Abstract
Together with protein tyrosine kinases (PTKs), protein tyrosine phosphatases (PTPs) serve as hallmarks in cellular signal transduction by controlling the reversible phosphorylation of their substrates. The human genome is estimated to encode more than 100 PTPs, which can be divided into eleven sub-groups according to their structural and functional characteristics. All the crystal structures of catalytic domains of sub-groups have been elucidated, enabling us to understand their precise catalytic mechanism and to compare their structures across all sub-groups. In this review, I describe the structure and mechanism of catalytic domains of PTPs in the structural context. [BMB Reports 2012; 45(12): 693-699]
Keywords: Classical PTP, Crystal structure, Dual specificity PTP, Eyes absent, Protein tyrosine phosphatase (PTP)
INTRODUCTION
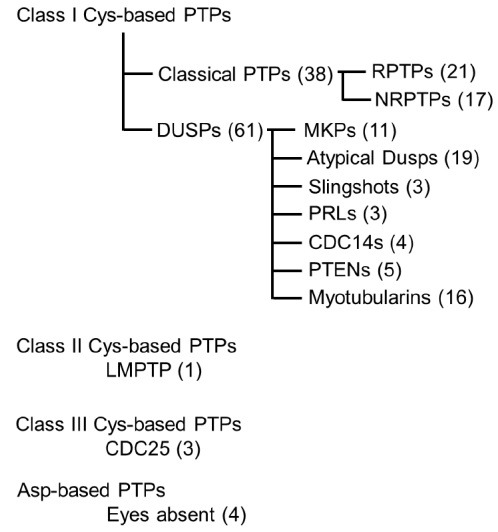
Reversible phosphorylation by the action of kinase and its counterpart phosphatase play important roles in cellular signal transduction processes. It is now apparent that phosphorylation-mediated signaling covers virtually the whole process including metabolism, cell growth, differentiation, immune response, oncogenesis and apoptosis (1). Thus aberrant phosphorylation has been linked to various diseases including cancer, diabetes, neurological disorder, and immune diseases (2). Protein phosphorylation occurs on serine, threonine or tyrosine residues. Although the extent of phosphorylation on tyrosine residues is found to be the least compared to the other two (3), tyrosine phosphorylation is a unique characteristic of eukaryote and multicellular signaling (4). Protein tyrosine phosphatases (PTPs) constitute a large protein family with more than ∼100 genes in human (5-7). Furthermore 90 are known to phosphorylate tyrosine residues, termed as protein tyrosine kinases (PTKs) in the human genome (4), suggesting that both PTPs and PTKs represent not only prominent roles in cellular signaling, but the importance of balanced action. PTPs largely comprise three classes of Cys-based PTPs [Class I; classical PTPs & dual specific protein tyrosine phosphatases (DSPs), Class II; low molecular PTP, Class III; CDC25] and one class of Asp-based PTP, which can be further divided into eleven sub-groups according to their substrate specificity and structural characteristics (Fig. 1) (5). The importance of PTPs led to make great efforts to understand the structure and underlying mechanism by determining three dimensional structures. Currently more than 50 unique PTP structures are deposited in the PDB (www.rcsb.org) and most of them are contributed by the New York SGX research center for structural genomics (http://www.nysgxrc.org), SGC (Structural Genomics Consortium; http://www.sgc.utoronto.ca) and KRIBB (Korea Research Institute of Bioscience and Biotechnology; https://www.kribb.re.kr) (8).
Fig. 1. Classification of human protein tyrosine phosphatome. Figure is adapted from Alonso et al. (5).
Comprehensive reviews have discussed on the structure and the catalytic mechanism of the cysteine-based PTP superfamily (9,10). However the number of PTP genes is still growing, which include unusual PTP such as eyes absent, which dephosphorylate tyrosyl phosphorylated substrates through an unusual mechanism (11-13). As the structural information of the entitre sub-group of PTPs including eyes absent are available now, it is the right time to present a comprehensive review of the structure and mechanism of PTPome in terms of the three-dimensional structure. In this short review, we discuss a comparative analysis of our understanding of the catalytic domain of PTP structures and their mechanism.
