) as compared to a p53MO only control (C). Quantification shows that the increase in pH3+ cells is significant in both larvae injected with tbl3MO alone or tbl3MO co-injected with p53MO at 3 dpf (E). Control larvae were injected with phenol red. Whole Z-stacks of dorsal view retinas were projected and the number of pH3+ cells were counted and then averaged to determine the average number of mitotic cells at 3 dpf (E, see methods for more details on quantification). Tbl3 morphants also have an increased number of cells in S-phase as compared to wild-type (F) indicating there are more proliferating cells at 3 dpf. The average number of BrdU positive cells in five lateral view sections was calculated for each retina (F, see methods for more details). The progression of cells from S-phase to G2/M is \Tbl3 morphants than control embryos as is indicated by the smaller percentage of cells co-labeled with BrdU and anti-pH3 over a 6 hour period of BrdU recovery (G). At 1 hpBrdU N = 4, 8, 9, 8 for control, p53MO, tbl3MO, and tbl3p53MO respectively. At 2 hpBrdU N = 4, 4, 5, 5. At 4 hpBrdU N = 7, 4, 4, 5. At 6 hpBrdU N = 9, 11, 8, 6. Error bars in (E - G) represent standard error of the mean. Scale Bars are 130μm.
Cell death is normal in tbl3 morphant zebrafish
Loss of Tbl3 results in smaller eyes despite the prolonged maintenance of the proliferative phase of retinal development. One potential reason for this observation is an increase in cell death. To test this possibility we stained tbl3 morphant retinas with TUNEL (Terminal deoxynucleotidyl transferase dUTP Nick End Labeling) and counted the number of labeled cells in the retina. We did not observe a change in cell death in tbl3 morphants (Table 1). These data indicate that loss of Tbl3 induces small eye size independent of cell death.
Table 1.
Average # of TUNEL positive cells
| Control | p53MO | tbl3MO | tbl3p53MO | |
|---|---|---|---|---|
| 3 dpf | 1.6+/−1.3 N.S. | 1.1+/−0.6 N.S. | 0.2+/−0.15 N.S. | 1.5+/−1.0 N.S. |
| # of larvae | 7 | 14 | 9 | 13 |
N.S. = not significant
Slowing of the cell cycle in larvae with reduced Tbl3
Cell cycle delay is another possible explanation for the reduction of eye size in the face of increased proliferation in Tbl3-deficient individuals. This hypothesis predicts, first, that the number of S-phase cells is increased. Second, in the absence of Tbl3 the transition from S to G2/M phase is slowed. To test the first prediction we first counted the number of BrdU+ retinal cells in 3 dpf retinas after a 30-minute exposure to BrdU followed by a one-hour recovery period (Figure 6F, Supplementary Figure 4F). The number of S-phase cells was increased in both cey mutants and tbl3 morphants compared to controls, reflecting a general increase of cells in the cell cycle (Figure 6F, Supplementary Figure 4F). To test the second prediction, we labeled cells with both BrdU and anti-pH3 and analyzed the dynamic emergence of double-labeled cells after a 1-, 2-, 4- or 6-hour recovery at 28°C. We reasoned that if the cell cycle in retinas with reduced Tbl3 was generally delayed compared to controls the percentage of double-labeled cells in mutants would increase at a lower rate than wild-type. At 1 hour post-BrdU the percentage of pH3+ cells co-labeled with BrdU was the same in control, tbl3 morphant and tbl3/p53 double morphant larvae (Figure 6G). However, at 2, 4 and 6 hours post BrdU treatment there were significantly fewer co-labeled cells in larvae with reduced Tbl3 as compared to controls (Figure 6G). This suggests that there is a p53 independent slowing of the cell cycle when Tbl3 is reduced, resulting in delay of retinal development and reduced eye size. These data are the first to show a role for Tbl3 regulating the cell cycle during retinal development.
DISCUSSION
We present the first data showing that Tbl3 regulates cell cycle length in a p53-independent manner during zebrafish development. Our data illustrates that tbl3 is maternally expressed and remains abundant in highly proliferative tissues. Absence of tbl3 affects the size of differentiated tissues but not their specification. The small organ size in the absence of Tbl3 is due to slowing of the cell cycle. These data lead us to propose that Tbl3 is required in highly proliferative tissues to maintain the correct cell cycle speed in a p53-independent manner.
