Abstract
The insect midgut epithelium is generally lined with a unique chitin and protein structure, the peritrophic membrane (PM), which facilitates food digestion and protects the gut epithelium. We used gel electrophoresis and mass spectrometry to identify the extracted proteins from the silkworm PM to obtain an in-depth understanding of the biological function of the silkworm PM components. A total of 305 proteins, with molecular weights ranging from 8.02 kDa to 788.52 kDa and the isoelectric points ranging from 3.39 to 12.91, were successfully identified. We also found several major classes of PM proteins, i.e. PM chitin-binding protein, invertebrate intestinal mucin, and chitin deacetylase. The protein profile provides a basis for further study of the physiological events in the PM of Bombyx mori. [BMB Reports 2012; 45(11): 665-670]
Keywords: Bombyx mori, Chitin-binding proteins, LTQ-Orbitrap, Peritrophic matrix, Proteome
INTRODUCTION
The peritrophic membrane (PM) is an invertebrate-unique semi-permeable structure that lines the midgut of an insect. The presence of this matrix in insects has been recognized for over two centuries. Lyonet (1) found a sheath encasing the food bolus in lepidopteran larvae in 1972. Balbiani (2) was the first to refer to this matrix as PM in 1980. The PM is mainly composed of chitin nanofibers embedded in the matrix of proteins, glycoproteins, and proteoglycans. Wang and Granados (3) provided the first molecular model of the lepidopteran PM. However, the functions and the chemical components of the matrix are still rudimentary. Sudha and Muthu (4) reported on the high abrasion in the midgut epithelium and the membranous bodies of an unusual Bombyx mori mutant that lacks a PM in the larval stage caused by food in 1988. Over the past decade, the development of molecular, biological, and biochemical approaches improved remarkably the awareness of the PM functions. The main biological functions of PM include the spatial organization of digestion, protection from ingested toxins, and serves as a physical barrier to pathogens.
PM proteins have at least two chitin-binding domains that are presumed to function in the cross-linking of the chitin fibrils. The peritrophins-44 and peritrophins-48 are the first integral PM glycoproteins reported from the larvae of Lucilia cuprina. These proteins contain a signal peptide, followed by five sequential CBDs, conforming to the CX13-20CX5-6CX9-19CX10-14CX4-14C peritrophin-A consensus. This multiple cysteine-rich domain enables the protein to bind with chitin, the major constituent of PM (5,6). Wang and Granados (7) identified an invertebrate intestinal mucin (IIM) from Trichoplusia ni that exhibits strong association with chitin and could degrade by baculovirus enhancement, but is highly resistant to proteolytic attack from endogenous midgut proteases. Ferreira et al. (8) found two peritrophins from Tenebrio molitor and Spodoptera frugiperda by cDNA library screenings with antibodies in 2007. The two peritrophins are immobilized on the midgut cell surface and concur in PM formation. A downregulated chitin deacetylase-like protein selected from a group of expression changes the Helicoverpa armigera genes after infection with H. armigera single nucleopolyhedrovirus, which might reduce the susceptibility of this bollworm to baculovirus by decreasing its PM permeability (9).
Various proteomic approaches has been utilized recently to identify the proteins from the PM of various insects. Ramos et al. (10) used a two-dimensional gel electrophoresis to analyze the black fly type I PM and found two PM1-specific proteins (66 and 61 kDa). Lehane et al. (11) showed that the PM contains a range of proteins, most of which require relatively harsh treatment for their solubilization. Moskalyk et al. (12) found approximately 20 major proteins from the Aedes aegypti PM and approximately 40 major proteins from the Anopheles gambiae PM through 2-D gel electrophoresis and lectin-binding affinity assays. Shotgun proteomics is a remarkably powerful technology for identifying complex samples of proteins on a global level. Two separate studies have recently focused on shotgun proteome analysis of PM. Forty-one proteins were identified from the PM in H. armigera using 1D-LC-MS/MS (13). Dinglasan et al. (14) described the complete PM proteome of the A. gambiae and identified 209 proteins by MudPIT analysis in 2009.
The PM not only plays important roles in facilitating food digestion and providing protection to the gut epithelium, but can also be a significant structural target for insect control. The proteins of the PM may also provide unique opportunities for the control of insect pests and vector borne diseases. The silkworm, Bombyx mori, is not only a domesticated insect for silk production, but is also a model lepidopteran insect for pest control. Therefore, we used the shotgun proteome technology to identify comprehensively the proteins from the PM of silkworm in the present study.