CLASSICAL PTPs; NON RECEPTOR TYPE & RECEPTOR-TYPE
38 Tyrosine specific PTPs (also termed as classical PTPs) can be divided into non-receptor (17 members) and receptor types (21 members) (5,6,14). The classical PTP domains are highly conserved modules in terms of primary and tertiary structures, which has ∼280 amino acids (14). They share a common architecture of a central, highly twisted nine β-stranded sheet (Fig. 2A). One side of β sheet is covered by six α helices and two short, antiparallel β-strands, and the other by the three α helices. Classical PTPs are defined by the active signature motif (HCX5R) and mobile general acid/base loop (WPD), in which cysteine residue acts as a nucleophile and is essential for catalysis (15,16). As shown in Fig. 2B and C, the phosphate group from the phosphor-tyrosine substrate is coordinated by main-chain amides of the PTP loop and side chain of arginine. The side chain of the active site cysteine is located beneath the PTP loop, in which it has a close proximity to main-chain amides and has an unusually low pKa value of ∼5.0 (17). Thus the active site cysteine exists as a thiolate ion in physiological condition and attacks the phosphorus atom of phosphor-tyrosyl residue. At the same time, the WPD loop move by ∼8Å toward the substrate in which aspartic acid is ideally positioned to act as a general acid to donate protons to phenolic oxygen of the substrate with consequent formation of a phosphor-cysteine intermediate. This intermediate is resolved by an activated water molecule aided by an asparate in the WPD loop and an adjacent glutamine residue.
Fig. 2. Structure and mechanism of classical PTPs. (A) Ribbon diagram of TP1B (pdb code: 2HNP). (B) Crystal structure of PTP1B in complex with phosphor-Tyr (pdb code: 1PTV). Consensus sequences (HCX5R), a general acid Asp181 and bound phosphor-Tyr are shown as stick models. (C) Twostep catalytic mechanism of classical PTP.

Receptor type PTP D1-D2
12 out of 21 receptor type PTP contain two catalytic domains, termed proximal and distal PTP domains, respectively. Most distal PTP domains of receptor type PTP lack important amino acids for catalysis and show very low catalytic activity. In particular the active site cysteine is changed to aspartate in a few cases and the general acid in the WPD loop is changed to other residue in all cases (14). The proximal and distal PTP domains of receptor PTPs are arranged in a tandem manner linked with 4-5 amino acids (18). Although the roles of these inactive distal PTPs are largely unknown, biochemical and structural analyses indicate that distal PTPs might play a role in regulating catalytic activities of proximal PTPs (19-22).
Dual specificity protein tyrosine phosphatases (Dusps)
Dusps constitutes the largest sub-group of protein tyrosine phosphatome that contains more than 60 species. Unlike classical PTPs, Dusps have a low level of sequence homology and several structural variations among species possibly due to granting a wide variety of substrate specificities (Fig. 3). While the substrate of classical PTPs is confined to phosphor-Tyr, Dusps can dephosphorylate substrates including phosphor-Ser/Thr, phosphor-Tyr, phosphatidyl inositol phosphates, phosphorylated carbohydrate and mRNA. The active site pocket shape appeared to be a main determinant for substrate specificity. The pocket of the classical PTP is narrow and deep so as to only accommodate phosphor-Tyr. However, Dusps have wide and shallow pockets, allowing all substrates to be dephosphorylated (16,23). The common fold of Dusps shares an essential motif of classical PTPs, with conserved active site signature motifs (HCX5R) (23). Although Dusps do not possess a WPD loop, they have conserved aspartate residue in the corresponding position, acting as a general acid/base during catalysis. Dusps can be classified into seven sub-groups according to their substrate specificities and modules (5). However, in terms of the structural aspects of catalytic domains, atypical Dusps, LMPTP and slingshots share essentially the same structural homology (24).
Fig. 3. Crystal structures of representative Dusps. All Dusps are drawn with the same point of view. The secondary structural elements in blue indicate the deviations from canonical Dusp structure.