Very little is known about the function of Tbl3 in other vertebrates, but there has been some characterization of the yeast homologue, otherwise known as Utp13. Utp13 was first identified as a U3 small nucleolar (sno) RNA-associated protein (Dragon et al., 2002; Watkins et al., 2004). Utp13 is localized to the nucleolus and, along with several other Utp proteins, is part of the small subunit (SSU) processome required for 18S rRNA biogenesis (Dosil and Bustelo, 2004; Dragon et al., 2002; Watkins et al., 2004). Mutations in two other utp genes have been identified in zebrafish: utp10, otherwise known as bap28 (Azuma et al., 2006) and utp21/wdr36 (Skarie and Link, 2008). Both mutants are characterized by defective18S rRNA biogenesis and increased cell death (Azuma et al., 2006; Skarie and Link, 2008). However, only wdr36 mutants have a corresponding increase in proliferation that is similar to cey mutants (Skarie and Link, 2008).
Wdr36 is a WD-repeat protein that is linked to glaucoma (Fan et al., 2006; Hauser et al., 2006; Monemi et al., 2005; Rao et al., 2011; Ray and Mookherjee, 2009). Similar to cey mutants, wdr36 mutants have a small eye phenotype and increased anti-pH3 labeling. In addition, the small eye phenotype in wdr36 morphants persists in the absence of p53, suggesting that the phenotype is p53-independent. In contrast to cey, the wdr36 mutants show evidence of S-phase arrest, rather than slowing of the cell cycle, leading Skarie et al. to conclude that the wdr36 morphant phenotype is due to defects in cell cycle exit and subsequent cell death induced by the truncated isoform of p53 - 113p53 (Skarie and Link, 2008). In contrast, cell death is unaffected in tbl3 morphants. In addition, the p53 mutant we used to inject tbl3MO has a premature stop (Parant et al., 2010) that would prevent formation of both p53 and p53- 113p53 transcripts, suggesting that the tbl3 morphant phenotype is p53 independent.
The similarities of the wdr36 and cey mutant phenotypes, as well as the evidence in yeast showing homologues of both proteins in the SSU processome, suggest that tbl3 and wdr36 interact during rRNA biogenesis or ribosomal assembly in zebrafish. However, their differential dependence on p53 suggests that they may also have distinct roles during development of specific tissues. In fact, we have directly tested the possibility that tbl3 may be involved in rRNA processing by Northern blot but have found no difference in rRNA species between cey mutant and wild-type siblings (Supplementary Figure 5). Whether tbl3 may be involved in other steps required for proper ribosome biogenesis is the subject of future studies.
In contrast to the RPCs, HSPCs are significantly reduced when Tbl3 is lost. However, the remaining HSPCs continue to proliferate and only some differentiated blood cell types are affected. T cells and erythrocytes are both severely reduced, while neutrophils and thrombocytes are normal. This is surprising because we would expect a slowing of the HSPC cell cycle to result in a reduction of all blood cell types. However, based on mRNA expression, maternal Tbl3 protein is presumably present in cey mutants during early development, providing a possible explanation for the absence of visible defects at 2 dpf. This idea is supported by the fact that the tbl3 ATG MO, affecting both maternal and zygotic transcripts, has a more severe and earlier phenotype than either the SB MO or cey mutants(data not shown). Previous analysis of a mutation in the Medaka wdr55 gene, encoding a WD repeat protein that regulates 5s rRNA biogenesis, showed that maternal Wdr55 protein was stable for up to 10 dpf, suggesting it could sustain larval development significantly past the onset of zygotic transcription (Iwanami et al., 2008). As maternal Tbl3 is progressively depleted, the cell cycle begins to slow, resulting in fewer differentiated cells. T cells are first visible in the thymus between 2 and 3 dpf (Kissa et al., 2008; Trede et al., 2001; Willett et al., 1999) suggesting they would be more severely affected by loss of zygotic Tbl3 than blood cell types that are born earlier, like neutrophils and thrombocytes. Primitive erythrocytes are generated beginning during the first day of life and are the only circulating erythroid cells until 4 dpf (de Jong and Zon, 2005; Weinstein et al., 1996). They do not decline significantly until 6 dpf (de Jong and Zon, 2005; Weinstein et al., 1996). Erythrocytes are generated in much larger quantities than other blood cell types and a decrease in HSPCs therefore should have a bigger impact on the number of erythrocytes during times of peak production. This is reflected by the decrease in E1-globin staining in cey at 6 dpf. Our data correlate with a slowing of the cell cycle in HSPCs and we propose that all highly proliferative cell types require Tbl3 to regulate their cell cycle speed.