RESULTS AND DISCUSSION
Overview of proteomic analysis
We characterized the silkworm larval PM proteins profile using the shotgun proteome technology. Several previously reported proteins detected in other insect studies were observed in the present study. However, the limited number of lepidopteran sequences available for PMF(Peptide Mass Fingerprinting)in database searches resulted in either in low coverage and probability scores or no significant matches. In the present work, the combined databases containing proteins sequences downloaded from NCBI and from silkDB allowed us to overcome some of these limitations and identify more protein homologues for the selected proteins. The confidence in the data was controlled by searching for the data in target-decoy database to estimate the false-positive rate and further validating the data using TPP.
The protein extracts from the PM of fifth instar actively feeding larvae were separated by SDS-PAGE, and the gel was cut into 7 slices for the LC–MS/MS analysis (Fig. 1A). The obtained 7 raw data sets were searched against a combined in-house database according to the procedure in the Materials and Methods section, and a total of 305 proteins were identified (Table S1). The number of proteins was higher than the previously reported number in other insect studies. Approximately 12.45% (38/305) of these proteins had a single unique peptide, whereas the others had more than one and even up to 145 unique peptides, such as the chlorophyllide A binding protein precursor. Approximately 43.28% (132/305) of the identified proteins had amino acid coverage of more than 20%.
Fig. 1. Protein Profiles of the silkworm PM in the 5th day of the fifth instar. (A): 1D SDS-PAGE pattern of PM proteins extracted from the B. mori. Each gel lane was cut into 7 bands (PM1 to PM7) followed by in-gel digestion used for shotgun analysis. The letters ‘M’ and ‘S’ represent protein marker and sample, respectively. (B): Distribution of isoelectric points (pI) and molecular weight (MW) for all of the proteins identified by the shotgun approach.
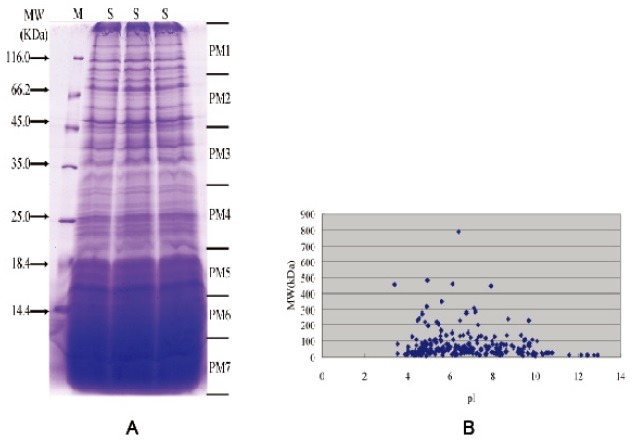
The distribution of molecular mass and isoelectric points (pI) of the identified proteins is shown in Fig. 1B. The molecular weights mostly ranged between 8.02 kDa and 788.52 kDa, except for BGIBMGA006856-PA (2002.29 kDa) and BGIBMGA-010471-PA (1538.87 kDa). A total of 79.34% (242/305) of the identified proteins were smaller than 100 kDa. The most acidic and basic proteins identified by LC-MS/MS had pIs of 3.39 and 12.91, respectively.
Chitin and chitin-binding proteins
The presence of multiple chitin-binding domains in PM proteins has been suggested to be a mechanism for PM formation. We found that several PM proteins have the chitin-binding domains of several invertebrate proteins. The BGIBMGA001504-PA is a chitin-binding protein, and the chitin-binding domains indicate a mechanism to bind chitin in the PM, homologous with CBP1 and CBP2 of T. ni, which might possess a proteinase-resistant mechanism against proteolytic degradation by the digestive proteinases (15). The mammalian mucins were likely to have a number of functions, i.e., lubrication of the passage of food through the gut, protection from invasion by bacteria, and protection of epithelial cells from digestive proteases (16). Invertebrate intestinal mucin (IIM) had many of the molecular properties of mammalian mucins. Wang and Granados (7) identified an IIM from T. ni that also exhibits a strong association with chitin and could degrade by a baculovirus enhancement, but is highly resistant to proteolytic attack from endogenous midgut proteases. Chitin deacetylase (CDA) plays an important role in the softening of the insect cuticle to allow easier mycelial penetration as well as evading lysozyme action in fungi and bacteria. The roles of insect chitin deacetylases are not well understood. Drosophila melanogaster mutants suggest that deacetylase activity increases the rigidity of chitin (17). HaCDA5a lacks a typical chitin-binding domain, but can bind with chitin, and has possible mechanism to reduce the susceptibility to baculovirus by decreasing the PM permeability (9).