Atypical Dusps, LMPTP and slingshots
They have a minimal basic motif of classical PTPs with smaller size (∼150 amino acids). They contain a central twisted five-stranded β sheet surrounded by five or six helices on or beneath the β sheet. Structural variations occur at the N-terminus and the C-terminus of the catalytic domain. VHR, TMDP and Dusp27 contain an additional α helix at the N-terminus, which are thought to play a role in substrate recognition (23,25,26). Dusp18 contains short antiparallel β-strands at the C-terminus (27). VHY contain an α helix at the C-terminus, which also occurs in PTP1B upon substrate or inhibitor binding (28,29). Although LMPTP shares the same homology with the members of Dusps, LMPTP has a different topological order where the HCX5R motif occurs near the N-terminus (30).
MKPs
Some of the catalytic domains of MKPs undergo drastic conformational changes upon substrate binding. In the structures of PAC-1, MKP-3 and MKP-4, an additional β strand is added to the central β and serious deviations from canonical PTP loops are observed with very low catalytic activities (31-33). The activation of those MKPs is achieved through the interactions of their MAPK binding domain at N-terminus with the cognate MAPK substrate, which represents the so-called substrate-induced activation (34). The substrate binding is thought to render catalytically important residues to be positioned in the proper positions. However, other MKPs do not follow substrate-induced activation and adopt canonically active conformation irrespective of substrate binding (35-37).
PRLs
PRL-3 is highly overexpressed in liver metastasis of colon cancer but not in non-metastatic nor in normal colorectal epithelium (38). Further, PRL-3 gene amplification is found in metastatic lesions from patients, and catalytic activity of PRL phosphatases contributes to promoting cell migration and invasion (39). Extensive structural studies show PRLs have several distinct differences compared to canonical Dusp structure (40-43). First, unlike other Dusps, PRLs have a WFPDD motif instead of a WPD-like loop present in classical PTPs. This loop adopts an open conformation in a substrate-free condition while a closed conformation is observed in the existence of a competitive inhibitor, reminiscent of a mobile WPD loop in classical PTP. Second, PRLs exist as a trimer both in vitro and in vivo, possibly due to increasing the possibility of targeting to membranes (41). Third, PRLs have alanine next to arginine of the PTP loop, which are strictly conserved as serine or threonine in other PTPs. The alcoholic group in this position is important for the breakdown of the phosphor-enzyme intermediate (44).
CDC14s
CDC14s are involved in dephosphorylation of phosphor-Thr in the activation loop of Cdk. As shown in the structure of kinase-associated phosphatase (KAP), a member of CDC14s, a short β-hair pin is inserted between the β2 strand and α2 helix, which cannot be seen in other classical PTPs and Dusps. It appeared to be important for the recognition of phosphorylated CDK2 substrate (45).
PTENs
PTEN is a hallmark of tumor suppression whose mutations are commonly found in most human cancer cells. ∼400 amino acid of PTEN enzyme contains the catalytic domain of Dusp. The catalytic domain of PTEN intensively interacts with the tensin homolog domain involved in targeting PTEN to a membrane. PTEN can dephosphorylate phosphatidylinositol (3,4,5)-trisphosphate (PIP3) at the D3 position, mediating negative regulation of Akt signaling (46, 47). Although PTEN shares a high degree of structural similarity with the )canonical Dusp structure, PTEN has a four-residue insertion between the β2 strand and α1 helix relative to VHR, which results in an extended pocket. This larger pocket accounts for the large size of the PIP3 substrate (49).
Myotubularins
Myotubularin enzymes (MTMRs) contain the largest catalytic domain (∼380 amino acids) among protein tyrosine phosphatomes. Further, its catalytic domain is highly associated with the GRAM domain at the N-terminus. The catalytic domain of the MTMR consists of a central seven-stranded β sheet flanked by 13 α helices (49). The structural superposition of MTMR with VHR shows good alignment in which most VHR structures are present in the MTMR structure. In addition, two strands and seven helices are unique features in the catalytic domain of MTMR. MTMRs dephosphorylate either PI (3,5)P2 or PI (3)P at the D3 position. Although MTMR shares a consensus sequence motif, CX5R, it contains no aspartate at the position of the general acid in other PTPs. Instead, aspartate preceding arginine of the PTP loop acts as a general acid, as presented by mutational analysis and the complex structure of MTMR:PI (3,5) P2 (50).