Tbl3 function in vertebrates is unclear, but a number of intriguing results suggest that Tbl3 may cause disease and act in signaling pathways potentially independent of rRNA biogenesis or ribosomal assembly. For example, human Tbl3 has been mapped to the autosomal dominant polycystic kidney disease (ADPKD) locus (Weinstat-Saslow et al., 1993). While no Tbl3 mutation has yet been linked with ADPKD, it raises the possibility that Tbl3 may play a role in maintaining tissue function beyond early developmental stages. Human enhanced S-cone syndrome (ESCS), a retinal degeneration disease, is another human disease with connections to Tbl3. Mouse Tbl3 associates with the co-repressor Ret-CoR complex that also binds photoreceptor cell-specific nuclear receptor (PNR) protein. PNR is spontaneously mutated in ESCS patients and regulates proliferation of S-cone cells in mice via transcriptional repression (Takezawa et al., 2007). Recent work in the mouse retina shows that PNR requires the co-repressor Ret-CoR and its associated proteins, including Tbl3, to effectively repress transcription of cell cycle dependent targets, such as cyclinD1 (Takezawa et al., 2007). Ret-CoR protein is differentially expressed during the cell cycle suggesting that cell cycle regulation is an important part of PNR proliferation regulation (Takezawa et al., 2007). These data suggest Tbl3 could regulate cell cycle rate through nuclear receptor signaling during zebrafish development, opening intriguing avenues for future investigations into the mechanism of Tbl3 function.
Supplementary Material
Highlights.
>We analyzed a zebrafish mutant with reduced tissue differentiation. >We show the defects in this mutant are due to a loss of the tbl3 gene. >Loss of tbl3 causes cell cycle slowing and reduced organ size independent of p53 and cell death. >We conclude that tbl3 regulates cell cycle rate during vertebrate development.
Acknowledgements
The authors wish to thank Bret Pearson, Kristen Kwan and Priya Choudhry for their insights and for proofreading the manuscript. Thank you to Brian Ciruna for support and reagents. Sarah Hutchinson was funded by a NIH institutional training grant (MCTRC) and postdoctoral fellowship from the American Cancer Society (PF-08-105-01-LIB). This work was also supported by grants from the NIH (NIH1R21HD060310), the Shwachman-Diamond Syndrome Foundation, the Huntsman Cancer Foundation (NST) and the Huntsman Cancer Institute core facilities (supported by grant P30 CA04201).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: SAH and NST conceived the experiments. SAH, ETL, TK and NST mapped and identified tbl3 as the gene affected in cey. SAH, ETL and TK performed WISH. JW and ST performed Northern blot. SAH performed embryo injections, IHC, imaging and data analysis and wrote the paper.
References
- Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annual review of cell and developmental biology. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiades MR, Cepko C. Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev Dyn. 1996;205:293–307. doi: 10.1002/(SICI)1097-0177(199603)205:3<293::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Austin CP, Feldman DE, Ida JA, Jr., Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- Azuma M, Toyama R, Laver E, Dawid IB. Perturbation of rRNA synthesis in the bap28 mutation leads to apoptosis mediated by p53 in the zebrafish central nervous system. J Biol Chem. 2006;281:13309–13316. doi: 10.1074/jbc.M601892200. [DOI] [PubMed] [Google Scholar]
- Batista MF, Jacobstein J, Lewis KE. Zebrafish V2 cells develop into excitatory CiD and Notch signalling dependent inhibitory VeLD interneurons. Dev Biol. 2008;322:263–275. doi: 10.1016/j.ydbio.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Baye LM, Link BA. The disarrayed mutation results in cell cycle and neurogenesis defects during retinal development in zebrafish. BMC Dev Biol. 2007;7:28. doi: 10.1186/1471-213X-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MW, Jordan CT. Leukemia stem cells in 2010: current understanding and future directions. Blood Rev. 2011;25:75–81. doi: 10.1016/j.blre.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Belliveau MJ, Cepko CL. Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development. 1999;126:555–566. doi: 10.1242/dev.126.3.555. [DOI] [PubMed] [Google Scholar]