Toxin binding and immunity
The aminopeptidase N (APN) serves a variety of functions mainly dependent on its location. This protein works in cooperation with endopeptidases and carboxypeptidases in the lepidopteran larval midgut to digest proteins in food. APN is also a major Bt Cry toxin receptor in the midgut of insects. A 120 kDa APN of silkworm binds Cry1Aa with a 7.6 nM affinity (18). APN also act as a mediator of Cry toxin susceptibility to distinguish between Cry-binding proteins and proteins that confer Cry toxin susceptibility. The first cadherin-like protein shown to interact with Cry toxins, BT-R1, is a 210 kDa glycoprotein identified in Manduca sexta (19). Nagamatsu et al. (20) found a 175 kDa glycoprotein, BtR175, that was identified as a Cry1Aa receptor in B. mori. The gene of BtR175 was added to Cry1Aa-resistant Sf9 cells in vitro, which made the Sf9 cells susceptible to the Cry1Aa toxin. Alkaline phosphatase (ALP) is also a Cry toxin receptor in various insect species. In Heliothis virescens, a membranebound form of ALP not only could bind to Cry1Ac, but the expression levels are also reduced in the resistant strain, suggesting a functional role in toxicity (21). ALP mediates Cry1Ac susceptibility and is important in binding toxin in M. sexta (22). A. aegypti ALP also exhibit properties similar to those of the lepidopteran ALPs (23).
Several other candidate receptor proteins were identified. Serine protease inhibitors not only play important roles in the biological immune systems, but can also potentiate the insecticidal activity of Cry toxins. They reduce the degradation of Cry or of toxin-receptors to enhance the insecticidal activity (24). The chlorophyllide A-binding protein (ChBP), identified from the midgut of B. mori, can bind to a derivative of chlorophyll (25). The ChBP was characterized by binding to Cry1Aa, Cry1Ab, and Cry1Ac under nondenaturing conditions, and probably has antimicrobial activity, similar to P252 (26).
Several identified proteins were related with immune response. The serine proteases (SP) are a large group of proteolytic enzymes that participate in the innate immune response in insects. Zhao et al. found 65 SP gene expression changes after the silkworm was infected by four different microorganisms (27). The lipase was isolated from the silkworm digestive juice and has been proved to have strong resistance against the BmNPV, and the expression was upregulated by the virus attack (28). Gram-negative bacteria-binding protein (GNBP) is a highly conserved innate immunity molecule, which recognizes bacteria. There are three GNBP genes contained in the genome of Drosophila, and two of these genes play a role in pattern recognition in the upstream of Toll signal pathway. GNBP can bind with β-1 and 3-glucan to activate cellar immune response in B. mori (29).
Functional annotation and classification
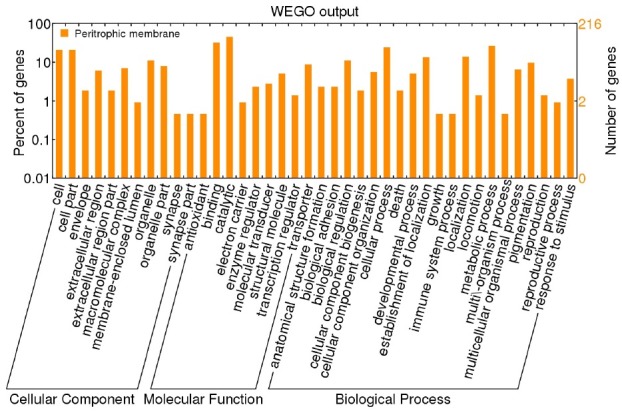
Gene Ontology tools were used to analyze the PM proteome to present an overall view on the functional categories of PM proteins. The results indicate that 216 of the 305 identified proteins show at least one matched GO annotation. The proteins were classified into cellular component, molecular function, and biological process according to the GO hierarchy using WEGO (Fig. 2). Majority of the proteins were assigned to the cell and cell part in terms of “cellular component.” The highest distribution was associated with binding and catalytic activity in terms of “molecular function.” The remaining proteins were linked to different activities, such as antioxidation, electron transport, and enzyme regulation, and among others. The proteins were classified according to different categories based on “biological process.” The highest number of proteins was mapped to proteins involved in metabolic process, followed by proteins associated with cellular process.