CDC25
Aside from having consensus sequences (HCX5R), CDC25 has no homology with the catalytic domain of protein tyrosine phosphatome. CDC25 has rather a rhodanase-like fold structurally and topologically (51). Structural and mutational analysis shows that a general acid for catalysis is positioned next to catalytic cysteine (52).
Eyes absent
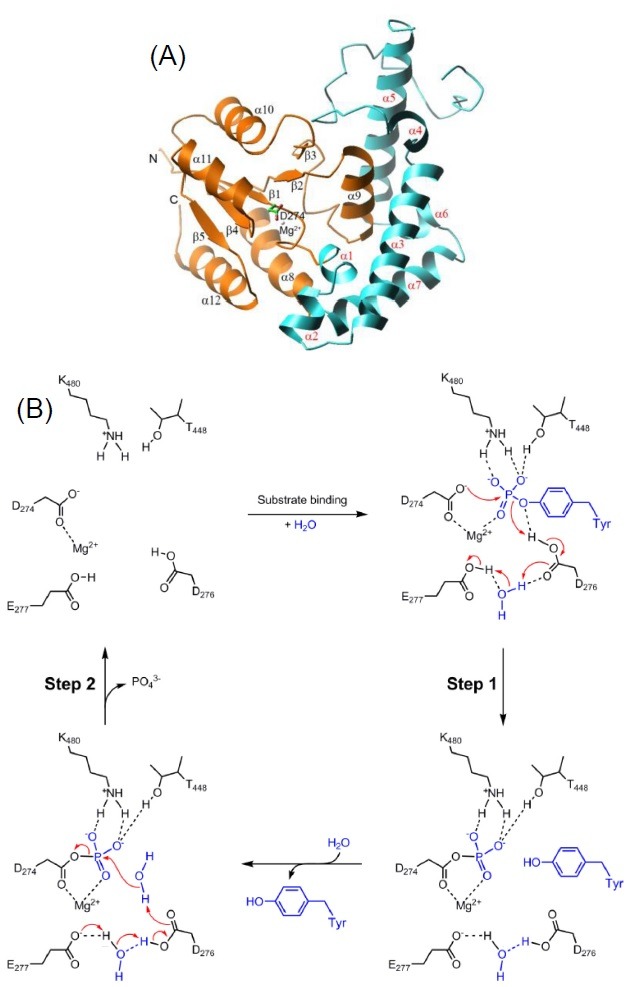
Eyes absent is recently identified as Asp-based PTPs (11-13). Unlike Cys-based PTPs, eyes absent uses aspartic acid as a nucleophile in a metal-dependent reaction. The crystal structure of the catalytic domain of eyes absent shows two domain arrangements: a halo-acid dehalogenase (HAD)–like catalytic domain and helix bundle motif (HBM) (Fig. 4A) (53,54). In contrast to other HAD members, HBM is elongated along the back of the catalytic site, resulting in th accommodation of large protein substrates. Eyes absent human homologue 2 shares three consensus sequence motifs and a bound magnesium ion with the members of the HAD family. As shown in Fig. 4B, Asp274 is a nucleophile and also anchored by a magnesium ion. Magnesium ion is further stabilized by the interactions with the side chain of Asp502, backbone carbonyl group of Asp276. Asp276 act as a general acid/base by stabilizing the leaving group during the first step and is then involved in activating water-mediated hydrolysis of the phosphoenzyme complex. Lys480 and Thr448 appear to play a role in substrate binding.
Fig. 4. Structure and catalytic mechanism of eyes absent. (A) Ribbon diagram of catalytic domain of eyes absent. Catalytic domain (orange) and HBM (cyan) are colored differently. The active site aspartate and magnesium ion are drawn as ball-and-stick models. (B) Catalytic mechanism of eyes absent. A proposed two-step catalytic mechanism for eyes absent is shown.
CONCLUSION
Given the importance of tyrosine phosphorylation, many PTPs and PTKs are predicted to be related to human diseases. In fact many specific inhibitors for the individual PTKs has been successfully developed in clinics (55). PTPs including PTP1B, CDC25 and PRL-3 have drawn much attention as therapeutic targets. Despite substantial effort, the search for clinically applicable modulators has rarely been successful because of difficulties in getting sufficient selectivity over other PTPs and good bio-availability. As the number of elucidated PTP structures grows, we have not only a better understanding about PTP in terms of substrate specificities, and its catalytic mechanism and regulation, but a better opportunity to develop specific modulators for the individual PTPs which may be used for clinics.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea government (No. 2011-0030027).