- Belliveau MJ, Young TL, Cepko CL. Late retinal progenitor cells show intrinsic limitations in the production of cell types and the kinetics of opsin synthesis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20:2247–2254. doi: 10.1523/JNEUROSCI.20-06-02247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemar F, Argenton F, Schmidtke R, Epperlein S, Peers B, Driever W. Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev Biol. 2001;230:189–203. doi: 10.1006/dbio.2000.0103. [DOI] [PubMed] [Google Scholar]
- Bilitou A, Ohnuma S. The role of cell cycle in retinal development: cyclin-dependent kinase inhibitors co-ordinate cell-cycle inhibition, cell-fate determination and differentiation in the developing retina. Dev Dyn. 2010;239:727–736. doi: 10.1002/dvdy.22223. [DOI] [PubMed] [Google Scholar]
- Bolande RP. Developmental pathology. Am J Pathol. 1979;94:623–683. [PMC free article] [PubMed] [Google Scholar]
- Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat Immunol. 2010;11:666–673. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasis JA, Mohandas N. Erythroblastic islands: niches for erythropoiesis. Blood. 2008;112:470–478. doi: 10.1182/blood-2008-03-077883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, Lin S. Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. British journal of haematology. 2011;152:217–228. doi: 10.1111/j.1365-2141.2010.08396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JL, Zon LI. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet. 2005;39:481–501. doi: 10.1146/annurev.genet.39.073003.095931. [DOI] [PubMed] [Google Scholar]
- DeHart SL, Heikens MJ, Tsai S. Jagged2 promotes the development of natural killer cells and the establishment of functional natural killer cell lines. Blood. 2005;105:3521–3527. doi: 10.1182/blood-2004-11-4237. [DOI] [PubMed] [Google Scholar]
- Dosil M, Bustelo XR. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J Biol Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, Hunt DF, Baserga SJ. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001;2:333–342. doi: 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- Ellett F, Lieschke GJ. Zebrafish as a model for vertebrate hematopoiesis. Current opinion in pharmacology. 2010;10:563–570. doi: 10.1016/j.coph.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Fan BJ, Wang DY, Lam DS, Pang CP. Gene mapping for primary open angle glaucoma. Clinical biochemistry. 2006;39:249–258. doi: 10.1016/j.clinbiochem.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Fibach E. Involvement of phosphatases in proliferation, maturation, and hemoglobinization of developing erythroid cells. J Signal Transduct. 2011;2011:860985. doi: 10.1155/2011/860985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ES, Stubbs JL, Levine EM. Genetic rescue of cell number in a mouse model of microphthalmia: interactions between Chx10 and G1-phase cell cycle regulators. Development. 2003;130:539–552. doi: 10.1242/dev.00275. [DOI] [PubMed] [Google Scholar]
- Hauser MA, Allingham RR, Linkroum K, Wang J, LaRocque-Abramson K, Figueiredo D, Santiago-Turla C, del Bono EA, Haines JL, Pericak-Vance MA, Wiggs JL. Distribution of WDR36 DNA sequence variants in patients with primary open-angle glaucoma. Investigative ophthalmology & visual science. 2006;47:2542–2546. doi: 10.1167/iovs.05-1476. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Hu M, Easter SS. Retinal neurogenesis: the formation of the initial central patch of postmitotic cells. Developmental biology. 1999;207:309–321. doi: 10.1006/dbio.1998.9031. [DOI] [PubMed] [Google Scholar]
- Iwanami N, Higuchi T, Sasano Y, Fujiwara T, Hoa VQ, Okada M, Talukder SR, Kunimatsu S, Li J, Saito F, Bhattacharya C, Matin A, Sasaki T, Shimizu N, Mitani H, Himmelbauer H, Momoi A, Kondoh H, Furutani-Seiki M, Takahama Y. WDR55 is a nucleolar modulator of ribosomal RNA synthesis, cell cycle progression, and teleost organ development. PLoS Genet. 2008;4:e1000171. doi: 10.1371/journal.pgen.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusuf PR, Almeida AD, Randlett O, Joubin K, Poggi L, Harris WA. Origin and determination of inhibitory cell lineages in the vertebrate retina. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:2549–2562. doi: 10.1523/JNEUROSCI.4713-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K, Murayama E, Zapata A, Cortes A, Perret E, Machu C, Herbomel P. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 2008;111:1147–1156. doi: 10.1182/blood-2007-07-099499. [DOI] [PubMed] [Google Scholar]
- Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA, Weissman IL. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- Laguerre L, Soubiran F, Ghysen A, Konig N, Dambly-Chaudiere C. Cell proliferation in the developing lateral line system of zebrafish embryos. Dev Dyn. 2005;233:466–472. doi: 10.1002/dvdy.20343. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel JP, Kanki JP, Zon LI, Look AT, Trede NS. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7369–7374. doi: 10.1073/pnas.0402248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Xu Q, Breitbart RE. A new tinman-related gene, nkx2.7, anticipates the expression of nkx2.5 and nkx2.3 in zebrafish heart and pharyngeal endoderm. Dev Biol. 1996;180:722–731. doi: 10.1006/dbio.1996.0341. [DOI] [PubMed] [Google Scholar]
- Li Z, Hu M, Ochocinska MJ, Joseph NM, Easter SS., Jr. Modulation of cell proliferation in the embryonic retina of zebrafish (Danio rerio) Dev Dyn. 2000;219:391–401. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1063>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Lin HF, Traver D, Zhu H, Dooley K, Paw BH, Zon LI, Handin RI. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 2005;106:3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME, Jacks T. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 2004;18:1681–1694. doi: 10.1101/gad.1203304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus RC, Delaney CL, Easter SS., Jr. Neurogenesis in the visual system of embryonic and adult zebrafish (Danio rerio). off. Vis Neurosci. 1999;16:417–424. doi: 10.1017/s095252389916303x. [DOI] [PubMed] [Google Scholar]
- Milewski WM, Duguay SJ, Chan SJ, Steiner DF. Conservation of PDX-1 structure, function, and expression in zebrafish. Endocrinology. 1998;139:1440–1449. doi: 10.1210/endo.139.3.5768. [DOI] [PubMed] [Google Scholar]
- Monemi S, Spaeth G, DaSilva A, Popinchalk S, Ilitchev E, Liebmann J, Ritch R, Heon E, Crick RP, Child A, Sarfarazi M. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Human molecular genetics. 2005;14:725–733. doi: 10.1093/hmg/ddi068. [DOI] [PubMed] [Google Scholar]
- Mudumana SP, Wan H, Singh M, Korzh V, Gong Z. Expression analyses of zebrafish transferrin, ifabp, and elastaseB mRNAs as differentiation markers for the three major endodermal organs: liver, intestine, and exocrine pancreas. Dev Dyn. 2004;230:165–173. doi: 10.1002/dvdy.20032. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Downing JR. Global genomic characterization of acute lymphoblastic leukemia. Semin Hematol. 2009;46:3–15. doi: 10.1053/j.seminhematol.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF, Handin RI, Herbomel P. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Ng A, Uribe RA, Yieh L, Nuckels R, Gross JM. Zebrafish mutations in gart and paics identify crucial roles for de novo purine synthesis in vertebrate pigmentation and ocular development. Development. 2009;136:2601–2611. doi: 10.1242/dev.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik EJ, Zon LI. Hematopoietic development in the zebrafish. The International journal of developmental biology. 2010;54:1127–1137. doi: 10.1387/ijdb.093042ep. [DOI] [PubMed] [Google Scholar]
- Parant JM, George SA, Holden JA, Yost HJ. Genetic modeling of Li-Fraumeni syndrome in zebrafish. Dis Model Mech. 2010;3:45–56. doi: 10.1242/dmm.003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen JT, Janson E, Utami KH, Pereboom TC, Essers PB, van Rooijen C, Zivkovic D, Macinnes AW. The nucleolar GTP-binding proteins Gnl2 and nucleostemin are required for retinal neurogenesis in developing zebrafish. Dev Biol. 2011;355:286–301. doi: 10.1016/j.ydbio.2011.04.028. [DOI] [PubMed] [Google Scholar]
- Payne EM, Bolli N, Rhodes J, Abdel-Wahab OI, Levine R, Hedvat CV, Stone R, Khanna-Gupta A, Sun H, Kanki JP, Gazda HT, Beggs AH, Cotter FE, Look AT. Ddx18 is essential for cell-cycle progression in zebrafish hematopoietic cells and is mutated in human AML. Blood. 2011;118:903–915. doi: 10.1182/blood-2010-11-318022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao KN, Nagireddy S, Chakrabarti S. Complex genetic mechanisms in glaucoma: an overview. Indian journal of ophthalmology. 2011;59(Suppl):S31–42. doi: 10.4103/0301-4738.73685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Mookherjee S. Molecular complexity of primary open angle glaucoma: current concepts. Journal of genetics. 2009;88:451–467. doi: 10.1007/s12041-009-0065-3. [DOI] [PubMed] [Google Scholar]