Fig. 2. GO categories of the identified proteins in the silkworm PM. The identified proteins were classified into cellular component, molecular function, and biological process by WEGO, according to the GO terms.
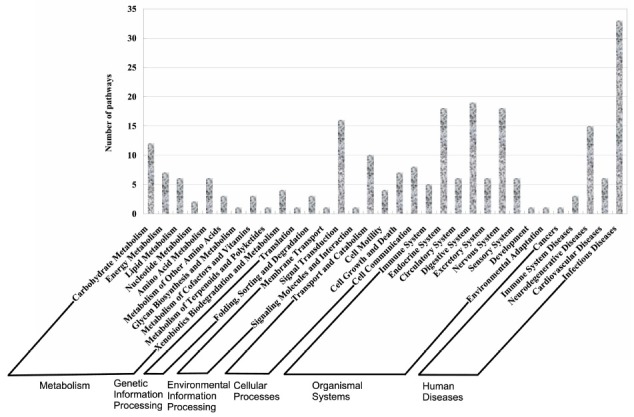
The PM proteins were mapped to the KEGG ortholog level for the KEGG analysis. Two hundred and thirty-four different pathways were linked to the PM. These pathways were classified into metabolism (45), genetic information processing (4), environmental information processing (18), cellular processes (29), organismal systems (80), and human diseases (58) (Fig. 3). Among the organismal systems, the digestive system was the most active. Moreover, five pathways were related to the immune system. This phenomenon is likely because the PM not only could help the midgut in coordinating the movement of enzymes and nutrient uptake, but also plays an important role in insect defense strategy.
Fig. 3. Categories of commonly related pathways in PM according to KEGG pathway taxonomy. The pathways were methodically clustered into metabolism, genetic information processing, environmental information processing, cellular processes, and human diseases.
The studies on the expression profiles of proteins in the PM can facilitate the identification of molecular targets that can be used for developing novel and environmentally benign controlling strategies. In the present work, we identified successfully 305 proteins from silkworm PM in the 5th day of the fifth instar by proteomic tools. We have characterized several PM proteins based on the proteomic analysis, and a holistic model that relates the PM structure to its function is presented. The PM proteins provide a basis for further study of the physiological events in the PM of Bombyx mori, and help us to understand the PM structure and the role in immune response.
MATERIALS AND METHODS
Sample preparation and gel electrophoresis
The B. mori strain DaZao was used and maintained at State Key Laboratory of Silkworm Genomics, Southwest University. The silkworm was reared using a mulberry under a 12 h/12 h light/dark photoperiod at 26̊ ± 1℃ with 70% to 85% relative humidity. The silkworms were pricked and dissected gently to collect the intact PMs from the 5th day of the fifth instar larval. The PMs were washed with Milli-Q water until no food debris remained. Approximately 100 PMs were accumulated and stored at −80℃.
The total protein of PMs was extracted in 400 μl lysis buffer (containing 2.5% SDS, 10% glycerin, 5% β-mercaptoethanol and 50 mM Tris-HCl pH 8.8), was homogenized with a tissue grinder, and then vortexed. The PMs were fully disintegrated after incubation with the lysis buffer, blended several times for 1 h at 4℃, and then centrifuged for 30 min at 12,000 rpm at 4℃. The total protein content in the supernate was determined using the method of Bradford (30). The sample was boiled for 10 min, and each 100 μg proteins was loaded into three lanes of one-dimensional (1D) SDS-PAGE, which was performed by standard method using 5% stacking gel and a 15% resolving gel. The electrophoresis was run on the Hoefer SE600 Ruby (Hoefer), and the gels were stained by Coomassie Brilliant Blue R250 (Sigma).