References
- 1.Graves J. D., Krebs E. G. Protein phosphorylation and signal transduction. Pharmacol. Ther. (1999);82:111–121. doi: 10.1016/S0163-7258(98)00056-4. [DOI] [PubMed] [Google Scholar]
- 2.Easty D., Gallagher W., Bennett D. C. Protein tyrosine phosphatases, new targets for cancer therapy. Curr. Cancer Drug Targets. (2006);6:519–532. doi: 10.2174/156800906778194603. [DOI] [PubMed] [Google Scholar]
- 3.Mustelin T., Feng G.-S., Bottini N., Alonso A., Kholod N., Birle D., Merlo J., Huynh H. Protein tyrosine phosphatases. Front. Biosci. (2002);7:85–142. doi: 10.2741/mustelin. [DOI] [PubMed] [Google Scholar]
- 4.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. Global, in vivo and site-specific phosphorylation dynamics in signaling networks. Cell. (2006);127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 5.Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. (2004);117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Tonks N. K. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell. Biol. (2006);7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 7.Cesareni G., Perfetto L., Castagnoli L., Sacco F. The human phosphatase interactome: An intricate family portrait. Febs Lett. (2012);586:2732–2739. doi: 10.1016/j.febslet.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almo S. C., Bonanno J. B., Sauder J. M., Emtage S., Teresa P., Dilorenzo T. P., VMalashkevich V., Wasserman S. R., S., Swaminathan S., Eswaramoorthy S., Agarwal R., Kumaran D., Madegowda M., Ragumani S., Patskovsky Y., Alvarado J., A., Ramagopal U. A., Faber-Barata J., Chance M. R., Sali A., Andras Fiser A., Zhang Lawrence D. S., Burley S. K. Structural genomics of protein phosphatases. J. Struct. Funct. Genomics. (2007);8:121–140. doi: 10.1007/s10969-007-9036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barford D., Das A. K., Egloff M.-P. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. (1998);27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z.-Y. Protein tyrosine phosphatases: structure and function, substrate specificity and inhibitor development. Annu. Rev. Pharmacol. Toxicol. (2002);42:209–234. doi: 10.1146/annurev.pharmtox.42.083001.144616. [DOI] [PubMed] [Google Scholar]
- 11.Rayapureddi J. P., Kattamuri C., Steinmetz B. D., Frankfort B. J., Ostrin E. J., Mardon G., Hedge R. S. Eyes absent represents a class of protein tyrosine phosphatases. Nature. (2003);426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- 12.Li X., Oghi K. A., Zhang J., Krones A., Bush K. T., Glass C. K., Nigam S. K., Aggarwal A. K., Maas R., Rose D. W., Rosenfeld M. G. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. (2003);426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 13.Tootle T. L., Silver S. J., Davies E. L., Newman V., Latek R. R., Mills I. A., Selengut J. D., Parlikar B. E. W., Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. (2003);426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- 14.Anderson J. N., Mortensen O. H., Peters G. H., Drake P. G., Iversen L. F., Olsen O. H., Jansen P. G., anderen H. S., Tonks N. K., Møller N. P. H. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. (2001);21:7117–7136. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barford D., Flint A. J., Tonks N. K. Crystal structure of human protein tyrosine phosphatase 1B. Science. (1994);263:1397–1404. doi: 10.1126/science.8128219. [DOI] [PubMed] [Google Scholar]