- Rhodes MM, Kopsombut P, Bondurant MC, Price JO, Koury MJ. Adherence to macrophages in erythroblastic islands enhances erythroblast proliferation and increases erythrocyte production by a different mechanism than erythropoietin. Blood. 2008;111:1700–1708. doi: 10.1182/blood-2007-06-098178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, Hopkins N, Lees JA. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 2006;20:3117–3129. doi: 10.1101/gad.1482106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarie JM, Link BA. The primary open-angle glaucoma gene WDR36 functions in ribosomal RNA processing and interacts with the p53 stress-response pathway. Hum Mol Genet. 2008;17:2474–2485. doi: 10.1093/hmg/ddn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa S, Yokoyama A, Okada M, Fujiki R, Iriyama A, Yanagi Y, Ito H, Takada I, Kishimoto M, Miyajima A, Takeyama K, Umesono K, Kitagawa H, Kato S. A cell cycle-dependent co-repressor mediates photoreceptor cell-specific nuclear receptor function. EMBO J. 2007;26:764–774. doi: 10.1038/sj.emboj.7601548. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Taylor AM, Zon LI. Modeling Diamond Blackfan anemia in the zebrafish. Seminars in hematology. 2011;48:81–88. doi: 10.1053/j.seminhematol.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Thompson MA, Ransom DG, Pratt SJ, MacLennan H, Kieran MW, Detrich HW, 3rd, Vail B, Huber TL, Paw B, Brownlie AJ, Oates AC, Fritz A, Gates MA, Amores A, Bahary N, Talbot WS, Her H, Beier DR, Postlethwait JH, Zon LI. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- Trede NS, Medenbach J, Damianov A, Hung LH, Weber GJ, Paw BH, Zhou Y, Hersey C, Zapata A, Keefe M, Barut BA, Stuart AB, Katz T, Amemiya CT, Zon LI, Bindereif A. Network of coregulated spliceosome components revealed by zebrafish mutant in recycling factor p110. Proc Natl Acad Sci U S A. 2007;104:6608–6613. doi: 10.1073/pnas.0701919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trede NS, Ota T, Kawasaki H, Paw BH, Katz T, Demarest B, Hutchinson S, Zhou Y, Hersey C, Zapata A, Amemiya CT, Zon LI. Zebrafish mutants with disrupted early T-cell and thymus development identified in early pressure screen. Dev Dyn. 2008;237:2575–2584. doi: 10.1002/dvdy.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trede NS, Zapata A, Zon LI. Fishing for lymphoid genes. Trends in immunology. 2001;22:302–307. doi: 10.1016/s1471-4906(01)01939-1. [DOI] [PubMed] [Google Scholar]
- Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Uechi T, Nakajima Y, Chakraborty A, Torihara H, Higa S, Kenmochi N. Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond-Blackfan anemia. Human molecular genetics. 2008;17:3204–3211. doi: 10.1093/hmg/ddn216. [DOI] [PubMed] [Google Scholar]
- Uribe RA, Gross JM. Id2a influences neuron and glia formation in the zebrafish retina by modulating retinoblast cell cycle kinetics. Development. 2010;137:3763–3774. doi: 10.1242/dev.050484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Weinstat-Saslow DL, Germino GG, Somlo S, Reeders ST. A transducin-like gene maps to the autosomal dominant polycystic kidney disease gene region. Genomics. 1993;18:709–711. doi: 10.1016/s0888-7543(05)80380-5. [DOI] [PubMed] [Google Scholar]
- Weinstein BM, Schier AF, Abdelilah S, Malicki J, Solnica-Krezel L, Stemple DL, Stainier DY, Zwartkruis F, Driever W, Fishman MC. Hematopoietic mutations in the zebrafish. Development. 1996;123:303–309. doi: 10.1242/dev.123.1.303. [DOI] [PubMed] [Google Scholar]
- Willett CE, Cortes A, Zuasti A, Zapata AG. Early hematopoiesis and developing lymphoid organs in the zebrafish. Dev Dyn. 1999;214:323–336. doi: 10.1002/(SICI)1097-0177(199904)214:4<323::AID-AJA5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Yang Q, Jeremiah Bell J, Bhandoola A. T-cell lineage determination. Immunol Rev. 2010;238:12–22. doi: 10.1111/j.1600-065X.2010.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gray J, Wu L, Leone G, Rowan S, Cepko CL, Zhu X, Craft CM, Dyer MA. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat Genet. 2004;36:351–360. doi: 10.1038/ng1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.