LC-MS/MS analysis
After visualization by Coomassie staining, each gel lane was manually cut into 7 bands from top to bottom. The gel bands were sliced into smaller pieces and subjected to in-gel tryptic digestion as described by Shevchenko et al. (31). Tryptic peptides from gel bands were analyzed by LC–MS/MS using a linear ion trap mass spectrometer (Finnigan LTQ, Thermo Finnigan). The peptide mixture was loaded onto a Zorbax 300SB-C18 peptide traps column (Agilent Technologies) by the autosampler and were desalted for 20 min, prior to separation by reverse-phase analytical column (0.15 × 150 mm [RP-C18], Column Technology Inc.). The mobile phases consisted of buffer A (0.1% methanoic acid in water) and buffer B (84% ACN, 0.1% methanoic acid in water). The peptides were eluted using buffer B linear gradient from 4% to 50% in 50 min, from 50% to 100% buffer B in 54 min, and buffer B maintained at 100% in 60 min. The data-dependent acquisition was performed on the LTQ-Orbitrap mass spectrometer in the positive ion mode. The spray voltage was 3.2 kV, and the capillary temperature was 200℃. Each full MS scan was followed by the MS/MS scans of the 20 most intense peaks in the MS spectrum with dynamic exclusion enabled. The m/z scan range was 100 to 2,000 for full mass range.
Database search
The databases were constructed using ‘silkworm’ as a keyword in searching from the NCBI (http://www.ncbi.nlm.nih.gov/) protein database. In addition, the open reading frames of silkworm were obtained from silkworm genome sequences, which were downloaded from silkDB (http://silkworm.swu.edu.cn/silkdb/doc/download.html). The combined databases containing 23,017 protein sequences (8,394 sequences from NCBI and 14,623 sequences from silkDB) were analyzed using MS/MS data with the algorithm of SEQUEST, which is a module of Biowork 3.0 (Thermo Finnigan) on a local server. The SEQUEST parameters were outlined as follows. The mass tolerances of precursor and fragmentation ions were set to 1 Da, full tryptic constraint allowing 1 missed cleavage, fixed modification (carboxamidomethyl) on cysteine, and variable modification (oxidation) on methionine. The search results were filtered with the following criteria: charge +1, Xcorr ≥ 1.9; charge +2, Xcorr ≥ 2.2; charge +3, Xcorr ≥ 3.75; DelCN ≥ 0.1. The datasets were searched against the target-decoy database under the same parameters stated above to estimate the false-positive rate. The target-decoy database is a combination of forward and reverse protein sequences (32). The confidence was controlled by filtering the initial identifications to FDR ≤1% for each sample class.
The database search results were further validated using TPP (v4.4 VUVUZELA rev 1) (http://localhost/tpp-bin/tpp_gui.pl), which was downloaded from the website (http://tools.proteomecenter.org/TPP.php) and installed with the default options. The peptide and protein probability thresholds for running PeptideProphet and ProteinProphet were set at 0.8 and 0.9, respectively.
Bioinformatics analysis
Gene Ontology (GO) IDs of the proteins identified in the present study were obtained from the Gene Ontology (http://www.geneontology.org/). GO classifications of these proteins were conducted using WEGO (http://wego.genomics.org.cn/) following the method described by Ye et al. (33). The EC numbers of the identified proteins were acquired (if available) with E-value ≤ e−15 by using KEGG GENES BLASTP (http://blast.genome.jp/), and then were subjected to search against the KEGG reference pathway database (http://www.genome.jp/kegg/tool/search_pathway.html). The pathways with at least three EC numbers were accepted and classified according to the definition of KEGG (http://www.genome.ad.jp/kegg/pathway.html).
Supporting information
Table S1. The detailed information about the identified proteins of silkworm PM. The data were searched in SEQUEST with a target-decoy database to estimate the false-positive rate and further validated using TPP.
Acknowledgments
The present work was supported by grants from the National Basic Research Program of China (Grant No. 2012CB114604), the National Hi-Tech Research and Development Program of China (Grant No. 2011AA100306), the National Natural Science Foundation (Grant No. 30972147), and the Doctorial Innovation Fund of Southwest University (Grant No. Kb2009003).