- 16.Jia Z., Barford D., Flint A. J., Tonks N. K. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. (1995);268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z. Y., Dixon J. E. Active site labelling of the Yersinia protein tyrosine phosphatase: the determination of the pKa of the active site cysteine and the function of the conserved histidine 402. Biochemistry. (1993);32:9340–9345. doi: 10.1021/bi00087a012. [DOI] [PubMed] [Google Scholar]
- 18.Nam H.-J., Poy F., Krueger N. X., Saito H., Frederick C. A. Crystal structure of the tandem phosphatase domains of RPTP LAR. Cell. (1999);97:449–457. doi: 10.1016/S0092-8674(00)80755-2. [DOI] [PubMed] [Google Scholar]
- 19.Jiang G., den Hertog J., Su J., Noel J., Sap J., Hunter T. Dimerization inhibits the activity of receptor-like protein-tyrosine phosphatase-α. Science. (1999);401:606–610. doi: 10.1038/44170. [DOI] [PubMed] [Google Scholar]
- 20.Bilwes A. M., den Hertog J., Hunter T., Noel J. P. Structural basis for inhibition of receptor protein-tyrosine phosphatase-a by dimerization. Nature. (1996);382:555–559. doi: 10.1038/382555a0. [DOI] [PubMed] [Google Scholar]
- 21.Groen A., Lemeer S., van der Wijk T., Overvoorde J., Heck A. J. R., Ostman A., Barford D., Slijper M., den Hertog J. Differential oxidation of protein-tyrosine phosphatases. J. Biol. Chem. (2005);280:10298–10304. doi: 10.1074/jbc.M412424200. [DOI] [PubMed] [Google Scholar]
- 22.Barr A. J., Ugochukwu E., Lee W. H., King O. N. F., Filippakopoulos P., Alfano I., Savitsky P., Burgess-Brown N. A., Müller S., Knapp S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell. (2009);136:352–363. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuvaniyama J., Denu J. M., Dixon J. E., Saper M. A. Crystal structure of the dual specificity protein phosphatase VHR. Science. (1996);272:1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 24.Jung S.-K., Jeong D. G., Yoon T.-S., Kim J. H., Ryu S. E., Kim S. J. Crystal Structure of human slingshot phosphatase 2. Proteins. (2007);68:408–412. doi: 10.1002/prot.21399. [DOI] [PubMed] [Google Scholar]
- 25.Kim S. J., Jeong D. G., Jeong S. K., Yoon T. S., Ryu S. E. Crystal structure of human TMDP, a testis-specific dual specificity protein phosphatase: implications for substrate specificity. Proteins. (2007);66:239–245. doi: 10.1002/prot.21197. [DOI] [PubMed] [Google Scholar]
- 26.Loutos G. T., Tropea J. E., Waugh D. S. Structure of human dual-specificity phosphatase 27 at 2.38 Å resolution. Acta. Crysallogr. D67:471–479. doi: 10.1107/S090744491100970X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong D. G., Cho Y. H., Yoon T.-S., Kim J. H., Son J. H., Ryu S. E., Kim S. J. Structure of human Dsp18, a member of the dual-specificity protein tyrosine phosphatase family. Acta Crysallogr. (2006);D62:582–588. doi: 10.1107/S0907444906010109. [DOI] [PubMed] [Google Scholar]
- 28.Yoon T. S., Jeong D. G., Kim J. H., Cho Y. H., Son J. H., Ryu S. E., Kim S. J. Crystal structure of the catalytic domain of human VHY, a dual specificity protein phosphatase. Proteins. (2005);61:694–697. doi: 10.1002/prot.20642. [DOI] [PubMed] [Google Scholar]
- 29.Wiesmann C., Barr K. H., Kung J., Zhu J., Erlanson D. A., Shen W., Fahr B. J., Zhong M., Randal M., McDowell R. S., Hansen S. K. Allosteric inhibition of protein tyrosine phosphatase 1B. Nat. Str. Mol. Biol. (2004);11:730–737. doi: 10.1038/nsmb803. [DOI] [PubMed] [Google Scholar]
- 30.Zhang M., Van Etten R. L., Stauffacher C. V. Crystal structure of bovine heart phosphotyrosyl phosphatase at 2.2 Å resolution. Biochemisty. (1994);33:11097–11105. doi: 10.1021/bi00203a006. [DOI] [PubMed] [Google Scholar]
- 31.Farooq A., Chaturvedi G., Mujtaba S., Plotnikova O., Zeng L., Dhalluin C., Ashton R., Zhou M.-M. Solution structure of Erk2 binding domain of MAPK phosphatase MKP3. Mol. Cell. (2001);7:387–399. doi: 10.1016/S1097-2765(01)00186-1. [DOI] [PubMed] [Google Scholar]
- 32.Farooq A., Plotnikova O., Chaturvedi G., Yan S., Zeng L., Zhang Q., Zhou M. M. Solution structure of the MAPK phosphatase PAC1 catalytic domain. Structure. (2003);11:155–164. doi: 10.1016/S0969-2126(02)00943-7. [DOI] [PubMed] [Google Scholar]
- 33.Jeong D. G., Yoon T. S., Jung S.-K., Park B. C., Park H. S., Ryu S. E., Kim S. J. Exploring binding sites other than catalytic core in the crystal structure of catalytic domain of MKP-4. Acta Crystallogr. (2011);D67:25–31. doi: 10.1107/S0907444910042381. [DOI] [PubMed] [Google Scholar]
- 34.Camps M., Nichols A., Gillieron C., Antonsson B., Muda M., Chabert C., Boschert U., Arkinstall S. Catalytic activation of the phosphatase MKP3 by ERK2 mitogen-activated protein kinase. Science. (1998);280:1262–1265. doi: 10.1126/science.280.5367.1262. [DOI] [PubMed] [Google Scholar]
- 35.Jeong D. G., Yoon T.-S., Kim J. H., Shim M. Y., Jung S.-K., Son J. H., Ryu S. E., Kim S. J. Crystal structure of the catalytic domain of human MAP Kinase phosphatase 5: structural insight into constitutively active phosphatase. J. Mol. Biol. (2006);360:946–955. doi: 10.1016/j.jmb.2006.05.059. [DOI] [PubMed] [Google Scholar]
- 36.Jeong D. G., Cho Y. H., Yoon T.-S., Kim J. H., Ryu S. E., Kim S. J. Crystal structure of the catalytic domain of human dusp5, a dual specificity MAP Kinase protein phosphatase. Proteins. (2007);66:253–258. doi: 10.1002/prot.21224. [DOI] [PubMed] [Google Scholar]
- 37.Jeong D. G., Jung S.-K., Yoon T.-S., Woo E.-J., Kim J. H., Park B. C., Ryu S. E., Kim S. J. Crystal structure of the catalytic domain of human MKP-2 reveals a 24-mer assembly. Proteins. (2009);76:763–767. doi: 10.1002/prot.22423. [DOI] [PubMed] [Google Scholar]
- 38.Saha S., Bardelli A., Buckhaults P., Velculescu V. E., Rago C., St Croix B., Romans K. E., Choti M. A., Lengauer C., Kinzler K. W., Vogelstein B. A phosphatase associated with metastasis of colorectal cancer. Science. (2001);284:1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 39.Zeng Q., Dong J. M., Guo K., Li J., Tan H. X., Koh V., Pallen C. J., Manser E., Hong W. PRL-3 and PRL-1 promote cell migration, invasion, and metastasis. Cancer Res. (2003);63:2716–2722. [PubMed] [Google Scholar]
- 40.Kozlov G., Cheng J., Ziomek E., Banvile D., Gehring K., Ekiel I. Structural insights into molecular function of the metastasis-associated phosphatase PRL-3. J. Biol. Chem. (2004);279:11882–11889. doi: 10.1074/jbc.M312905200. [DOI] [PubMed] [Google Scholar]
- 41.Jeong D. G., Kim S. J., Kim J. H., Son J. H., Park M. R., Lim S. M., Yoon T.-S., Ryu S. E. Trimeric structure of PRL-1 phosphatase reveals an active enzyme conformation and regulation mechanisms. J. Mol. Biol. (2005);345:401–413. doi: 10.1016/j.jmb.2004.10.061. [DOI] [PubMed] [Google Scholar]
- 42.Kim K.-A., Song J.-S., Jee J., Sheen M. R., Lee C., Lee T. G., Ro S., Cheong C. Structure of human PRL-3, the phosphatase associated cancer metastasis. FEBS Lett. (2004);565:181–187. doi: 10.1016/j.febslet.2004.03.062. [DOI] [PubMed] [Google Scholar]