References
- 1.Lyonet P. Trait´e Anatomique de la Chenille qui ronge le bois de Saule. Gosse and Pinet; La Haye, Holland: (1762). [Google Scholar]
- 2.Balbiani E. G. Etudes anatomiques et histologiques sur le tube digestif des Crytops. Arch. Zool. Exp. Gen. (1980);8:1–82. [Google Scholar]
- 3.Wang P., Granados R. R. Molecular structure of the peritrophic membrane (PM): identification of potential PM target sites for insect control. Arch. Insect. Biochem. Physiol. (2001);47:110–118. doi: 10.1002/arch.1041. [DOI] [PubMed] [Google Scholar]
- 4.Sudha P. M., Muthu S. P. Damage to the midgut epithelium caused by food in the absence of peritrophic membrane. Curr. Sci. (1988);57:624–625. [Google Scholar]
- 5.Elvin C. M., Vuocolo T., Pearson R. D., East I. J., Riding G. A., Eisemann C. H., Tellam R. L. Characterization of a major peritrophic membrane protein, peritrophin-44, from the larvae of Lucilia cuprina. cDNA and deduced amino acid sequences. J. Biol. Chem. (1996);271:8925–8935. doi: 10.1074/jbc.271.15.8925. [DOI] [PubMed] [Google Scholar]
- 6.Schorderet S., Pearson R. D., Vuocolo T., Eisemann C., Riding G. A., Tellam R. L. cDNA and deduced amino acid sequences of a peritrophic membrane glycoprotein, 'peritrophin-48', from the larvae of Lucilia cuprina. Insect. Biochem. Mol. Biol. (1998);28:99–111. doi: 10.1016/S0965-1748(97)00103-3. [DOI] [PubMed] [Google Scholar]
- 7.Wang P., Granados R. R. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc. Natl. Acad. Sci. U.S.A. (1997);94:6977–6982. doi: 10.1073/pnas.94.13.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira A. H., Cristofoletti P. T., Lorenzini D. M., Guerra L. O., Paiva P. B., Briones M. R., Terra W. R., Ferreira C. Identification of midgut microvillar proteins from Tenebrio molitor and Spodoptera frugiperda by cDNA library screenings with antibodies. J. Insect. Physiol. (2007);53:1112–1124. doi: 10.1016/j.jinsphys.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Jakubowska A. K., Caccia S., Gordon K. H., Ferre J., Herrero S. Downregulation of a chitin deacetylase-like protein in response to baculovirus infection and its application for improving baculovirus infectivity. J. Virol. (2010);84:2547–2555. doi: 10.1128/JVI.01860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos A., Mahowald A., Jacobs-Lorena M. Peritrophic matrix of the black fly Simulium vittatum: formation, structure, and analysis of its protein components. J. Exp. Zool. (1994);268:269–281. doi: 10.1002/jez.1402680403. [DOI] [PubMed] [Google Scholar]
- 11.Lehane M. J., Allingham P. G., Weglicki P. Composition of the peritrophic matrix of the tsetse fly, Glossina morsitans morsitans. Cell Tissue Res. (1996);283:375–384. doi: 10.1007/s004410050548. [DOI] [PubMed] [Google Scholar]
- 12.Moskalyk L. A., Oo M. M., Jacobs-Lorena M. Peritrophic matrix proteins of Anopheles gambiae and Aedes aegypti. Insect Mol. Biol. (1996);5:261–268. doi: 10.1111/j.1365-2583.1996.tb00100.x. [DOI] [PubMed] [Google Scholar]
- 13.Campbell P. M., Cao A. T., Hines E. R., East P. D., Gordon K. H. Proteomic analysis of the peritrophic matrix from the gut of the caterpillar, Helicoverpa armigera. Insect Biochem. Mol. Biol. (2008);38:950–958. doi: 10.1016/j.ibmb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Dinglasan R. R., Devenport M., Florens L., Johnson J. R., McHugh C. A., Donnelly-Doman M., Carucci D. J., Yates J. R. 3rd, Jacobs-Lorena M. The Anopheles gambiae adult midgut peritrophic matrix proteome. Insect Biochem. Mol. Biol. (2009);39:125–134. doi: 10.1016/j.ibmb.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P., Li G., Granados R. R. Identification of two new peritrophic membrane proteins from larval Trichoplusia ni: structural characteristics and their functions in the protease rich insect gut. Insect Biochem. Mol. Biol. (2004);34:215–227. doi: 10.1016/j.ibmb.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Tellam R. L., Wijffels G., Willadsen P. Peritrophic matrix proteins. Insect Biochem. Mol. Biol. (1999);29:87–101. doi: 10.1016/S0965-1748(98)00123-4. [DOI] [PubMed] [Google Scholar]
- 17.Luschnig S., Batz T., Armbruster K., Krasnow M. A. serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. (2006);16:186–194. doi: 10.1016/j.cub.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 18.Yaoi K., Kadotani T., Kuwana H., Shinkawa A., Takahashi T., Iwahana H., Sato R. Aminopeptidase N from Bombyx mori as a candidate for the receptor of Bacillus thuringiensis Cry1Aa toxin. Eur. J. Biochem. (1997);246:652–657. doi: 10.1111/j.1432-1033.1997.t01-1-00652.x. [DOI] [PubMed] [Google Scholar]
- 19.Vadlamudi R. K., Ji T. H., Bulla L. A. Jr. A specific binding protein from Manaduca sexta for the insecticidal toxin of Bacillus thuringiensis subsp. berliner. J. Biol. Chem. (1993);268:12334–12340. [PubMed] [Google Scholar]
- 20.Nagamatsu Y., Koike T., Sasaki K., Yoshimoto A., Furukawa Y. The cadherin-like protein is essential to specificity determination and cytotoxic action of the Bacillus thuringiensis insecticidal CryIAa toxin. FEBS Lett. (1999);460:385–390. doi: 10.1016/S0014-5793(99)01327-7. [DOI] [PubMed] [Google Scholar]
- 21.Jurat-Fuentes J. L., Adang M. J. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur. J. Biochem. (2004);271:3127–3135. doi: 10.1111/j.1432-1033.2004.04238.x. [DOI] [PubMed] [Google Scholar]
- 22.McNall R. J., Adang M. J. Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect. Biochem. Mol. Biol. (2003);33:999–1010. doi: 10.1016/S0965-1748(03)00114-0. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez L. E., Aimanova K. G., Gill S. S., Bravo A., Soberon M. A GPI-anchored alkaline phosphatase is a functional midgut receptor of Cry11Aa toxin in Aedes aegypti larvae. Biochem. J. (2006);394:77–84. doi: 10.1042/BJ20051517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardo-Lopez L., Munoz-Garay C., Porta H., Rodriguez-Almazan C., Soberon M., Bravo A. Strategies to improve the insecticidal activity of Cry toxins from Bacillus thuringiensis. Peptides. (2009);30:589–595. doi: 10.1016/j.peptides.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mauchamp B., Royer C., Garel A., Jalabert A., Da Rocha M., Grenier A. M., Labas V., Vinh J., Mita K., Kadono K., Chavancy G. Polycalin (chlorophyllid A binding protein): a novel, very large fluorescent lipocalin from the midgut of the domestic silkworm Bombyx mori L. Insect. Biochem. Mol. Biol. (2006);36:623–633. doi: 10.1016/j.ibmb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Pandian G. N., Ishikawa T., Togashi M., Shitomi Y., Haginoya K., Yamamoto S., Nishiumi T., Hori H. Bombyx mori midgut membrane protein P252, which binds to Bacillus thuringiensis Cry1A, is a chlorophyllide-binding protein, and the resulting complex has antimicrobial activity. Appl. Environ. Microbiol. (2008);74:1324–1331. doi: 10.1128/AEM.01901-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao P., Wang G. H., Dong Z. M., Duan J., Xu P. Z., Cheng T. C., Xiang Z. H., Xia Q. Y. Genome-wide identification and expression analysis of serine proteases and homologs in the silkworm Bombyx mori. BMC Genomics. (2010);11:405. doi: 10.1186/1471-2164-11-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponnuvel K. M., Nakazawa H., Furukawa S., Asaoka A., Ishibashi J., Tanaka H., Yamakawa M. A lipase isolated from the silkworm Bombyx mori shows antiviral activity against nucleopolyhedrovirus. J. Virol. (2003);77:10725–10729. doi: 10.1128/JVI.77.19.10725-10729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee W. J., Lee J. D., Kravchenko V. V., Ulevitch R. J., Brey P. T. Purification and molecular cloning of an inducible gram-negative bacteria-binding protein from the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. (1996);93:7888–7893. doi: 10.1073/pnas.93.15.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. (1976);72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. (2006);1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 32.Reidegeld K. A., Eisenacher M., Kohl M., Chamrad D., Korting G., Bluggel M., Meyer H. E., Stephan C. An easy-to-use decoy database builder software tool, implementing different decoy strategies for false discovery rate calculation in automated MS/MS protein identifications. Proteomics. (2008);8:1129–1137. doi: 10.1002/pmic.200701073. [DOI] [PubMed] [Google Scholar]
- 33.Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L. WEGO: a web tool for plotting GO annotations. Nucleic. Acids. Res. (2006);34:W293–297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]