- 43.Sun J.-P., Wang W.-Q., Yang H., Liu S., Liang F., Fedorov A. A., Almo S. C., Zhang Z.-Y. Structure and biochemical properties of PRL-1, a phosphatase implicated in cell growth, differentiation and tumor invasion. Biochemistry. (2005);44:12009–12021. doi: 10.1021/bi0509191. [DOI] [PubMed] [Google Scholar]
- 44.Denu J. M., Lohse D. L., Vijayalakshmi J., Saper M. A., Dixon J. E. Visualization of intermediate and transition-state structures in protein tyrosine phosphatase catalysis. Proc. Natl. Acad. Sci. U.S.A. (1996);93:2493–2498. doi: 10.1073/pnas.93.6.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song H., Hanlon N., Brown N. R., Noble M. E. M., Johnson L. N., Barford D. Phosphoprotein- protein interactions revealed by the crystal structure of kinase- associated phosphatase in complex with phosphoCDK2. Mol. Cell. (2001);7:615–626. doi: 10.1016/S1097-2765(01)00208-8. [DOI] [PubMed] [Google Scholar]
- 46.Li D. M., Sun H. PTEN/MMAC1/TEP1 suppresses the tumorigenicity and induce G1 cell cycle arrest in human glioblastoma cells. Proc. Natl. Acad. Sci. U.S.A. (1998);95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon I.-S., Lee K.-H., Choi J. W., Ahn J.-Y. PI(3,4,5)P3 regulates the interaction between Akt and B23 in the nucleus. BMB Rep. (2010);43:127–132. doi: 10.5483/BMBRep.2010.43.2.127. [DOI] [PubMed] [Google Scholar]
- 48.Lee J.-O., Yang H., Georgescu M.-M., Cristofano A. D., Maehama T., Shi Y., Dixon J. E., Pandolfi P., Pavletich N. P. Crystal structure of the PTEN tumer suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. (1999);99:323–334. doi: 10.1016/S0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 49.Begley M. J., Taylor G. S., Kim S.-A., Veine D. M., Dixon J. E., Stuckey J. A. Crystal structure of a phophoinositide phosphatase, MTMR2: insights into myotubular myopathy and charcot-marie-tooth syndrome. Mol. Cell. (2003);12:1391–1402. doi: 10.1016/S1097-2765(03)00486-6. [DOI] [PubMed] [Google Scholar]
- 50.Begley M. J., Taylor G. S., Brock M. A., Ghosh P., Woods V. L., Dixon J. E. Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase. Proc. Natl. Acad. Sci. U.S.A. (2006);103:927–932. doi: 10.1073/pnas.0510006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fauman E. B., Cogswell J. P., Lovejoy B., Rocque W. J., Holmes W., Montana V. G., Piwnica-Worms H., Rink M. J., Saper M. A. Crystal structure of the catalytic domain of the human cell cycle control phosphatase, Cdc25A. Cell. (1998);93:617–625. doi: 10.1016/S0092-8674(00)81190-3. [DOI] [PubMed] [Google Scholar]
- 52.McCain D. F., Catrina I. E., Hengge A. C., Zhang Z.-Y. The catalytic mechanism of Cdc25A phosphatase. J. Biol. Chem. (2002);277:11190–11200. doi: 10.1074/jbc.M109636200. [DOI] [PubMed] [Google Scholar]
- 53.Krishinan N., Jeong D. G., Jung S.-K., Ryu S. E., Xiao A., Allis C. D., Kim S. J., Tonks N. K. Dephosphorylation of the C-terminal tyrosyl residue of the DNA damage-related histone H2A.X is mediated by the protein phosphatase Eyes Absent. J. Biol. Chem. (2009);284:16066–16070. doi: 10.1074/jbc.C900032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jung S.-K., Jeong D. G., Chung S. J., Kim J. H., Park B. C., Tonks N. K., Ryu S. E., Kim S. J. Crystal structure of ED-eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor. FASEB J. (2010);24:560–569. doi: 10.1096/fj.09-143891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D.-H., Sim T. Chemical kinomics: a powerful strategy for target deconvolution. BMB Rep. (2010);43:711–719. doi: 10.5483/BMBRep.2010.43.11.711. [DOI] [PubMed] [Google Scholar]


